Highlights
-
•
Constitutive signals indicate homeostatic dysregulation but their effect on signal transduction remains largely unexplored.
-
•
A theoretical approach is undertaken to examine how oxidative stress may affect redox signal transduction.
-
•
Constitutive signals can result in a ‘molecular habituation’ effect that interferes with information transmission.
-
•
The robustness of such a theoretical observation to the underlying methodology hints at the generality of this principle.
Keywords: Oxidative stress, Redox signalling, Ageing, Information theory, Systems modelling
Abstract
The ability of reactive oxygen species (ROS) to cause molecular damage has meant that chronic oxidative stress has been mostly studied from the point of view of being a source of toxicity to the cell. However, the known duality of ROS molecules as both damaging agents and cellular redox signals implies another perspective in the study of sustained oxidative stress. This is a perspective of studying oxidative stress as a constitutive signal within the cell. In this work, we adopt a theoretical perspective as an exploratory and explanatory approach to examine how chronic oxidative stress can interfere with signal processing by redox signalling pathways in the cell. We report that constitutive signals can give rise to a ‘molecular habituation’ effect that can prime for a gradual loss of biological function. This is because a constitutive signal in the environment has the potential to reduce the responsiveness of a signalling pathway through the prolonged activation of negative regulators. Additionally, we demonstrate how this phenomenon is likely to occur in different signalling pathways exposed to persistent signals and furthermore at different levels of biological organisation.
1. Introduction
Whilst reactive oxygen species (ROS) are known to be deleterious and unavoidable products of cellular metabolism, it is apparent that these molecules mediate essential signalling functions within cells (Winterbourn, 2015, Wang and Hai, 2016). Just a few examples of processes mediated by redox signalling include the modulation of insulin signalling (Besse-Patin and Estall, 2014), the stress response (Jiang et al., 2011), cell survival (Trachootham et al., 2008) and tissue regeneration (Sen and Roy, 2008). The elucidation of redox signalling pathways occurred in parallel to the accumulation of evidence that various tissues displayed markers of oxidative stress in various pathologies (Besse-Patin and Estall, 2014, Barbieri and Sestili, 2012, De Marchi et al., 2013, Kim et al., 2015, Sosa et al., 2013, Brioche and Lemoine-Morel, 2016, Lepetsos and Papavassiliou, 2016) and the ageing process (Sanz, 2016, Kirkwood and Kowald, 2012). The established double-edged nature of ROS raises questions as to how cells move from a state of controlled ROS production to a state of oxidative stress.
Oxidative stress is defined as a cellular state involving a mismatch between the abundance of oxidant molecules and the antioxidant capacity of the cell, favouring the former (Sies, 2015). The resulting elevation in the intracellular levels of oxidant can be transient or constitutive (Pickering et al., 2013). Transient (acute) oxidative stress is associated with redox signalling. Constitutive oxidative stress is associated with a prolonged state of elevated oxidant levels. Constitutive or chronic oxidative stress thus involves longer time-scales as is the case in chronic diseases, age-related diseases and the ageing process. Oxidative stress has drawn considerable attention due to the intrinsic reactivity of ROS (Winterbourn, 2015). This chemical property confers these molecules the capacity to cause molecular damage, consequently flagging them as potential causal agents of observed homeostatic disruptions in age and disease.
The perspective of studying oxidative stress as a constitutive signal within the cell has generated some insights: for example, how chronic oxidative stress can become a constant inhibitory signal in calcium signalling (Gorlach et al., 2015, Roedding et al., 2013) and T cell activation (Fulop et al., 2014). However, it remains unclear how redox signalling within cells may be affected by sustained oxidative stress. In other words, how will redox signalling pathways respond to an acute ROS signal on top of a constitutively elevated basal oxidant level in the cellular environment. This is of physiological significance, since redox signalling pathways have been shown to become dysfunctional in a variety of tissues in contexts where oxidative stress is also present in the cell (Sohal and Orr, 2012, McDonagh et al., 2014, Cobley et al., 2015, Vasilaki et al., 2006, Claflin et al., 2015, Jackson, 2016, Zhang et al., 2015, Done et al., 2016).
The problem becomes whether the constitutive presence of a signal in the environment affects a signalling pathway’s ability to transduce a subsequent acute pulse of the same signal. Whilst the reactivity of reactive oxygen species limits the resolution of current experimental methods (Woolley et al., 2013, Ribou, 2016), very few studies have looked at the effects of long-term exposure of cells to controlled oxidant levels (Covas et al., 2013, Tan et al., 2015, Millonig et al., 2012, Sobotta et al., 2013), with even fewer explicitly examining what effect this exposure would have on a subsequent acute redox signal fed through the system. Work published by Pickering et al. seems to indicate that a chronic exposure of cells to elevated oxidant levels can blunt redox-mediated adaptive responses (Pickering et al., 2013).
Testing all of the potential mechanisms via which chronic oxidative stress could affect physiological redox signalling would be a time-consuming endeavour. In this work, we adopt a theoretical perspective as an exploratory and explanatory approach to examine how chronic oxidative stress can interfere with signal processing by redox signalling pathways in the cell. We report that a constitutive signal in the environment has the ability to reduce the responsiveness of the signalling pathway through the prolonged activation of negative regulators. Additionally, we demonstrate how this phenomenon is likely to occur in different signalling pathways exposed to persistent signals and furthermore at different levels of biological organisation.
2. A rationale for a generic redox model
The major redox stress response pathways in the cell, i.e. NFκB, Nrf2, ASK1, HIF1 and HSF1, reveal conserved topological features. In all pathways cellular stress will interfere with an inhibitor-activator complex (Soga et al., 2012, Hoesel and Schmid, 2013, Tebay et al., 2015, Masoud and Li, 2015, Jiang et al., 2015): IκB − NFκB (Hoesel and Schmid, 2013); Keap1–Nrf2 (Tebay et al., 2015); Thioredoxin1–ASK1 (Soga et al., 2012); VHL − HIF1α (Masoud and Li, 2015) or HSP70/90–HSF1 (Jiang et al., 2015). Oxidant molecules will directly disrupt the inhibition of the activator molecule in the case of the Nrf2 and ASK1 responses, arguably the NFκB response (Oliveira-Marques et al., 2009, Morgan and Liu, 2011) and also the HIF1 response (Nanduri et al., 2015, Chandel et al., 2000). The case of HSF1 differs in that oxidative stress is likely to be sensed indirectly through the abundance of unfolded proteins or the activation of other pathways (Yoo et al., 2014, Swan and Sistonen, 2015). In any case, all pathways have been reported to be activated in response to an oxidant stimulus. It follows from this, that in all pathways the activator molecule must undergo a binding event with, or a modification by, a second molecule to be stabilised and perform a function. These are other ASK1 molecules in case of the ASK1 pathway or co-factors in the case of the Nrf2, NFκB, HIF1, HSF1 transcription factors. Additionally, a stabilising phosphorylation step has been reported for all molecules. Eventually the response must be terminated and these pathways must return to their original state: an activator being actively bound by an inhibitor to form an inactive complex. This requires the complex to be regenerated. Such regeneration occurs through the post-translational modification of the inhibitor, e.g. reduction, or through its de novo synthesis.
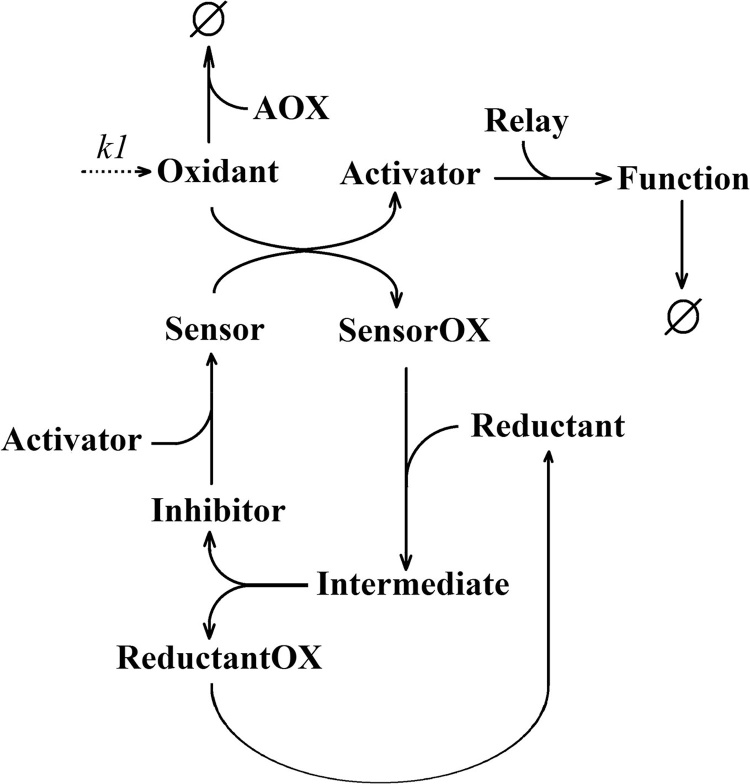
Based on these observations we defined a core generic redox model (Fig. 1), hereafter referred to as Model 1. Such core model consists of ‘Sensor’ molecules that can react with ‘Oxidant’ molecules which can additionally be scavenged by antioxidant molecules (AOX). Oxidation of ‘Sensor’ molecules to yield ‘SensorOX’ will cause the release of an ‘Activator’ molecule, which must first bind a ‘Relay’ molecule in order to be stabilised and be able to perform a function. The ‘Function’ molecule is used as a readout of the activity of the stabilised ‘Activator’ molecules. Oxidised sensor molecules (SensorOX) can be reduced through a two-step process involving the binding to a ‘Reductant’ molecule to form an ‘Intermediate’ complex which is then resolved. The reduced form of the sensor that is not bound to an ‘Activator’ molecule is termed as an ‘Inhibitor’ species since it can bind ‘Activator’ molecules to reform the ‘Sensor’ complex.
Fig. 1.
Network diagram of generic redox signalling Model 1. The steady-state level of the ‘Oxidant’ signal is modulated through the rate constant k1 which determines the flux of oxidant generation. An oxidation reaction will result in the ‘Activator’ escaping inhibition and performing a ‘Function’ after being stabilised by a binding event with a ‘Relay’ molecule. Meanwhile the inhibitor will undergo a two-step regeneration process before it is able to bind the ‘Activator’ into an inhibitory complex (Sensor). AOX=Antioxidant. OX suffix = oxidized. Slashed circle = degraded.
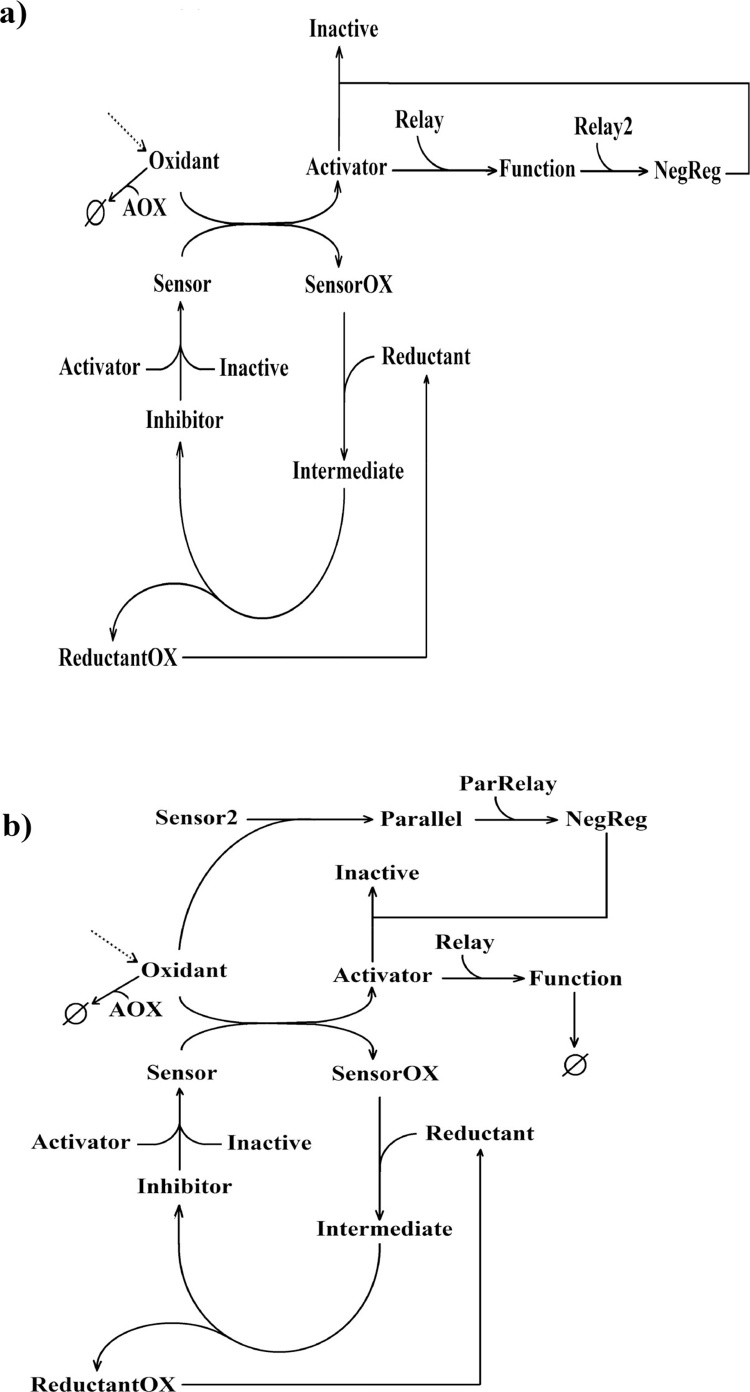
Model 1 ignores feedback mechanisms within the pathways. However, in the major redox stress response pathways outlined above, there are significant uncertainties with regards to the number of negative regulators in each pathway and their relative importance. Despite this, all the pathways have been reported to contain a negative feedback loop occurring through the activator-mediated transcription of inhibitor genes. The negative regulator that mediates the negative feedback loop is able to destabilise the activator in a first step and this results in the subsequent formation of the inhibitor-activator complex. This would correspond to the disruption of the transcriptional complexes and the subsequent nuclear export of NFκB, Nrf2, HSF1 and HIF1 transcription factors or the destabilisation of the ASK1 signalosome. Model 2 is an expansion of Model 1 that includes this element of negative feedback in a simplified time-scale (Fig. 2a). Within Model 2, a negative regulator entity ‘NegReg’ is introduced into the system downstream of the functional activity of the stabilised activator molecules. This is modelled as ‘NegReg’ being formed by the reaction between the functional readout molecule (Function) and a second relay molecule (Relay2). Negative feedback occurs by the ability of the ‘NegReg’ molecules to react with the ‘Activator’ molecules to render them ‘Inactive’.
Fig. 2.
Network diagrams of generic redox signalling models incorporating a) a negative feedback loop (Model 2) or b) a negative feedforward loop (Model 3). AOX = Antioxidant. Slashed circle = degraded.
In the case of the Nrf2 pathway, which is the major regulator of the antioxidant response in cells (Tebay et al., 2015), the main mechanism behind the response shutdown occurs through the delayed activation of GSK3β (Tebay et al., 2015, Cuadrado, 2015). Delayed activation does not depend on Nrf2 activity and occurs through a slower parallel branch to the activation of Nrf2 by oxidant molecules (Cuadrado, 2015, Kaspar et al., 2009). This is an example of a negative feedforward loop. In contrast to the negative feedback loop which follows an in-series structure, the parallel topology of a negative feedforward loop allows the negative regulator to be introduced independently from the activator molecule. Such pathway structure is incorporated into Model 3 as an expansion of Model 1 (Fig. 2b). Model 3 follows the same principle with regards to how the ‘NegReg’ species enters the system, with the key difference that it does not depend on the activity of stabilised ‘Activator’ molecules. In this model, ‘NegReg’ is introduced by a parallel signalling branch that is activated by the oxidation of a second sensor molecule (Sensor2) by ‘Oxidant’ molecules. The result of this oxidation is the formation of a species arbitrarily named ‘Parallel’ which will introduce the negative regulator after a time delay modelled by its reaction with a relay molecule (ParRelay). Note that for simplicity, the two sensor molecules in Model 3 are not modelled to compete for oxidant species. The equations and parameters of Models 1–3 can be found in Supplementary Tables 1–9. With Models 2 and 3 as generalised representations of redox signalling pathways (exemplified in Fig. S1), we set to investigate how a redox signal would feed into the system in the presence and absence of basally elevated oxidant levels.
3. Results
3.1. Pathway responsiveness to an acute redox signal is reduced when oxidant levels are constitutively elevated
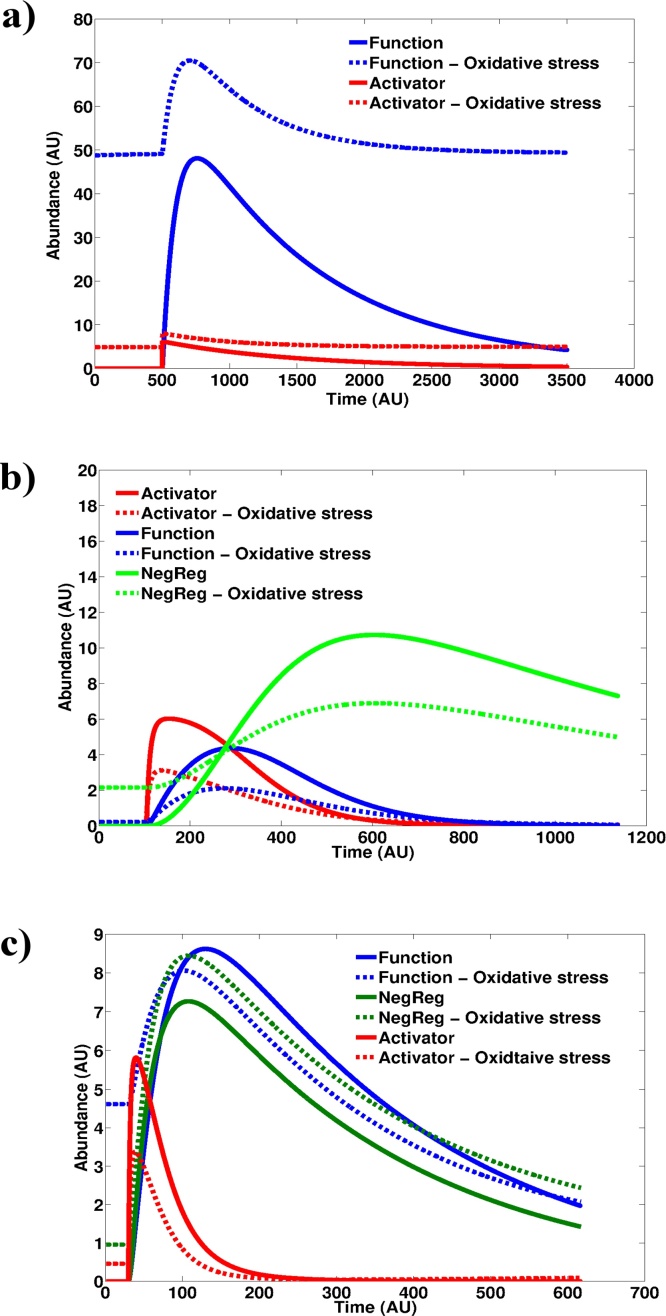
The pathway activation profiles for both Model 2 and Model 3 displayed a lower response magnitude to the same redox signal when parameter k1, controlling the basal generation of oxidant, is increased (Fig. 3). A parameter scan reveals that with a step-wise increase in the rate of oxidant production, the response peaks in Models 2 and 3 trail off (Fig. S2a-d). This is indicative of the dampening effect being dependent on the relative values of k1 and the stimulation strength, both of which are arbitrarily assigned in the models. The only exception to this observation is the negative regulator in Model 3, which displays an increased activation magnitude at higher values of k1. This is significant since the negative regulator in Model 3 is independent of the levels of the ‘Activator’ molecule suggesting the reduced sensitivity could stem from a reduced signal processing at the level of the activator. An increased steady-state level of oxidant is furthermore seen to result in a constitutively elevated level of the ‘Activator’, ‘Function’ and ‘NegReg’ molecules (Fig. 3). When we performed the same parameter scan on Model 1 however, we obtained typical saturation profiles (Fig. 3a) with the magnitude of pathway activation by the redox signal increasing with increasing k1.
Fig. 3.
Oxidative stress reduces the response magnitude to an acute redox stimulus. Model 1 (a), Model 2 (b) and Model 3 (c) were simulated at different values of basal oxidant levels. Absence of oxidative stress corresponds to a k1 value of 0 whilst oxidative stress corresponds to a k1 value of 1. Stimulus strength = 100.
A possible explanation for the observed loss in responsiveness is that the continuous flux of oxidant in the model is stabilising a new steady state which displays a reduced sensitivity at a whole-network scale (Dalle Pezze et al., 2014). To test for this we performed sensitivity analysis on all three models (Fig. S3). No obvious network-scale reduction in sensitivity is observed in any of the models. In fact, a gain in sensitivity to the rate of reduction of the oxidized inhibitor (k6 in Model 2 and k7 in Models 1&3) is seen in all models under the presence of a basally elevated level of oxidant. Upon a closer inspection of the sensitivity analysis data we observed that, under conditions of oxidative stress, ‘Activator’ and ‘Function’ molecules displayed the highest sensitivity to parameters directly involving the negative regulation process. These are parameters k10/k12 in Model 2 and k10/k11/k12 in Model 3.
3.2. The constitutive elevation of the basal levels of negative regulators by oxidative stress drives the loss of pathway responsiveness
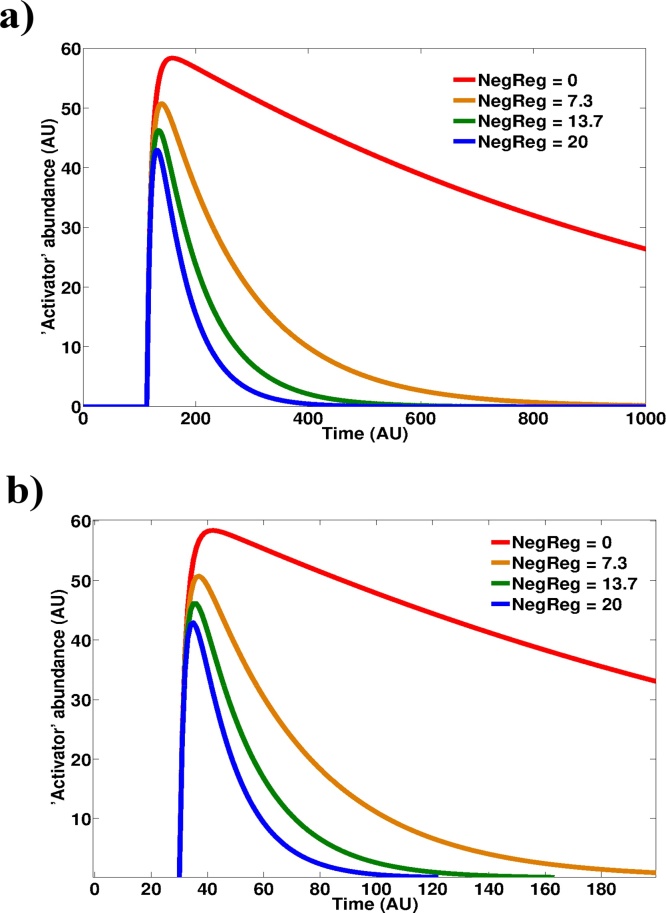
The main difference between Model 1 and Models 2 and 3 is that the latter two contain a negative regulator. It thus seems apparent that the basally elevated level of negative regulator stabilised by a sustained oxidant flux could provide a constant source of dampening of the redox signal. Should this be the case then the same response-blunting observation should be observed if, instead of altering the steady state levels of oxidant, the steady state levels of the negative regulator were clamped to fixed values and the system was stimulated with the same redox signal. When we performed this experiment in silico we observed a reduced activation magnitude with increasing levels of negative regulator (Fig. 4). We additionally checked that the blunting behaviour observed with increasing levels of oxidant production was robust to parameters controlling negative regulation strength and additionally independent on how this negative regulation was modelled, i.e. mass-action kinetics vs competitive inhibition kinetics (data not shown).
Fig. 4.
An increase in the basal levels of negative regulators reproduces the response dampening observed under conditions of oxidative stress. Simulations of pathway activation in Model 2 (a) and Model 3 (b) by an acute stimulus were run at different steady state levels of negative regulator (NegReg) molecules. Plots are shown for ‘Activator’ species. Rate constant k1 was set to zero and ‘NegReg’ abundance clamped at 4 different abundances. Stimulus strength = 100.
The behaviours displayed by our kinetic models are determined by the underlying ordinary differential equations. Thus, there is an implicit assumption that our adopted modelling framework is a good representation of the underlying biological process, and we can model cellular signalling pathways as molecular fluxes distributed across reaction branches. In order to be confident that our finding was robust to the modelling framework employed to model the signalling pathways, we simulated Models 2 and 3 as particles undergoing Brownian motion within an enclosed container in a 3D cellular automaton model. The aim being to confirm our findings in a modelling framework that does not rely on solving differential equations but where simulated profiles arise from first principles, that is, the Brownian movement of particles in three dimensional space. These simulations showed the same behaviour involving a reduced pathway activation alongside the presence of elevated negative regulator levels under conditions of oxidative stress (Fig. S4).
3.3. The observed behaviour is robust to variations in model topology
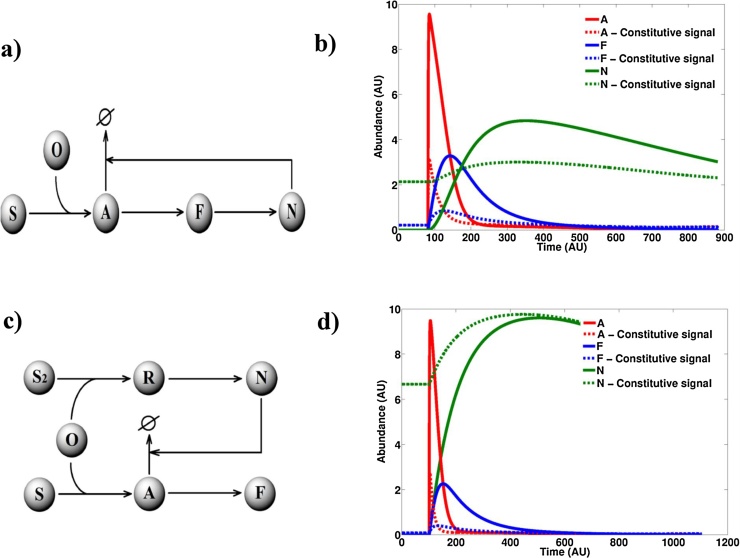
When formalising computational models of cellular signalling pathways there is always a degree of uncertainty regarding the representativeness of the underlying network structure. In order to check the robustness of the blunting behaviour to the model structures we further abstracted Models 2 and 3 to Models 4 and 5 respectively (Fig. 5a&c). Such a further abstraction could provide clues as to whether the reported effect of constitutive signals could affect generic circuits of negative regulation that are not necessarily part of redox signalling pathways. These simplified representations of a negative feedback loop and a negative feedforwards loop still displayed the same loss in responsiveness to an acute signal when the same signal was constitutively present in the environment (Fig. 5). This is, again, attributable to a steady state with basally elevated levels of the negative regulator N.
Fig. 5.
Generic circuits of negative regulation display a reduced response to an acute stimulus under the presence of a constitutive signal. a) Network diagram of Model 4. b) Deterministic simulation of Model 4 under k1 value of 0 (continuous line) and 0.02 (dashed line). c) Network diagram of Model 5. d) Deterministic simulation of Model 5 under k1 value of 0 (continuous line) and 0.02 (dashed line). k1 is the rate constant for O generation. Slashed circle = degraded. Stimulus strength = 100.
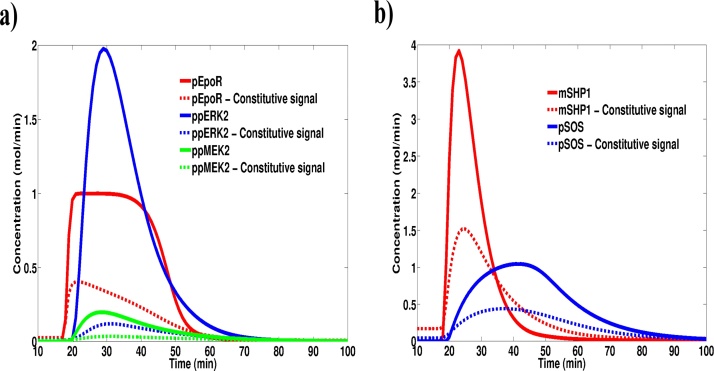
Sensitivity analysis of Models 4 and 5 showed a similar shift in model sensitivities as Models 2 and 3 (Fig. S5). Additionally, it is of interest to look for this blunting behaviour in models that are more complex, calibrated with experimental data and from an entirely different pathway. We performed a minor alteration to the extracellular-signal regulated kinase (ERK) signalling model published by Schilling et al. (Schilling et al., 2009) involving the addition of a reaction for Erythropoietin (Epo) synthesis and a reaction for Epo degradation. This minor modification constituted an elevated steady state level of this molecule on top of which the signalling event within the model would occur. Once again, we observed a reduced activation magnitude for this signalling pathway resulting from consistently elevated levels of the negative regulator molecules (Fig. 6).
Fig. 6.
The dampening effect of constitutive signals is not unique to redox signalling systems. The ERK signalling model published by Shilling et al. (48) was simulated in the presence and absence of a constitutive Epo signal. a) Activator molecules in the pathway. b) Inhibitor molecules in the pathway. The constitutive signal corresponds to a rate constant of Epo generation of 0.1. The modification of the original model only involved the addition of two reactions; i) Epo generation following zero-order kinetics and ii) Epo degradation occurring through a first order mass action reaction with a rate constant of 0.1.
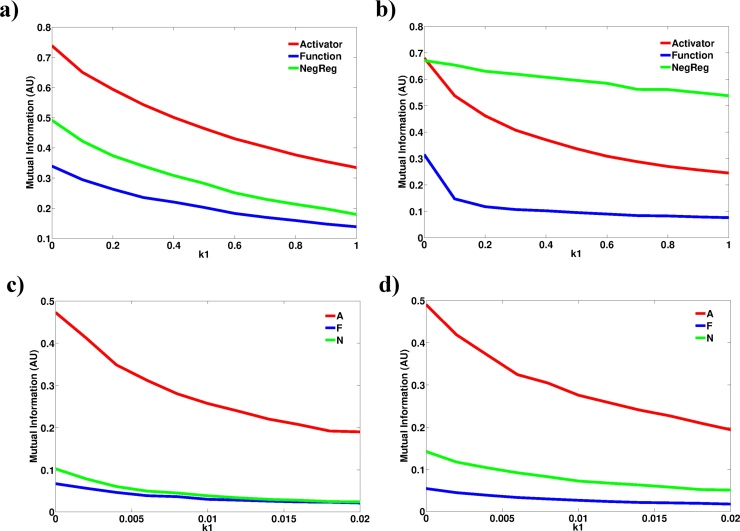
3.4. The constitutive presence of a signal will cause a reduced information flow through the signalling pathway
What is the functional consequence of the observed reduction in pathway responsiveness? This is a non-trivial question since although the activation magnitude may be reduced it is unclear if the resulting magnitude would still be large enough to trigger a response in the cell. Conceptually, a cell will trigger a response to a stimulus once the most downstream element of the signalling pathway undergoes a sufficient change from its basal abundance. The functional response thus depends on how well the downstream signalling molecule (Y) maps to the changes that occur in the upstream signalling molecule (X). How much information the state of Y provides about the state of X can be quantified through the use of mutual of information (Shannon, 2001). Mutual information analysis has been extensively used to quantify information transmission by biological signalling pathways (Waltermann and Klipp, 2011, Rhee et al., 2012, McMahon et al., 2015, Uda and Kuroda, 2016, Bowsher and Swain, 2014). Analysis of the mutual information between the signal molecule and the downstream signalling molecules in all models reveals that an increasing basal level of the signal will reduce the information flow through the signalling pathways (Fig. 7). The functional consequence of this reduced information flow can be appreciated in the simulated dose-response curves (Fig. S6), where stimulation magnitudes can be less clearly mapped to abundance values for molecule F in the presence of a constitutive signal. Thus, the presence of a constitutive signal can be a source of uncertainty to the cell regarding the actual level of the signal in the environment and consequently whether a response should be mounted or not. Such signalling dysregulation at the molecular level could impact at the tissue scale as populations of cells that fail to respond appropriately to a physiological signal.
Fig. 7.
Constitutive signals reduce information flow through signalling pathways. Mutual information between model species and the input signal was quantified in Model 2 (a), Model 3 (b), Model 4 (c) and Model 5 (d) at different k1 values. ‘A’, ‘F’ and ‘N’ refer to molecular species in Models 4 and 5. Mutual information was calculated from the molecule distributions at the response peak derived from 1000 stochastic simulations. Stimulus strength = 100.
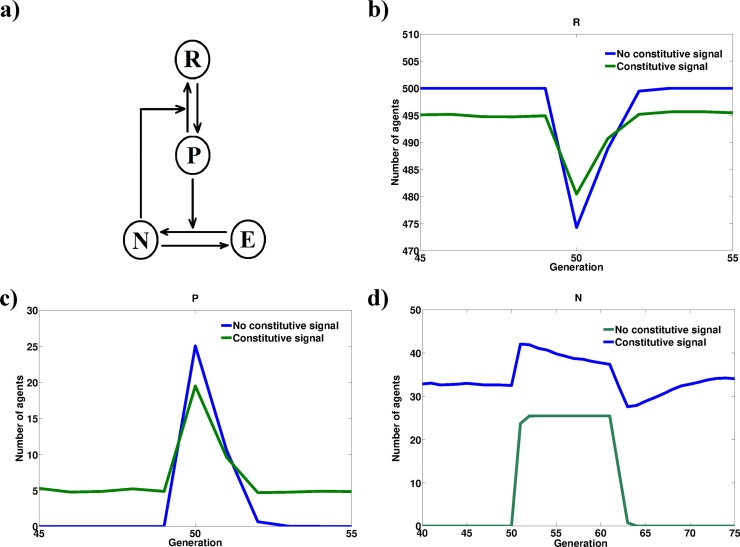
3.5. Reduced responsiveness can occur across levels of biological organisation
The level of abstraction in Models 4 and 5 suggest that the observed reduction in system response as a result of constitutively present negative regulators may not be restricted to molecular systems involved in intracellular signalling. Indeed, the mathematical formalisms behind Models 4 and 5 are sufficiently abstract to represent a system of interacting cellular or animal populations. To test this, we developed a 3D agent-based model through a cellular automaton framework. In this model agents begin at a resting state (R) and can transition to a perturbed state (P) which leads to the recruitment of agents (N) that induce a return to the resting state and become non-existent (E) from the simulation after a defined number of generations (Fig. 8a). All state transitions are probabilistic except for the lifetime of the N state. A transient change in the probability of transition from state R to state P causes a transient shift in the agent populations which is smaller in magnitude when the basal transition probability is higher (Fig. 8b–d). This is observed alongside an increased steady state population of N agents. Not only do these results re-inforce the independency of the blunting phenomenon to the underlying computational framework and interaction topology, but it also suggests that it could be observed at higher levels of biological organisation such as inter-cellular signalling.
Fig. 8.
The response dampening by persistent inputs could occur at higher levels of biological organisation. Interacting agents were simulated in a three dimensional cellular automaton grid. a) Interaction map between the modelled agents. b) Time course for R agents. c) Time course for P agents. d) Time course for N agents. Constitutive signal corresponds to a basal induction probability Pind value of 0.01. Stimulation is modelled as an increase in Pind to 0.05 for a single generation. The absence of a constitutive signal corresponds to a basal Pind of 0. Simulation run 1000 times at Gi = 10, ni = 1, Prec = 1, Pres = 0.9 and average agent populations plotted.
4. Discussion
The ageing process and indeed many other chronic and acute pathological conditions are associated with a loss of homeostasis that often manifests as a broad range of signalling dysfunctionalities. Within the ageing process, it is still unclear how the loss of homeostasis may arise. However, it is known that oxidative stress is a common consequence of such loss of homeostasis, with the potential to cause molecular damage and drive the ageing process. In this work we show how oxidative stress can be viewed as a constitutive signal in the cellular environment that ultimately hinders the cell’s ability to mount a response to a physiological redox signal. Our observations seem to transcend the case of oxidative stress alone and be applicable to other pathways and levels of biological organisation. For instance, the same loss in responsiveness would be expected to be seen in the case of chronic inflammation, where inflammatory signals are constantly present in the cellular environment. In this context, the cells within a tissue would mount physiological responses which would be likely to be of insufficient strength, at an altered time and of an altered duration. Such state of quasi-functionality would be expected to lead to a gradual accumulation of damage and functional decline.
The basis behind the reduced responsiveness in biological systems exposed to a constant environmental signal is due to an increased basal level of negative regulator entities. It has been shown that cells are able to modulate pathway responsiveness through the abundance of negative regulator molecules (Toyoshima et al., 2012). Furthermore, the sustained presence of negative regulators has been associated with physiological habituation responses (Lee et al., 2013, Ramaswami, 2014, Grissom and Bhatnagar, 2009, Herman, 2013). Additional evidence comes from work in Candida albicans where the sustained activation of SAPKs stabilise a new steady state of sub-maximal Hog1 activation through an increase in the basal levels of the PTP negative regulators (Day et al., 2017). Dues et al. (Dues et al., 2016) systematically tested the ability of C. elegans to activate stress responses to a variety of stresses. The authors report a loss in responsiveness of all the stress responses in C. elegans, albeit not accompanied by an increase in their basal activation, with age (Dues et al., 2016). It is thus a possibility that the loss in responsiveness could serve an adaptive function which results in adverse effects when the constitutive signal persists for too long.
Such a possibility seems in line with the concept of hormesis, where a constant exposure to a low-level stress reduces the susceptibility to further perturbation (Yun and Finkel, 2014). A case has been made that the phenotypic culprit of biological ageing arises from the persistent presence of antagonistic hallmarks (Lopez-Otin et al., 2013). Antagonistic hallmarks, like cellular senescence, are a response to damage that initially serve a protective function but then become deleterious when constitutively present (Lopez-Otin et al., 2013). The reported effect of constitutive signals on both pathway responsiveness and information transmission appears to be in accordance with this notion. Supporting the idea of a short term benefit of pathway dampening by constitutive signals is the observation that the extended presence of negative regulators can be used as a refractory mechanism in cellular signalling (Adamson et al., 2016, Vizan et al., 2013).
Negative feedback loops have been shown to be able to lock biological systems in stable intermediate states under conditions of weak activation (Rahi Sahand et al., 2016). Their importance in shaping cellular signalling responses could lie behind the observation that negative regulators can correlate better with lifespan than the activator molecules they regulate (Lewis et al., 2015). In accordance with this is the observation within our simulations that the negative regulator provides a higher mutual information about the signal molecule than the ‘Function’ or ‘F’ molecules in Models 2–5. In the case of ageing and chronic disease, it thus seems relevant that the constitutively elevated levels of signalling molecules could be actively maintaining a state of dysfunctional signalling. It is worth noting that constitutive signals could potentially stabilise a new less responsive steady state through the sustained activation of positive feedback loops or other network sub-structures that display bistability (Shiraishi et al., 2010). Constitutive signals could in such way draw the system closer to a critical transition (Scheffer et al., 2009). In fact it could be speculated that the stochastic activation of these network sub-structures with the potential for bistability could be the source of the constitutive signal in the first place (Faucon et al., 2014). It would be an interesting possibility that these structures could provide an architectural weak-point within biological networks.
Whilst it is not always trivial to predict the consequences of a fractional loss in the responsiveness of a given biological system, an interesting perspective has been put forward by Dalle Pezze et al. (Dalle Pezze et al., 2014) who demonstrate how a reduction in network sensitivity can result in a loss of functionality since ‘the global decrease in sensitivity upon kinetic rate constants indicated that the semantics of these model parameters, e.g. promoter or inhibitor, became more uncertain. As a consequence, this uncertainty increased system noise and decreased network robustness, which, in the context of a cell, translated into weak signalling regulations and therefore poor intervention effectiveness’.
A finding of particular interest is the ability of constitutive signals to reduce information flow through signalling pathways. Meaning that downstream effectors in signalling pathways will be less able to accurately reflect changes in the upstream signalling molecules. This translates into an increased heterogeneity in the cellular responses to physiological signals and therefore a greater proportion of cells that fail to respond appropriately. This increase in heterogeneity has been associated with a reduced system-level responsiveness and reduced effectiveness of interventions in senescent cells (Dalle Pezze et al., 2014). Interestingly, we found that the loss of information transmission through Models 2–5 with increasing basal levels of the constitutive signal closely resembles the loss in synaptic transmission efficacy of the snail Helix aspersa during habituation (Prescott and Chase, 1999). However, it should be noted that the models analysed do not display ultrasensitive behaviour. The mapping of an input signal to the on/off state of a downstream effector instead of a range of abundance values could mean the cell is more robust to losses in information transmission.
Importantly, a loss in information flow through signalling pathways means that cells will likely respond to signals with an altered magnitude, duration and/or timing. This state of quasi-functionality will likely be sufficient to maintain function but sub-optimal responses will also be likely to prime the cell for further damage and dysregulation and potentially drive a gradual loss of function typical of ageing processes. Such a state of quasi-functionality can result in subtle cellular- and tissue- level changes in the short term that only develop into an obvious loss of homeostasis in the long term. This is a new perspective on the propagation of signalling dysfunctionality across biological networks during the ageing process.
Other age-related changes that may affect the cellular regulatory machinery, such as altered expression of key sensor molecules or altered intermolecular binding affinities, are also likely to reduce information flow through signalling pathways. This suggests that information theory is an intuitive framework from which to understand loss of function and regulation during the process of biological ageing. An information-theoretic perspective has so far been overlooked and is likely to apply to be applicable to a wide range of biological systems that display age-related alterations. We believe such framework can mechanistically bridge the gap between the concept of stochastic damage in an organism lifetime and the observed gradual homeostatic decline with age.
It is important to note, especially in the case of redox signalling, that the loss of pathway responsiveness through the constitutive presence of negative regulators is likely to be acting in concert with other phenomena that would also affect pathway responsiveness. Examples could include the reduced expression of key sensor proteins, altered binding affinities that can result from genetic mutations or a reduced gradient across a membrane. In the case of oxidative stress, a redox signal generated in the mitochondria may encounter a reduced gradient due to a more oxidized cytosolic environment and so the same ROS pulse would experience a lower flux through the mitochondrial membrane. A constitutive low-level cysteine hyperoxidation could also be expected to blunt the responsiveness of redox signalling pathways to an acute redox stimulus.
A criticism that may arise from kinetic computational models is that they are often too simplistic and indeed appear “insulated” from the myriad of interactions that occur within the dense biological interaction networks. Questions also arise regarding how much of the parameter space supports the blunting behaviour within a given model topology. This criticism is the same as that for the analysis of network motifs (Ingram et al., 2006). We argue that, as is the case with network motifs, some observed biological dysregulation will be able to be mapped to this phenomenon in certain contexts and others maybe not so. It is also important to consider that should a biological system display perfect adaptation or dynamic compensation (Karin et al., 2016), it would be expected that constitutive signals might not be a source of dysfunctionality unless they interfered with the ability of the system to display such behaviours. For example, if the constitutive signal feeds into the system through crosstalk.
The loss of biological homeostasis in ageing and disease is expected to manifest as a constitutive elevation in signalling molecules, even if such a process arises from the constitutive downregulation of other molecules. This work is indicative of how a local loss in homeostasis in a cellular sub-system can spread through large portions of biological networks, potentially promoting a systemic dysregulation. Overall, it is informative to bear in mind that constitutive signals can lock signalling pathways into less responsive states. This is relevant for some diseases such as cancer where recognisable constitutive signals drive or stabilise a phenotype. It is also applicable to the ageing process, so often accompanied with the appearance of chronically elevated signals such as calcium (Decuypere et al., 2011), hormone imbalances (Maggio et al., 2013), inflammatory factors (Franceschi and Campisi, 2014) and oxidative stress.
In summary, this work reports a process of ‘molecular habituation’ within biological signalling pathways as a potential mechanism of signalling dysregulation during biological ageing. Such a phenomenon should be recognised by the seemingly paradoxical observation of a chronic activation of a signalling pathway occurring alongside an elevation in the levels of at least some of the negative regulators of such pathway.
5. Materials and methods
5.1. Signalling pathway simulation and analysis
The signalling pathway computational models are all constituted entirely by reactions following mass action kinetics unless specified otherwise. All deterministic simulations were run in COPASI (Hoops et al., 2006) with the LSODA parameters for relative and absolute tolerance set to 1e-06 and 1e-12 respectively and maximum internal steps set to 10000. Parameter scans were performed in COPASI. All stochastic simulations were run in Matlab’s Simbiology toolbox using the SSA solver. Note that in all types of simulations, an acute stimulus was simulated as an Event at a time point following the previous equilibration of the system to a steady state. All models have been deposited in the BioModels database (Chelliah et al., 2015) under the following identifiers; Model 1 − MODEL1710260000, Model 2 − MODEL1710260001, Model 3 − MODEL1710260002, Model 4 − MODEL1710260003, Model 5 − MODEL1710260004.
5.2. Molecular dynamics (MD)
Cellular automaton models can be used to simulate signalling pathways (Wurthner et al., 2000) and reaction kinetics (Schnell and Turner, 2004). A simple 3D Lattice Gas Cellular Automaton (LGCA) (Hatzikirou and Deutsch, 2010) simulator was developed in Matlab. The lattice-size is derived from a user-defined percentage occupancy of all of the specified initial molecule abundances. Individual cells in the lattice are treated as equal-sized molecules or empty space. User-defined molecular abundances are uniformly seeded at random in the lattice space. Molecular motion is simulated by a single-unit step-size random walk in a Moore neighbourhood. Upon encounter in space molecules react with a user-defined probability provided they have been defined as substrates. The outcome of a reaction is the replacement of the reactant molecules with product molecules and/or empty space. Thus, substrates that are not utilized in reactions (modifiers) are defined as both reactants and products of the reaction.
De novo synthesis reactions are simulated by the randomised uniform seeding of a user-defined number of molecules into the grid, with a user-defined probability, every generation. A signal is simulated as an assignment rule (Event) in the simulation where a user-defined molecular quantity is randomly distributed across the lattice at a specified generation. Encounter of non-substrate molecules or movement towards a cell outside of the grid will result in an 180° perfectly elastic collision.
5.3. Mutual information
A Matlab script was devised to calculate the mutual information (Shannon, 2001) between variables within the different computational models. The mutual information between two discrete random variables X and Y is defined as:
| (1) |
However, in practice, the joint distribution is not a direct observable. Thus we can derive:
Substitute into Eq. (1) so then mutual information becomes:
| (2) |
Consider X to be the upstream signalling molecule and Y to be the downstream protein effector. The calculation of the theoretical mutual information requires a simulated dose-response where, for each individual value x of the signal, the computational model must be stochastically simulated enough times to derive a consistent frequency tabulation of all possible abundance values of Y at a single time point or time window. Hence, p(y|x) is derived for each value of X and p(y) can be obtained from the marginal probabilities across all values of X. p(x) can be assumed to be a uniform distribution (Uda and Kuroda, 2016).
5.4. Agent model
A 3D cellular automaton model was constructed to simulate a regular lattice 50% occupied by agents seeded randomly across the grid. All agents begin in a resting state (R) and can transition to a series of other states throughout the simulation, namely, to a perturbed state (P), to a negative regulator state (N) or to a state of non-existence (E). At each generation, each randomly-selected agent can undergo the following rules:
-
i)
If state is (R) then transition to (P) with probability Pind
-
ii)
If state is (P) then ni neighbouring (E) agents transition to (N) with probability Prec
-
iii)
If state is (N) then neighbouring (P) agents transition to (R) with probability Pres
-
iv)
If state is (N) and state has existed for Gi generations then (N) transitions to (E)
Note that neighbouring agents are defined as those within the Moore neighbourhood of the selected agent. ni encodes the strength of negative regulation. Rule Gi represents the duration of the presence of a negative regulatory state.
5.5. Sensitivity analysis
Local sensitivity analysis was carried out in COPASI. This method systematically and sequentially alters parameter values by a user-defined magnitude (as a percentage of the parameter value) and examines how simulation output is changed by such alteration (Kent et al., 2013). Normalised sensitivities were plotted in Matlab (MathWorks Inc., Natick, MA, 2016). Sensitivity analysis was performed at steady state and configured with parameter values of 0.001 and 1e-12 for the delta factor and the delta minimum respectively. Sensitivity analysis was performed on all kinetic rate constant parameters.
Author summary
Ageing and disease can be understood as a loss of biological homeostasis. This commonly manifests as the constitutive elevation of biological signals. However, how such chronic elevation in the basal level of biological signals may affect regulatory pathways remains largely unexplored.
In this work we undertake a theoretical approach to explore how constitutive signals may affect the ability of biological systems to mount appropriate responses. We take oxidative stress feeding into redox signalling pathways as a working example.
We report that constitutive signals can reduce the responsiveness of biological signalling pathways by increasing the basal levels of negative regulator entities. The robustness of such a theoretical observation to the underlying methodology hints at the generality of this principle.
Acknowledgments
This work was funded by MRC-Arthritis Research UK Centre for Integrated research into Musculoskeletal Ageing (CIMA) (grant number MR/K006312/1).
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.mad.2017.11.010.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Adamson A., Boddington C., Downton P., Rowe W., Bagnall J., Lam C. Signal transduction controls heterogeneous NF-(B dynamics and target gene expression through cytokine-specific refractory states. Nat. Commun. 2016;7:12057. doi: 10.1038/ncomms12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri E., Sestili P. Reactive oxygen species in skeletal muscle signaling. J. Signal Trans. 2012;2012:982794. doi: 10.1155/2012/982794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse-Patin A., Estall J.L. An intimate relationship between ROS and insulin signalling: implications for antioxidant treatment of fatty liver disease. Int. J. Cell Biol. 2014;2014:519153. doi: 10.1155/2014/519153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowsher C.G., Swain P.S. Environmental sensing, information transfer, and cellular decision-making. Curr. Opin. Biotechnol. 2014;28:149–155. doi: 10.1016/j.copbio.2014.04.010. [DOI] [PubMed] [Google Scholar]
- Brioche T., Lemoine-Morel S. Oxidative stress, sarcopenia, antioxidant strategies and exercise: molecular aspects. Curr. Pharm. Des. 2016;22(18):2664–2678. doi: 10.2174/1381612822666160219120531. [DOI] [PubMed] [Google Scholar]
- Chandel N.S., McClintock D.S., Feliciano C.E., Wood T.M., Melendez J.A., Rodriguez A.M. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J. Biol. Chem. 2000;275(33):25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- Chelliah V., Juty N., Ajmera I., Ali R., Dumousseau M., Glont M. BioModels: ten-year anniversary. Nucleic Acids Res. 2015;43(D1):D542–D548. doi: 10.1093/nar/gku1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claflin D.R., Jackson M.J., Brooks S.V. Age affects the contraction-induced mitochondrial redox response in skeletal muscle. Front. Physiol. 2015;6:21. doi: 10.3389/fphys.2015.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobley J.N., Moult P.R., Burniston J.G., Morton J.P., Close G.L. Exercise improves mitochondrial and redox-regulated stress responses in the elderly: better late than never! Biogerontology. 2015;16(2):249–264. doi: 10.1007/s10522-014-9546-8. [DOI] [PubMed] [Google Scholar]
- Covas G., Marinho H.S., Cyrne L., Antunes F. Activation of Nrf2 by H2O2: de novo synthesis versus nuclear translocation. Methods Enzymol. 2013;528:157–171. doi: 10.1016/B978-0-12-405881-1.00009-4. [DOI] [PubMed] [Google Scholar]
- Cuadrado A. Structural and functional characterization of Nrf2 degradation by glycogen synthase kinase 3/beta-TrCP. Free Radic. Biol. Med. 2015;88(Pt B):147–157. doi: 10.1016/j.freeradbiomed.2015.04.029. [DOI] [PubMed] [Google Scholar]
- Dalle Pezze P., Nelson G., Otten E.G., Korolchuk V.I., Kirkwood T.B., von Zglinicki T. Dynamic modelling of pathways to cellular senescence reveals strategies for targeted interventions. PLoS Comput. Biol. 2014;10(8):e1003728. doi: 10.1371/journal.pcbi.1003728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A.M., Smith D.A., Ikeh M.A., Haider M., Herrero-de-Dios C.M., Brown A.J. Blocking two-component signalling enhances Candida albicans virulence and reveals adaptive mechanisms that counteract sustained SAPK activation. PLoS Pathog. 2017;13(1):e1006131. doi: 10.1371/journal.ppat.1006131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marchi E., Baldassari F., Bononi A., Wieckowski M.R., Pinton P. Oxidative stress in cardiovascular diseases and obesity: role of p66Shc and protein kinase C. Oxid. Med. Cell. Longevity. 2013;2013:564961. doi: 10.1155/2013/564961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decuypere J.P., Monaco G., Missiaen L., De Smedt H., Parys J.B., Bultynck G. IP(3) receptors, mitochondria, and Ca signaling: implications for aging. J. Aging Res. 2011;2011:920178. doi: 10.4061/2011/920178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Done A.J., Gage M.J., Nieto N.C., Traustadottir T. Exercise-induced Nrf2-signaling is impaired in aging. Free Radical Biol. Med. 2016;96:130–138. doi: 10.1016/j.freeradbiomed.2016.04.024. [DOI] [PubMed] [Google Scholar]
- Dues D.J., Andrews E.K., Schaar C.E., Bergsma A.L., Senchuk M.M., Van Raamsdonk J.M. Aging causes decreased resistance to multiple stresses and a failure to activate specific stress response pathways. Aging (Albany NY). 2016;8(4):777–795. doi: 10.18632/aging.100939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucon P.C., Pardee K., Kumar R.M., Li H., Loh Y.H., Wang X. Gene networks of fully connected triads with complete auto-activation enable multistability and stepwise stochastic transitions. PLoS One. 2014;9(7):e102873. doi: 10.1371/journal.pone.0102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C., Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. Series A, Biol. Sci. Med. Sci. 2014;69(Suppl 1):S4–S9. doi: 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- Fulop T., Le Page A., Fortin C., Witkowski J.M., Dupuis G., Larbi A. Cellular signaling in the aging immune system. Curr. Opin. Immunol. 2014;29:105–111. doi: 10.1016/j.coi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Gorlach A., Bertram K., Hudecova S., Krizanova O. Calcium and ROS. a mutual interplay. Redox Biol. 2015;6:260–271. doi: 10.1016/j.redox.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissom N., Bhatnagar S. Habituation to repeated stress: get used to it. Neurobiol. Learn. Mem. 2009;92(2):215–224. doi: 10.1016/j.nlm.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzikirou H., Deutsch A. Lattice-Gas cellular automaton modeling of emergent behavior in interacting cell populations. In: Kroc J., Sloot P.M.A., Hoekstra A.G., editors. Simulating Complex Systems by Cellular Automata. Springer Berlin Heidelberg; Berlin, Heidelberg: 2010. pp. 301–331. [Google Scholar]
- Herman J.P. Neural control of chronic stress adaptation. Front. Behav. Neurosci. 2013;7:61. doi: 10.3389/fnbeh.2013.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoesel B., Schmid J.A. The complexity of NF-kappaB signaling in inflammation and cancer. Mol. Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops S., Sahle S., Gauges R., Lee C., Pahle J., Simus N. COPASI?a COmplex PAthway SImulator. Bioinformatics. 2006;22(24):3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- Ingram P.J., Stumpf M.P., Stark J. Network motifs: structure does not determine function. BMC Genomics. 2006;7:108. doi: 10.1186/1471-2164-7-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M.J. Reactive oxygen species in sarcopenia: should we focus on excess oxidative damage or defective redox signalling. Mol. Aspects Med. 2016;50:33–40. doi: 10.1016/j.mam.2016.05.002. [DOI] [PubMed] [Google Scholar]
- Jiang F., Zhang Y., Dusting G.J. NADPH oxidase-mediated redox signaling: roles in cellular stress response, stress tolerance, and tissue repair. Pharmacol. Rev. 2011;63(1):218–242. doi: 10.1124/pr.110.002980. [DOI] [PubMed] [Google Scholar]
- Jiang S., Tu K., Fu Q., Schmitt D.C., Zhou L., Lu N. Multifaceted roles of HSF1 in cancer. Tumor Biol. 2015;36(7):4923–4931. doi: 10.1007/s13277-015-3674-x. [DOI] [PubMed] [Google Scholar]
- Karin O., Swisa A., Glaser B., Dor Y., Alon U. Dynamical compensation in physiological circuits. Mol. Syst. Biol. 2016;12(11) doi: 10.15252/msb.20167216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar J.W., Niture S.K., Jaiswal A.K. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radical Biol. Med. 2009;47(9):1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent E., Neumann S., Kummer U., Mendes P. What can we learn from global sensitivity analysis of biochemical systems? PLoS One. 2013;8(11):e79244. doi: 10.1371/journal.pone.0079244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G.H., Kim J.E., Rhie S.J., Yoon S. The role of oxidative stress in neurodegenerative diseases. Exp. Neurobiol. 2015;24(4):325–340. doi: 10.5607/en.2015.24.4.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood T.B.L., Kowald A. The free-radical theory of ageing −older, wiser and still alive. Bioessays. 2012;34(8):692–700. doi: 10.1002/bies.201200014. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Tsunada J., Cohen Y.E. A model of the differential representation of signal novelty in the local field potentials and spiking activity of the ventrolateral prefrontal cortex. Neural Comput. 2013;25(1):157–185. doi: 10.1162/NECO_a_00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepetsos P., Papavassiliou A.G. ROS/oxidative stress signaling in osteoarthritis. Biochim. Biophys. Acta. 2016;1862(4):576–591. doi: 10.1016/j.bbadis.2016.01.003. [DOI] [PubMed] [Google Scholar]
- Lewis K.N., Wason E., Edrey Y.H., Kristan D.M., Nevo E., Buffenstein R. Regulation of Nrf2 signaling and longevity in naturally long-lived rodents. Proc. Natl. Acad. Sci. U. S. A. 2015;112(12):3722–3727. doi: 10.1073/pnas.1417566112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio M., Colizzi E., Fisichella A., Valenti G., Ceresini G., Dall'Aglio E. Stress hormones, sleep deprivation and cognition in older adults. Maturitas. 2013;76(1):22–44. doi: 10.1016/j.maturitas.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Masoud G.N., Li W. HIF-1alpha pathway: role, regulation and intervention for cancer therapy. Acta Pharma. Sin. B. 2015;5(5):378–389. doi: 10.1016/j.apsb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonagh B., Sakellariou G.K., Jackson M.J. Application of redox proteomics to skeletal muscle aging and exercise. Biochem. Soc. Trans. 2014;42(4):965–970. doi: 10.1042/BST20140085. [DOI] [PubMed] [Google Scholar]
- McMahon S.S., Lenive O., Filippi S., Stumpf M.P. Information processing by simple molecular motifs and susceptibility to noise. J. R. Soc. Interface. 2015;12(110):0597. doi: 10.1098/rsif.2015.0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millonig G., Ganzleben I., Peccerella T., Casanovas G., Brodziak-Jarosz L., Breitkopf-Heinlein K. Sustained submicromolar H2O2 levels induce hepcidin via signal transducer and activator of transcription 3 (STAT3) J. Biol. Chem. 2012;287(44):37472–37482. doi: 10.1074/jbc.M112.358911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M.J., Liu Z.G. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 2011;21(1):103–115. doi: 10.1038/cr.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanduri J., Vaddi D.R., Khan S.A., Wang N., Makarenko V., Semenza G.L. HIF-1alpha activation by intermittent hypoxia requires NADPH oxidase stimulation by xanthine oxidase. PLoS One. 2015;10(3):e0119762. doi: 10.1371/journal.pone.0119762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Marques V., Marinho H.S., Cyrne L., Antunes F. Role of hydrogen peroxide in NF-kappaB activation: from inducer to modulator. Antioxidants Redox Signaling. 2009;11(9):2223–2243. doi: 10.1089/ars.2009.2601. [DOI] [PubMed] [Google Scholar]
- Pickering A.M., Vojtovich L., Tower J., Davies K.J.A. Oxidative stress adaptation with acute, chronic, and repeated stress. Free Radical Biol. Med. 2013;55:109–118. doi: 10.1016/j.freeradbiomed.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott S.A., Chase R. Sites of plasticity in the neural circuit mediating tentacle withdrawal in the snail Helix aspersa: implications for behavioral change and learning kinetics. Learn. Mem. 1999;6(4):363–380. [PMC free article] [PubMed] [Google Scholar]
- Rahi Sahand J., Pecani K., Ondracka A., Oikonomou C., Cross Frederick R. The CDK-APC/C oscillator predominantly entrains periodic cell-Cycle transcription. Cell. 2016;165(2):475–487. doi: 10.1016/j.cell.2016.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami M. Network plasticity in adaptive filtering and behavioral habituation. Neuron. 2014;82(6):1216–1229. doi: 10.1016/j.neuron.2014.04.035. [DOI] [PubMed] [Google Scholar]
- Rhee A., Cheong R., Levchenko A. The application of information theory to biochemical signaling systems. Phys. Biol. 2012;9(4) doi: 10.1088/1478-3975/9/4/045011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribou A.C. Synthetic sensors for reactive oxygen species detection and quantification: a critical review of current methods. Antioxidants Redox Signaling. 2016;25(9):520–533. doi: 10.1089/ars.2016.6741. [DOI] [PubMed] [Google Scholar]
- Roedding A.S., Tong S.Y., Au-Yeung W., Li P.P., Warsh J.J. Chronic oxidative stress modulates TRPC3 and TRPM2 channel expression and function in rat primary cortical neurons: relevance to the pathophysiology of bipolar disorder. Brain Res. 2013;1517:16–27. doi: 10.1016/j.brainres.2013.04.025. [DOI] [PubMed] [Google Scholar]
- Sanz A. Mitochondrial reactive oxygen species: do they extend or shorten animal lifespan? Biochim. Biophys. Acta. 2016;1857(8):1116–1126. doi: 10.1016/j.bbabio.2016.03.018. [DOI] [PubMed] [Google Scholar]
- Scheffer M., Bascompte J., Brock W.A., Brovkin V., Carpenter S.R., Dakos V. Early-warning signals for critical transitions. Nature. 2009;461(7260):53–59. doi: 10.1038/nature08227. [DOI] [PubMed] [Google Scholar]
- Schilling M., Maiwald T., Hengl S., Winter D., Kreutz C., Kolch W. Theoretical and experimental analysis links isoform-specific ERK signalling to cell fate decisions. Mol. Syst. Biol. 2009;5:334. doi: 10.1038/msb.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell S., Turner T.E. Reaction kinetics in intracellular environments with macromolecular crowding: simulations and rate laws. Prog. Biophys. Mol. Biol. 2004;85(2–3):235–260. doi: 10.1016/j.pbiomolbio.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Sen C.K., Roy S. Redox signals in wound healing. Biochim. Biophys. Acta. 2008;1780(11):1348–1361. doi: 10.1016/j.bbagen.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon C.E. A mathematical theory of communication. ACM SIGMOBILE Mobile Comput. Commun. Rev. 2001;5(1):3–55. [Google Scholar]
- Shiraishi T., Matsuyama S., Kitano H. Large-scale analysis of network bistability for human cancers. PLoS Comput. Biol. 2010;6(7):e1000851. doi: 10.1371/journal.pcbi.1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sies H. Oxidative stress: a concept in redox biology and medicine. Redox Biol. 2015;4:180–183. doi: 10.1016/j.redox.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobotta M.C., Barata A.G., Schmidt U., Mueller S., Millonig G., Dick T.P. Exposing cells to H2O2: a quantitative comparison between continuous low-dose and one-time high-dose treatments. Free Radical Biol. Med. 2013;60:325–335. doi: 10.1016/j.freeradbiomed.2013.02.017. [DOI] [PubMed] [Google Scholar]
- Soga M., Matsuzawa A., Ichijo H. Oxidative stress-Induced diseases via the ASK1 signaling pathway. Int. J. Cell Biol. 2012;2012:439587. doi: 10.1155/2012/439587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal R.S., Orr W.C. The redox stress hypothesis of aging. Free Radical Biol. Med. 2012;52(3):539–555. doi: 10.1016/j.freeradbiomed.2011.10.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa V., Moline T., Somoza R., Paciucci R., Kondoh H., ME L.L. Oxidative stress and cancer: an overview. Ageing Res. Rev. 2013;12(1):376–390. doi: 10.1016/j.arr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Swan C.L., Sistonen L. Cellular stress response cross talk maintains protein and energy homeostasis. EMBO J. 2015;34(3):267–269. doi: 10.15252/embj.201490757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan P.L., Shavlakadze T., Grounds M.D., Arthur P.G. Differential thiol oxidation of the signaling proteins Akt, PTEN or PP2A determines whether Akt phosphorylation is enhanced or inhibited by oxidative stress in C2C12 myotubes derived from skeletal muscle. Int. J. Biochem. Cell Biol. 2015;62:72–79. doi: 10.1016/j.biocel.2015.02.015. [DOI] [PubMed] [Google Scholar]
- Tebay L.E., Robertson H., Durant S.T., Vitale S.R., Penning T.M., Dinkova-Kostova A.T. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radical Biol. Med. 2015;88(Pt B):108–146. doi: 10.1016/j.freeradbiomed.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima Y., Kakuda H., Fujita K.A., Uda S., Kuroda S. Sensitivity control through attenuation of signal transfer efficiency by negative regulation of cellular signalling. Nat. Commun. 2012;3:743. doi: 10.1038/ncomms1745. [DOI] [PubMed] [Google Scholar]
- Trachootham D., Lu W., Ogasawara M.A., Nilsa R.D., Huang P. Redox regulation of cell survival. Antioxidants Redox Signaling. 2008;10(8):1343–1374. doi: 10.1089/ars.2007.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uda S., Kuroda S. Analysis of cellular signal transduction from an information theoretic approach. Seminars Cell Dev. Biol. 2016;51:24–31. doi: 10.1016/j.semcdb.2015.12.011. [DOI] [PubMed] [Google Scholar]
- Vasilaki A., McArdle F., Iwanejko L.M., McArdle A. Adaptive responses of mouse skeletal muscle to contractile activity: the effect of age. Mech. Ageing Dev. 2006;127(11):830–839. doi: 10.1016/j.mad.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Vizan P., Miller D.S., Gori I., Das D., Schmierer B., Hill C.S. Controlling long-term signaling: receptor dynamics determine attenuation and refractory behavior of the TGF-beta pathway. Sci. Signal. 2013;6(305):ra106. doi: 10.1126/scisignal.2004416. [DOI] [PubMed] [Google Scholar]
- Waltermann C., Klipp E. Information theory based approaches to cellular signaling. Biochim. Biophys. Acta. 2011;1810(10):924–932. doi: 10.1016/j.bbagen.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Wang X., Hai C. Novel insights into redox system and the mechanism of redox regulation. Mol. Biol. Rep. 2016;43(7):607–628. doi: 10.1007/s11033-016-4022-y. [DOI] [PubMed] [Google Scholar]
- Winterbourn C.C. Are free radicals involved in thiol-based redox signaling? Free Radical Biol. Med. 2015;80:164–170. doi: 10.1016/j.freeradbiomed.2014.08.017. [DOI] [PubMed] [Google Scholar]
- Woolley J.F., Stanicka J., Cotter T.G. Recent advances in reactive oxygen species measurement in biological systems. Trends Biochem. Sci. 2013;38(11):556–565. doi: 10.1016/j.tibs.2013.08.009. [DOI] [PubMed] [Google Scholar]
- Wurthner J.U., Mukhopadhyay A.K., Peimann C.J. A cellular automaton model of cellular signal transduction. Comput. Biol. Med. 2000;30(1):1–21. doi: 10.1016/s0010-4825(99)00020-7. [DOI] [PubMed] [Google Scholar]
- Yoo H.J., Im C.N., Youn D.Y., Yun H.H., Lee J.H. Bis is induced by oxidative stress via activation of HSF1. Korean J. Physiol. Pharmacol. 2014;18(5):403–409. doi: 10.4196/kjpp.2014.18.5.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J., Finkel T. Mitohormesis Cell metabolism. 2014;19(5):757–766. doi: 10.1016/j.cmet.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Davies K.J., Forman H.J. Oxidative stress response and Nrf2 signaling in aging. Free Radical Biol. Med. 2015;88(Pt B):314–336. doi: 10.1016/j.freeradbiomed.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.