Abstract
The human Y-chromosome has proven to be a powerful tool for tracing the paternal history of human populations and genealogical ancestors. The human Y-chromosome haplogroup Q is the most frequent haplogroup in the Americas. Previous studies have traced the origin of haplogroup Q to the region around Central Asia and Southern Siberia. Although the diversity of haplogroup Q in the Americas has been studied in detail, investigations on the diffusion of haplogroup Q in Eurasia and Africa are still limited. In this study, we collected 39 samples from China and Russia, investigated 432 samples from previous studies of haplogroup Q, and analyzed the single nucleotide polymorphism (SNP) subclades Q1a1a1-M120, Q1a2a1-L54, Q1a1b-M25, Q1a2-M346, Q1a2a1a2-L804, Q1a2b2-F1161, Q1b1a-M378, and Q1b1a1-L245. Through NETWORK and BATWING analyses, we found that the subclades of haplogroup Q continued to disperse from Central Asia and Southern Siberia during the past 10,000 years. Apart from its migration through the Beringia to the Americas, haplogroup Q also moved from Asia to the south and to the west during the Neolithic period, and subsequently to the whole of Eurasia and part of Africa.
Electronic supplementary material
The online version of this article (doi:10.1007/s00438-017-1363-8) contains supplementary material, which is available to authorized users.
Keywords: Y-chromosome, Haplogroup Q, Multidirectional migrations, Eurasia, Han Chinese
Introduction
In recent decades, the human Y-chromosome has proven to be a powerful tool for tracing the paternal history of human populations and genealogical ancestors. The human Y-chromosome haplogroup Q (also named Q-M242 in accordance with its defining mutation) probably originated in Central Asia and Southern Siberia during the time period of 15–25 KYA (1000 years ago) (Karafet et al. 2002, 2008; Bortolini et al. 2003; Seielstad et al. 2003), then subsequently diffused in the eastward, westward and southward directions (Zhong et al. 2011; Di Cristofaro et al. 2013; Sandoval et al. 2013; Liu et al. 2014; Rasmussen et al. 2014). Haplogroup Q has several subclades defined by single nucleotide polymorphisms (SNPs), and it reaches its highest frequency of 70–100% in the Americas (Bortolini et al. 2003; Seielstad et al. 2003; Zhong et al. 2011; Rasmussen et al. 2014). Although the diversity of haplogroup Q in the Americas has been studied in detail (Bisso-Machado et al. 2011; Toscanini et al. 2011, 2016; Jota et al. 2011; Malyarchuk et al. 2011; Dulik et al. 2012b; Battaglia et al. 2013; Lardone et al. 2013; Melton et al. 2013; Regueiro et al. 2013; Noguera et al. 2014; Sala and Corach 2014; Torres et al. 2015), investigations on the diffusion of haplogroup Q in Eurasia and Africa are still limited. Consequently, we studied samples of haplogroup Q in Eurasia to explore how it expanded from Central Asia and Southern Siberia during the Neolithic period.
The ancestors of present-day Native Americans migrated to the Americas from Siberia via the Beringia around 16 KYA (Raghavan et al. 2015; Llamas et al. 2016). Q1a2a1-L54 and its subclade Q1a2a1a1-M3 are the two predominant subclades of haplogroup Q found on both sides of the Bering Strait. Q1a2a1-L54 has spread throughout Northern Asia, the Americas, and Western and Central Europe (Raff and Bolnick 2014; Rasmussen et al. 2014). An ancient individual of the Clovis culture belonged to Q1a2a1-L54 (xQ1a2a1a1-M3) (O’Rourke and Raff 2010; Rasmussen et al. 2014). Q1a2a1a1-M3, one of the most thoroughly studied subclades within haplogroup Q, is frequent both in the Chukotka Peninsula of Siberia (close to Alaska) and the Americas (Lell et al. 2002). Previous studies indicated that Q1a2a1a1-M3 migrated from Siberia to the Americas and partially returned to Siberia (Hammer et al. 1997; Lell et al. 1997; Bortolini et al. 2003; Pakendorf et al. 2007). The estimated time of Q1a2a1a1-M3 is 13-22 KYA (Dulik et al. 2012a). Q1a2a1a1a-M19, a subclade of Q1a2a1a1-M3, remained in Southern America and has a similarly diversified pattern with its upstream lineage. The age of Q1a2a1a1a-M19 is approximately 7–8 KYA (Bortolini et al. 2003; Jota et al. 2011).
The frequencies of haplogroup Q range from 0 to 94% in Eurasia (approximately 5% on average) (Seielstad et al. 2003; Varzari et al. 2013). Haplogroup Q reaches its highest frequencies in Siberia, especially in Kets (90–94%) and Selkups (66–71%), and is rarely seen in Western, Southern and South-eastern Asia (Wells et al. 2001; Zerjal et al. 2002; Tambets et al. 2004; Sengupta et al. 2006; Sharma et al. 2007; Haber et al. 2011a, b; Dulik et al. 2011; Grugni et al. 2012; Di Cristofaro et al. 2013; Rasmussen et al. 2014). Subclade Q1a1a1-M120 appears almost only in Eastern Asia, and its diversity implies that haplogroup Q has migrated from north to south with the ancestors of current Han Chinese during the Neolithic period (Su et al. 2000; Wells et al. 2001; Tarazona-Santos et al. 2001; Wen et al. 2004; Gayden et al. 2007; Nonaka et al. 2007; Zhong et al. 2011; Zhao et al. 2015). Subclades Q1a1b-M25 and Q1a2-M346 have spread widely in Eurasia. Q1a1b-M25 reaches its highest frequency in Turkmen (34–43%) and shows low frequencies in other Eurasian populations (Underhill et al. 2000; Malyarchuk et al. 2011; Zhong et al. 2011), while Q1a2-M346 appears in Central, Western and Southern Asia, and most parts of Europe (Sengupta et al. 2006; Sharma et al. 2007; Abu-Amero et al. 2009; Bailliet et al. 2009).
Haplogroup Q has also appeared in other parts of the world. For instance, an ancient DNA study of a Saqqaq individual in Greenland suggests that haplogroup Q1a-MEH2 was frequent in Siberian and Native American populations (Karafet et al. 2008; Rasmussen et al. 2010; Raghavan et al. 2015). A few subclades of haplogroup Q have been identified in the Comoros population in Africa (Q1a2-M346) and the Polynesian islands in Oceania (Q1a2a1a1c-M199) (Hurles et al. 2003; Msaidie et al. 2010).
Nowadays, the distribution of haplogroup Q in the Americas has been studied thoroughly, but we know little about its dispersals on western and southern routes. In this study, we present an analysis of some SNP subclades of haplogroup Q, including Q1a1a1-M120, Q1a2a1-L54, Q1a1b-M25, Q1a2-M346, Q1a2a1a2-L804, Q1a2b2-F1161, Q1b1a-M378, and Q1b1a1-L245. Based on NETWORK and BATWING analyses of haplogroup Q, we were able to better understand its dispersals on western and southern routes, and their impacts on Eurasian populations.
Materials and methods
Ethic statement
This study was conducted after the approval of the Ethical Committee of the School of Life Sciences, Fudan University (Shanghai, China) and the ethical committee of the Lomonosov Moscow State University (Moscow, Russia). All donors of samples were completely informed and signed informed consent forms before sample collection.
Population samples
In this study, a total of 471 unrelated male samples were analyzed. We collected blood samples of 1757 healthy and unrelated volunteers from five populations in China, including 700 Hui, 64 Bao-An, 109 Dong-Xiang, 90 Li-Qian, and 794 Shao-Xing individuals. In addition, we collected saliva samples of 30 healthy and unrelated volunteers from 3 populations in Russia, including 4 Enets, 19 Ket, and 7 Selkup individuals. After genotyping all samples, we confirmed that 16 samples of China and 23 samples of Russia belonged to haplogroup Q, which were further investigated in this study. Furthermore, data from previous studies were also analyzed (Bailliet et al. 2009; Zhong et al. 2011; Lacau et al. 2012; Dulik et al. 2012; Di Cristofaro et al. 2013; Sandoval et al. 2013; Varzari et al. 2013; Hollard et al. 2014; Liu et al. 2014; Family Tree DNA). The populations were categorized in accordance with the location of residence as follows: from Gansu province of China: Bao-An, one individual from Ji-Shi Mountain; Dong-Xiang, two individuals from Dong-Xiang county, Hui Autonomous Prefecture of Lin-Xia; Li-Qian, four individuals from Yong-Chang county, Jin-Chang city; from Zhejiang Province of China: Shao-Xing, nine individuals from Shao-Xing city. In the Krasnoyarsk Region of Russia: Enets—two individuals from Potapovo; Ket—one individual from each of Farkovo, Sulomai/Bor, Sumarokovo, Turukhansk, and Verkhneimbatsk, two individuals from each of Bakhta, Baklanikha and Kellog, and five individuals from Sulomai; Selkup—three individuals from Farkovo, and two individuals from Turukhansk. These three populations are considered minorities in Russia according to the 2002 All-Russia Population Census (ESM_3). Enets (named Entses in ESM_3) has 237 individuals; Ket has 1494 individuals; Selkup has 4249 individuals.
Y-chromosome markers
Genomic DNA was extracted from the blood samples using the DP-318 Kit (Tiangen Biotechnology, Beijing, China), and the DNA extraction protocol for the saliva samples was adapted from the high-salt DNA extraction method (Quinque et al. 2006). The samples were typed as the most recent Y-chromosome phylogenetic tree (ISOGG 2017). The selected samples belonged to several subclades of haplogroup Q.
Binary markers were hierarchically genotyped by SNaPshot (ABI SNaPshot Multiplex Kit, Carlsbad, CA, USA) and fluorescent allele-specific PCR. The PCR products were electrophoresed on a 3730xl Genetic Analyzer (Applied Biosystems, Carlsbad, CA, USA). Seventeen Y-chromosomal STRs (DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS385a, DYS385b, DYS438, DYS439, DYS437, DYS448, DYS456, DYS458, DYS635 and YGATAH4) were amplified using the AmpFlSTR Yfiler PCR amplification kit (Applied Biosystems). The amplified products were separated and identified using a 3730xl Genetic Analyzer (Applied Biosystems) according to the protocol recommended by the manufacturer. The data were analyzed using a Gene-Mapper ID v3.2 (Applied Biosystems). In the analyses, DYS389II was calculated by subtracting the DYS389I allele size.
Statistical analyses
Networks of Y-chromosomal STR data were constructed by the reduced-median method using NETWORK v. 5.0.0.1 (http://www.fluxus-engineering.com) with haplogroups Q1a1a1-M120, Q1a2a1-L54, Q1a1b-M25, Q1a2-M346, Q1a2a1a2-L804, Q1a2b2-F1161, Q1b1a-M378, and Q1b1a1-L245. Because we collected samples from different studies, we had to adjust the number of loci used in our study to match those of other studies. The network of Q1a1a1-M120 was constructed with seven loci: DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, and DYS393. The network of Q1a2a1-L54 was constructed with 15 loci: DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS456, DYS458, DYS635 and GATA H4. The networks of Q1a1b-M25, Q1a2-M346, Q1a2a1a2-L804, Q1a2b2-F1161, Q1b1a-M378, and Q1b1a1-L245 were each constructed with ten loci: DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438 and DYS439.
We used the Markov chain Monte Carlo (MCMC) approach (Wilson et al. 2003) incorporated into the program BATWING to estimate the time to the most recent common ancestor (TMRCA) and the expansion time of the aforementioned Q subclades. Time estimates for subclades of haplogroup Q were made using seven to fifteen of the STRs listed above. A model of exponential growth from an initially constant-sized population was employed in BATWING for obtaining the time estimates. Four sets of widely used Y-STR mutation rates were applied in the time estimates as Wei et al. (2013): evolutionary mutation rate (EMR) (Zhivotovsky et al. 2004), two observed genealogical mutation rates (OMRB and OMRS) (Shi et al. 2010; Burgarella and Navascués 2011), and a genealogical mutation rate adjusted for population variation using a logistic model (lmMR) (Wilson et al. 2003). A generation time of 30 years was used to produce a time estimate in years (Tremblay and Vézina 2000). We applied weakly informative prior distribution parameters in BATWING estimations to analyze populations individually. For the initial effective population size (N), we used a broad prior gamma (1, 0.0001) (mean = 10,000, SD = 10,000). For population growth rate per generation (α), we also used the broad prior distribution gamma (2, 400) (mean = 0.005, SD = 0.0035). For the time in coalescent units when exponential growth (β) began we used gamma (2, 1) (mean = 2, SD = 1.41) (Xue et al. 2006). A total of 104 samples of the program’s output representing 106 MCMC cycles were taken after discarding the first 3 × 103 samples as “burn-in” (Xue et al. 2006), and convergence was confirmed by examining longer runs for all populations and finding the same posterior distributions. The TMRCA was calculated using the product of the estimated population size N and the height of the tree T (in coalescent units).
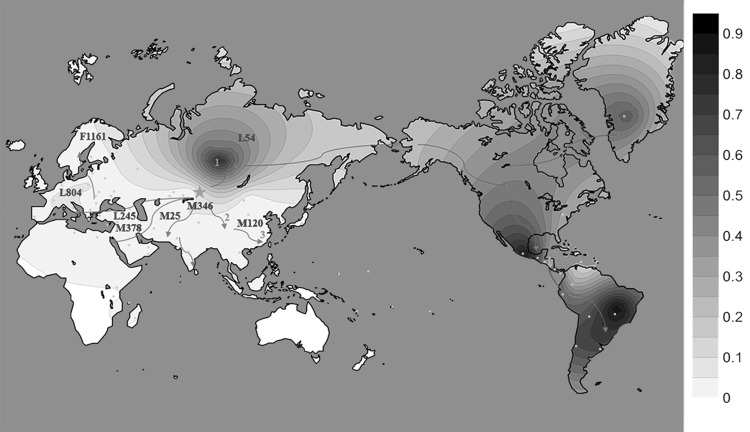
A contour map for the frequencies of haplogroups Q-M242 was generated using the Kriging procedure with the aid of the Golden Software Surfer 11 (Golden Software Inc., CO, USA) (Fig. 1). Since the frequency data were obtained from many sources, the identified subclades of haplogroup Q were different. To show all frequencies in one figure, we integrated the frequencies of different subclades into frequencies of Q-M242. The raw frequency data and references are shown in ESM_2.
Fig. 1.
Worldwide distribution of haplogroup Q-M242. The blue star is the original place of haplogroup Q-M242, around Central Asia and Siberia. The brown number one is Russian sample location in the Krasnoyarsk Region. The brown number two is Chinese sample location in Gansu province. The brown number three is Chinese sample location in Zhejiang province. The red arrows are the expansion routes of haplogroup Q-M242. The purple words show the locations of subclades of haplogroup Q used in this study. The orange points represent the sample locations collected from published studies (ESM_2) (color figure online)
Results
Worldwide distribution of haplogroup Q-M242
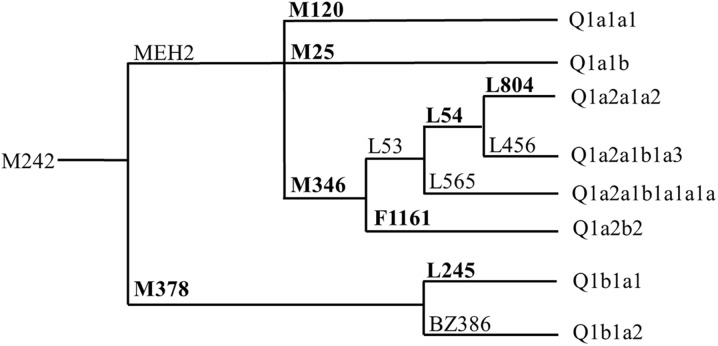
We calculated the frequencies of our samples and collected the frequency data from previous studies (ESM_2). As can be seen in Fig. 1, the frequencies of haplogroup Q-M242 are low in most of the world, except for the Americas and a small part of Siberia, which matches previously published observations on the distribution of haplogroup Q (Balanovsky et al. 2017). Moreover, we represented the migration routes of haplogroup Q-M242 based on our results and previous studies (Fig. 1, ESM_2). We also marked the main distribution regions of the subclades studied in this research (Fig. 1). We have constructed a phylogenetic tree within haplogroup Q to easily identify the downstream subclades (Fig. 2).
Fig. 2.
Phylogenetic tree of Y-chromosome haplogroup Q-M242. The haplogroup labeling is in agreement with the ISOGG conventions and recent updates (ISOGG 2017). The used subclades are showed in bold
The network of haplogroup Q subclades
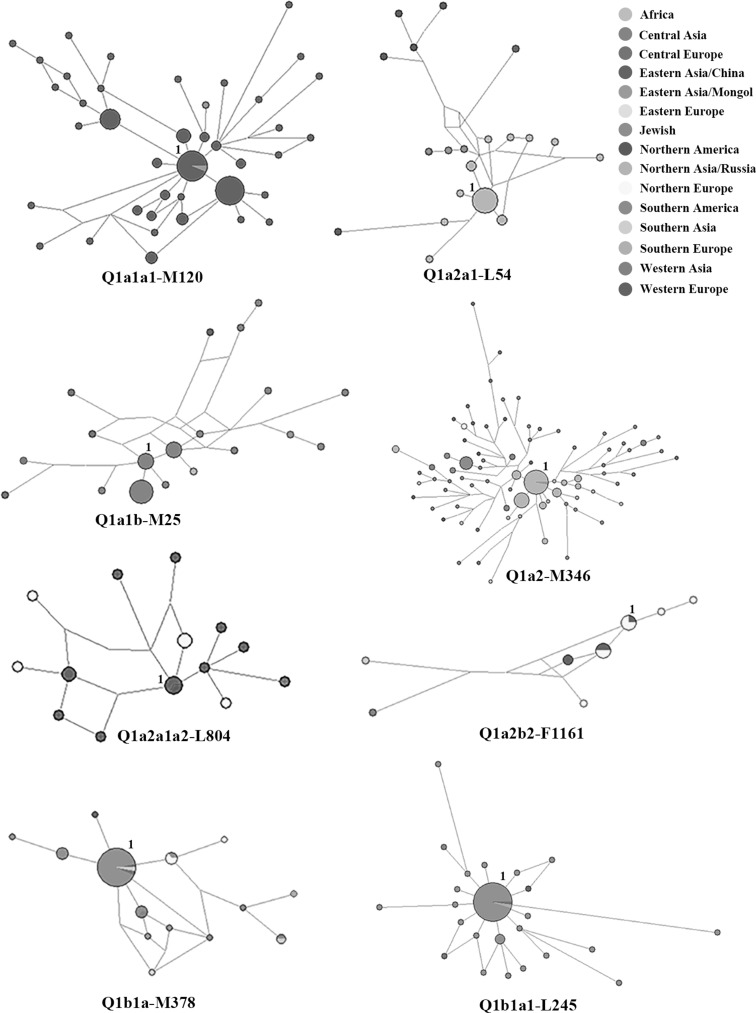
To reveal the detailed structures for subclades of haplogroup Q, we conducted a network analysis combining the SNP and the STR haplotype data for 471 individuals (Fig. 3). The network of Q1a1a1-M120 included most samples from China along with a small number of Mongolian samples. The network of Q1a2a1-L54 contained most samples from Siberia (Northern Asia/Russia), and few samples from Mongolia, China and Northern America. The network of Q1a1b-M25 consisted of samples from Central Asia with a small number of Eastern Asian/Mongolian, Western Asian, Central and Western European samples. The network of Q1a2-M346 mostly contained samples from Northern Asia/Russia and Mongolia, and a few samples from Asia, Europe, the Americas, and even Africa. The network of Q1a2a1a2-L804 had a central cluster shared by Western European and Northern American samples with other branches of Western, Northern and Central Europe. The network of Q1a2b2-F1161 was mainly composed of samples from Northern and Western Europe with two single branches of Southern and Western Asia. The network of Q1b1a-M378 was mainly composed of Jewish samples and a small number of Southern and Central Asian, Western, Northern and Southern European samples. The network of Q1b1a1-L245 had a star-like shape of Jewish samples and a small amount of European and Western Asian samples. We did not discuss the origins and migrations of samples from the Americas because we focused on the dispersals of haplogroup Q in Eurasia and just used samples from the Americas to construct the network.
Fig. 3.
Network of Y-STR haplotypes within haplogroup Q-M242. Q1a1a1-M120: cluster 1 is shared by 16 Eastern Asian/China samples and one Eastern Asian/Mongolia. Q1a2a1-L54: cluster 1 is shared by 11 Northern Asian/Russian samples. Q1a1b-M25: cluster 1 is shared by five Central Asian samples. Q1a2-M346: cluster 1 is shared by 36 Northern Asian samples and 1 Eastern Asian/Mongolia. Q1a2a1a2-L804: cluster 1 is shared by two Western European samples and one Northern American sample. Q1a2b2-F1161: cluster 1 is shared by three Northern European samples and one Western European sample. Q1b1a-M378: cluster 1 is shared by 29 Jewish samples (14 from Central Europe; eleven from Eastern Europe; three from Southern Europe; one from Western Asia), one Western European sample, one Central European sample and one Southern Asian samples. Q1b1a1-L245: cluster 1 is shared by 37 Jewish samples (16 from Central Europe; 14 from Eastern Europe; four from Western Asia; two from Southern Europe; one from Western Europe), one Western European sample and one Southern European sample. Samples included in every cluster 1 are colored by purple in ESM_1
Time estimates for haplogroup Q
We used BATWING to estimate the TMRCA and the expansion time for the subclades of haplogroup Q. As seen in Table 1, the three genealogical mutation rates had approximately similar results, while using the evolutionary mutation rate resulted in a much older TMRCA. The genealogical mutation rates were more reliable when we analyzed a large number of loci and closely related individuals, whereas the evolutionary mutation rate tended to be more effective for estimates on a smaller number of loci and genetically distant individuals (Wang et al. 2014; Wang and Li 2015). Since we used from seven to fifteen loci in the time estimates, and the used populations belonged to the same subclades of haplogroup Q, we decided to use the results of the three genealogical mutation rates.
Table 1.
The TMRCA and expansion times of haplogroup Q subclades (KYA)
| Branch (region or population) | EMR (TMRCA/expansion time) | OMRB (TMRCA/expansion time) | OMRS (TMRCA/expansion time) | lmMR (TMRCA/expansion time) |
|---|---|---|---|---|
| Q1a1a1-M120 | 38.5/16.2 | 11.1/6.4 | 11.1/6.4 | 9.8/5.8 |
| Q1a1a1-M120 DYS391 allele 6 (Han Chinese) | 20.7/13.0 | 6.5/5.5 | 6.7/5.6 | 5.6/5.0 |
| Q1a1a1-M120 DYS391 allele 9 (Han Chinese) | 24.2/13.7 | 7.6/5.8 | 7.8/5.9 | 6.5/5.2 |
| Q1a2a1-L54 (Yenisei basin) | 10.4/9.2 | 2.5/3.4 | 2.4/3.3 | 0.3/0.4 |
| Q1a1b-M25 (Turkic) | 10.9/5.3 | 3.3/2.2 | 4.6/2.7 | 3.1/2.0 |
| Q1a2-M346 (Turkic) | 13.4/8.0 | 4.5/3.8 | 5.5/4.4 | 4.0/3.6 |
| Q1a2-M346 (Western Asia) | 15.1/11.4 | 4.4/4.6 | 5.8/5.5 | 4.0/4.3 |
| Q1a2-M346 (Europe) | 14.0/10.0 | 4.1/4.1 | 5.2/4.9 | 3.8/3.9 |
| Q1a2a1a2-L804 | 19.0/12.6 | 5.8/5.5 | 7.1/6.3 | 5.3/5.2 |
| Q1a2b2-F1161 | 8.6/6.4 | 2.6/2.6 | 3.3/3.1 | 2.3/2.4 |
| Q1b1a-M378 (Jews) | 4.9/4.0 | 1.5/1.6 | 1.8/1.8 | 1.4/1.5 |
| Q1b1a-M378 (Europe) | 6.0/5.2 | 1.8/2.1 | 2.4/2.6 | 1.7/1.9 |
| Q1b1a-M378 (Southern Asia) | 4.0/7.0 | 1.1/3.5 | 1.5/4.1 | 1.0/3.4 |
| Q1b1a1-L245 (Jews) | 23.2/9.1 | 9.8/5.0 | 10.9/5.4 | 8.6/4.8 |
| Q1b1a1-L245 (Western Asia) | 14.6/11.4 | 4.1/4.5 | 5.6/5.5 | 3.9/4.3 |
| Q1b1a1-L245 (Southern Asia) | 7.1/6.1 | 2.1/2.5 | 2.7/2.9 | 1.9/2.3 |
TMRCA the time to the most recent common ancestor, KYA thousand years ago, EMR evolutionary mutation rate (EMR) (Zhivotovsky et al. 2004), OMRB observed genealogical mutation rate (Shi et al. 2010), OMRS observed genealogical mutation rate (Burgarella and Navascués 2011), lmMR a genealogical mutation rate adjusted for population variation using logistic model (Wilson et al. 2003)
Discussion
Subclade Q1a1a1-M120 was found specifically in the Han Chinese with a low frequency (Zhong et al. 2011). Our results suggested that subclade Q1a1a1-M120 had migrated from Mongolia to China during the Neolithic period, and spread over China with the ancestors of Han Chinese (Fig. 3; Table 1; ESM_1). Previous studies showed that Q1a1a1-M120 had migrated from north-western China to the Central Plain as nomads, and merged into the northern Han Chinese farmers at approximately 2.5–3 KYA (Zhao et al. 2010, 2014, 2015; Yan et al. 2014). Therefore, we supposed that the ancient nomads with Q1a1a1-M120 had migrated to south-eastward from north-western China and were assimilated by the Han Chinese farmers (Zhao et al. 2015).
Subclade Q1a2a1-L54 was mainly found in Yeniseian (Ket) and Samoyedic (Enets and Selkup) speakers (ESM_1). Genetic evidence showed that Yeniseian and Samoyedic speakers had genetic affinities to northern Altaians with high frequencies of haplogroup Q-M242 (xL54), while southern Altaians had many L54 samples and showed similarities with Turkic-speaking populations (Dulik et al. 2012b; Battaglia et al. 2013; Flegontov et al. 2016). However, Yeniseian and Samoyedic samples in this study belonged to L54, which was different from the results of previous studies (xL54). In view of the time estimates (Table 1), we postulated that Q1a2a1-L54 had migrated from the southern Altai region and was assimilated into Yeniseian- and Samoyedic-speaking populations during a recent historical period.
Both Q1a1b-M25 and Q1a2-M346 subclades were frequent in Turkic-speaking populations, and their time estimates were at approximately 3-5 KYA (ESM_1; Table 1). According to Fig. 3 and Table 1, Q1a1b-M25 had spread from Central Asia to Western Asia and to Hungary in Central Europe (ESM_1); Q1a2-M346 had migrated from Southern Siberia (Malyarchuk et al. 2011) to most parts of Eurasia and the Comoros Islands of Africa. The results coincided with Turkic nomadic migrations from Southern Siberia and Mongolia to Central and Western Asia, Caucasus, and Eastern Europe (Yunusbayev et al. 2015). Therefore, we suggested that Q1a1b-M25 and Q1a2-M346 probably migrated with Turkic nomads from Southern Siberia to most parts of Eurasia. A few Q1a1b-M25 and Q1a2-M346 samples in Mongolic-speaking populations probably indicated that Turkic nomads had overlapped with Mongolic-speaking populations when they lived in the present Mongolian territory (Yunusbayev et al. 2015). An ancient DNA study showed that the Hungarians probably originated from Central Asia–Southern Siberia at approximately 4 KYA (Neparáczki et al. 2016), which was consistent with our time estimates (Table 1). Therefore, we proposed that Q1a1b-M25 and Q1a2-M346 had migrated from Central Asia–Southern Siberia to Central Europe at least 4 KYA. Three individuals of Africa (the Comoros Islands) that belonged to Q1a2-M346 reaffirmed that Middle Eastern populations had a genetic influence on the Comoros Islands (Gourjon et al. 2011).
Subclades Q1a2a1a2-L804 and Q1a2b2-F1161 were the downstream of Q1a2-M346 (Fig. 2), both of which mainly distributed in Western and Northern Europe (Fig. 3). Q1a2a1a2-L804 arrived in Western and Northern Europe as early as 5-7 KYA (Table 1). Ancient DNA studies showed that first European farmers migrated from Central Europe to Western and Northern Europe between 5 and 7.5 KYA (Haak et al. 2005, 2010; Bramanti et al. 2009; Malmström et al. 2009). Therefore, we supposed that Q1a2a1a2-L804 had spread from Central Europe to Western and Northern Europe with European early Neolithic farmers. The time estimate for Q1a2b2-F1161 was one thousand years later than its upstream clade Q1a2-M346 (Table 1), which seemed to be unrelated to the Neolithic transition of Europe (Haak et al. 2010). Since Q1a2-M346 spread across Europe at that time, it probably brought Q1a2b2-F1161 to Western and Northern Europe, and even to Western and Southern Asia (Khurana et al. 2014; Yunusbayev et al. 2015).
Subclades Q1b1a-M378 and Q1b1a1-L245 were correlated with the Jewish people, both of which probably represented that some of the Jewish Diaspora populations had expanded into Europe within historical times (Table 1; Fig. 3). As seen in Fig. 3, the central clusters of Q1b1a-M378 and Q1b1a1-L245 mainly consisted of samples from Central and Eastern Europe. The results reaffirmed that some Jewish Diaspora populations had migrated from Central and Eastern Europe, and finally settled in other parts of Europe (Nogueiro et al. 2010; Zoossmann-Diskin 2010). Previous Y-chromosome studies showed that haplogroups J, R and Q3a1 had certain proportions in Jewish populations and spread over Europe (Nogueiro et al. 2010; Chaubey et al. 2016; Balanovsky et al. 2017). Subclades Q1b1a-M378 and Q1b1a1-L245 probably spread over Europe with haplogroups J, R and Q3a1. The Q1b1a-M378 samples from Southern Asia might represent the descendants of Ashkenazi Jewish populations because its upstream haplogroup Q-P36 was regarded as minor Ashkenazi Jewish founding lineages in Southern Asia (Lee et al. 2014).
Our study of the human Y-chromosome haplogroup Q in Eurasia revealed a clear pattern of its migration routes during the past 10,000 years, especially in Han Chinese, Yeniseian-, Samoyedic-, Turkic- speaking and Jewish populations. It is clear that a higher resolution database will be helpful to draw more conclusions on the origins, migrations, and ethno-linguistic affiliations of haplogroup Q.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Y-chromosomal STR data haplotypes used within haplogroup Q (XLSX 59 kb)
Frequencies of Haplogroup Q in the world (XLSX 47 kb)
National composition of population and citizenship in Russia (XLSX 25 kb)
Acknowledgements
This study was funded by NSFC for Excellent Young Scholar (31222030), NSFC (31671297), MOST (2016YFC0900300), MOE Scientific Research Project (113022A), Shanghai Shuguang Project (14SG05), and NSFC (31271338). C.C.W was supported by the Max Planck Society and Nanqiang Outstanding Young Talents Program of Xiamen University. L.H.W was supported by Future Scientists Project of China Scholarship Council. We also thank all the sample donors and the laboratory assistants. We thank both unknown reviewers for their constructive comments and suggestions and Ati Rosselet for the English editing.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Research involving human participants and/or animals
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s00438-017-1363-8) contains supplementary material, which is available to authorized users.
References
- Abu-Amero KK, Hellani A, González AM, Larruga JM, Cabrera VM, Underhill PA. Saudi Arabian Y-chromosome diversity and its relationship with nearby regions. BMC Genet. 2009;10:59. doi: 10.1186/1471-2156-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailliet G, Ramallo V, Muzzio M, Garcia A, Santos MR, Alfaro EL, Dipierri J, Salceda S, Carnese FR, Bravi CM, Bianchi NO, Demarchi DA. Brief communication: restricted geographic distribution for Y-Q* paragroup in South America. Am J Phys Anthropol. 2009;140:578–582. doi: 10.1002/ajpa.21133. [DOI] [PubMed] [Google Scholar]
- Balanovsky O, Gurianov V, Zaporozhchenko V, Balaganskaya O, Urasin V, Zhabagin M, Grugni V, Canada R, Al-Zahery N, Raveane A, Wen S-Q, Yan S, Wang X, Zalloua P, Marafi A, Koshel S, Semino O, Tyler-Smith C, Balanovska E. Phylogeography of human Y-chromosome haplogroup Q3-L275 from an academic/citizen science collaboration . BMC Evol Biol. 2017;17:18. doi: 10.1186/s12862-016-0870-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia V, Grugni V, Perego UA, Angerhofer N, Gomez-Palmieri JE, Woodward SR, Achilli A, Myres N, Torroni A, Semino O. The first peopling of South America: new evidence from Y-chromosome haplogroup Q. PLoS One. 2013 doi: 10.1371/journal.pone.0071390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisso-Machado R, Jota MS, Ramallo V, Paixão-Côrtes VR, Lacerda DR, Salzano FM, Bonatto SL, Santos FR, Bortolini MC. Distribution of Y-chromosome Q lineages in native Americans. Am J Hum Biol. 2011;23:563–566. doi: 10.1002/ajhb.21173. [DOI] [PubMed] [Google Scholar]
- Bortolini M-C, Salzano FM, Thomas MG, Stuart S, Nasanen SPK, Bau CHD, Hutz MH, Layrisse Z, Petzl-Erler ML, Tsuneto LT, Hill K, Hurtado AM, Castro-de-Guerra D, Torres MM, Groot H, Michalski R, Nymadawa P, Bedoya G, Bradman N, Labuda D, Ruiz-Linares A. Y-chromosome evidence for differing ancient demographic histories in the Americas. Am J Hum Genet. 2003;73:524–539. doi: 10.1086/377588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramanti B, Thomas MG, Haak W, Unterlaender M, Jores P, Tambets K, Antanaitis-Jacobs I, Haidle MN, Jankauskas R, Kind C-J, Lueth F, Terberger T, Hiller J, Matsumura S, Forster P, Burger J. Genetic discontinuity between local hunter-gatherers and Central Europe’s first farmers. Science. 2009;326:137–140. doi: 10.1126/science.1176869. [DOI] [PubMed] [Google Scholar]
- Burgarella C, Navascués M. Mutation rate estimates for 110 Y-chromosome STRs combining population and father-son pair data. Eur J Hum Genet. 2011;19:70–75. doi: 10.1038/ejhg.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubey G, Singh M, Rai N, Kariappa M, Singh K, Singh A, Singh DP, Tamang R, Selvi D. Genetic affinities of the Jewish populations of India. Nat Publ Gr. 2016 doi: 10.1038/srep19166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofaro J, Pennarun E, Mazieres S, Myres NM, Lin AA, Temori SA, Metspalu M, Metspalu E, Witzel M, King RJ, Underhill PA, Villems R, Chiaroni J. Afghan Hindu Kush: where Eurasian sub-continent gene flows converge. PLoS One. 2013 doi: 10.1371/journal.pone.0076748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulik MC, Osipova LP, Schurr TG. Y-chromosome variation in Altaian Kazakhs reveals a common paternal gene pool for Kazakhs and the influence of Mongolian expansions. PLoS One. 2011;6:e17548. doi: 10.1371/journal.pone.0017548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulik MC, Owings AC, Gaieski JB, Vilar MG, Andre A, Lennie C, Mackenzie MA, Kritsch I, Snowshoe S, Wright R, Martin J, Gibson N, Andrews TD, Schurr TG, The Genographic Consortium Y-chromosome analysis reveals genetic divergence and new founding native lineages in Athapaskan- and Eskimoan-speaking populations. Proc Natl Acad Sci. 2012;109:8471–8476. doi: 10.1073/pnas.1118760109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulik MC, Zhadanov SI, Osipova LP, Askapuli A, Gau L, Gokcumen O, Rubinstein S, Schurr TG. Mitochondrial DNA and y chromosome variation provides evidence for a recent common ancestry between Native Americans and indigenous Altaians. Am J Hum Genet. 2012;90:229–246. doi: 10.1016/j.ajhg.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegontov P, Changmai P, Zidkova A, Logacheva MD, Altınışık NE, Flegontova O, Gelfand MS, Gerasimov ES, Khrameeva EE, Konovalova OP, Neretina T, Nikolsky YV, Starostin G, Stepanova VV, Travinsky IV, Tříska M, Tříska P, Tatarinova TV. Genomic study of the Ket: a Paleo-Eskimo-related ethnic group with significant ancient North Eurasian ancestry. Sci Rep. 2016;6:20768. doi: 10.1038/srep20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayden T, Cadenas AM, Regueiro M, Singh NB, Zhivotovsky LA, Underhill PA, Cavalli-Sforza LL, Herrera RJ. The Himalayas as a directional barrier to gene flow. Am J Hum Genet Am J Hum Genet. 2007;8080:884–894. doi: 10.1086/516757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourjon G, Boëtsch G, Degioanni A. Gender and population history: sex bias revealed by studying genetic admixture of Ngazidja population (Comoro Archipelago) Am J Phys Anthropol. 2011;144:653–660. doi: 10.1002/ajpa.21474. [DOI] [PubMed] [Google Scholar]
- Grugni V, Battaglia V, Hooshiar Kashani B, Parolo S, Al-Zahery N, Achilli A, Olivieri A, Gandini F, Houshmand M, Sanati MH, Torroni A, Semino O. Ancient migratory events in the middle east: new clues from the Y-chromosome variation of modern Iranians. PLoS One. 2012 doi: 10.1371/journal.pone.0041252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak W, Forster P, Bramanti B, Matsumura S, Brandt G, Tanzer M, Villems R, Renfrew C, Gronenborn D, Alt KW, Burger J. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science. 2005;310:1016–1018. doi: 10.1126/science.1118725. [DOI] [PubMed] [Google Scholar]
- Haak W, Balanovsky O, Sanchez JJ, Koshel S, Zaporozhchenko V, Adler CJ, der Sarkissian CSI, Brandt G, Schwarz C, Nicklisch N, Dresely V, Fritsch B, Balanovska E, Villems R, Meller H, Alt KW, Cooper A. Ancient DNA from European early Neolithic farmers reveals their near eastern affinities. PLoS Biol. 2010 doi: 10.1371/journal.pbio.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber M, Platt DE, Badro DA, Xue Y, El-Sibai M, Bonab MA, Youhanna SC, Saade S, Soria-Hernanz DF, Royyuru A, Wells RS, Tyler-Smith C, Zalloua PA, Adhikarla S, The Genographic Consortium Influences of history, geography, and religion on genetic structure: the Maronites in Lebanon. Eur J Hum Genet. 2011;19:334–340. doi: 10.1038/ejhg.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber M, Platt DE, Khoury S, Badro DA, Abboud M, Tyler-Smith C, Zalloua PA. Y-chromosome R-M343 African lineages and sickle cell disease reveal structured assimilation in Lebanon. J Hum Genet. 2011;56:29–33. doi: 10.1038/jhg.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Spurdle AB, Karafet T, Bonner MR, Wood ET, Novelletto A, Malaspina P, Mitchell RJ, Horai S, Jenkins T, Zegura SL. The geographic distribution of human Y chromosome variation. Genetics. 1997;145:787–805. doi: 10.1093/genetics/145.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollard C, Keyser C, Giscard PH, Tsagaan T, Bayarkhuu N, Bemmann J, Crubézy E, Ludes B. Strong genetic admixture in the Altai at the Middle Bronze Age revealed by uniparental and ancestry informative markers. Forensic Sci Int Genet. 2014;12:199–207. doi: 10.1016/j.fsigen.2014.05.012. [DOI] [PubMed] [Google Scholar]
- Hurles ME, Maund E, Nicholson J, Bosch E, Renfrew C, Sykes BC, Jobling MA. Native American Y chromosomes in Polynesia: the genetic impact of the Polynesian slave trade. Am J Hum Genet. 2003;72:1282–1287. doi: 10.1086/374827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISOGG (2017) ISOGG Y-DNA haplogroup Q and its Subclades-2017. In: ISOGG. http://www.isogg.org/tree/ISOGG_HapgrpQ.html. Accessed 20 Apr 2017
- Jota MS, Lacerda DR, Sandoval JR, Vieira PPR, Santos-Lopes SS, Bisso-Machado R, Paixão-Cortes VR, Revollo S, Paz-Y-Miño C, Fujita R, Salzano FM, Bonatto SL, Bortolini MC, Santos FR. A new subhaplogroup of native American Y-chromosomes from the Andes. Am J Phys Anthropol. 2011;146:553–559. doi: 10.1002/ajpa.21519. [DOI] [PubMed] [Google Scholar]
- Karafet TM, Osipova LP, Gubina MA, Posukh OL, Zegura SL, Hammer MF. High levels of Y-chromosome differentiation among native Siberian populations and the genetic signature of a boreal hunter-gatherer way of life. Hum Biol. 2002;74:761–789. doi: 10.1353/hub.2003.0006. [DOI] [PubMed] [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana P, Aggarwal A, Mitra S, Italia YM, Saraswathy KN, Chandrasekar A, Kshatriya GK. Y chromosome haplogroup distribution in Indo-European speaking tribes of Gujarat, Western India. PLoS One. 2014;9:1–12. doi: 10.1371/journal.pone.0090414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacau H, Gayden T, Regueiro M, Chennakrishnaiah S, Bukhari A, Underhill PA, Garcia-Bertrand RL, Herrera RJ. Afghanistan from a Y-chromosome perspective. Eur J Hum Genet. 2012;20:1063–1070. doi: 10.1038/ejhg.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardone MC, Marengo A, Parada-Bustamante A, Cifuentes L, Piottante A, Ebensperger M, Valdevenito R, Castro A. Greater prevalence of Y chromosome Q1a3a haplogroup in Y-microdeleted Chilean men: a case–control study. J Assist Reprod Genet. 2013 doi: 10.1007/s10815-013-9950-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EY, Shin KJ, Rakha A, Sim JE, Park MJ, Kim NY, Yang WI, Lee HY. Analysis of 22 Y chromosomal STR haplotypes and Y haplogroup distribution in Pathans of Pakistan. Forensic Sci Int Genet. 2014;11:111–116. doi: 10.1016/j.fsigen.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Lell JT, Brown MD, Schurr TG, Sukernik RI, Starikovskaya YB, Torroni A, Moore LG, Troup GM, Wallace DC. Y-chromosome polymorphisms in native American and Siberian populations: identification of native American Y-chromosome haplotypes. Hum Genet. 1997;100:536–543. doi: 10.1007/s004390050548. [DOI] [PubMed] [Google Scholar]
- Lell JT, Sukernik RI, Starikovskaya YB, Su B, Jin L, Schurr TG, Underhill PA, Wallace DC. The dual origin and Siberian affinities of native American Y chromosomes. Am J Hum Genet. 2002;70:192–206. doi: 10.1086/338457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liao L, Gu M, Ye Y. Population genetics for 17 Y-STR loci in a Chinese Han population sample from Mudanjiang city, Northeast China. Forensic Sci Int Genet. 2014;13:e16–e17. doi: 10.1016/j.fsigen.2014.05.009. [DOI] [PubMed] [Google Scholar]
- Llamas B, Fehren-Schmitz L, Valverde G, Soubrier J, Mallick S, Rohland N, Nordenfelt S, Valdiosera C, Richards SM, Rohrlach A, Romero MIB, Espinoza IF, Cagigao ET, Jimenez LW, Makowski K, Reyna ISL, Lory JM, Torrez JAB, Rivera MA, Burger RL, Ceruti MC, Reinhard J, Wells RS, Politis G, Santoro CM, Standen VG, Smith C, Reich D, Ho SYW, Cooper A, Haak W. Ancient mitochondrial DNA provides high-resolution time scale of the peopling of the Americas. Sci Adv. 2016;2:e1501385. doi: 10.1126/sciadv.1501385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmström H, Gilbert MTP, Thomas MG, Brandström M, Storå J, Molnar P, Andersen PK, Bendixen C, Holmlund G, Götherström A, Willerslev E. Ancient DNA reveals lack of continuity between Neolithic hunter-gatherers and contemporary Scandinavians. Curr Biol. 2009;19:1758–1762. doi: 10.1016/j.cub.2009.09.017. [DOI] [PubMed] [Google Scholar]
- Malyarchuk B, Derenko M, Denisova G, Maksimov A, Wozniak M, Grzybowski T, Dambueva I, Zakharov I. Ancient links between Siberians and Native Americans revealed by subtyping the Y chromosome haplogroup Q1a. J Hum Genet. 2011;56:583–588. doi: 10.1038/jhg.2011.64. [DOI] [PubMed] [Google Scholar]
- Melton PE, Baldi NF, Barrantes R, Crawford MH. Microevolution, migration, and the population structure of five amerindian populations from nicaragua and costa rica. Am J Hum Biol. 2013;25:480–490. doi: 10.1002/ajhb.22382. [DOI] [PubMed] [Google Scholar]
- Msaidie S, Ducourneau A, Boetsch G, Longepied G, Papa K, Allibert C, Yahaya AA, Chiaroni J, Mitchell MJ. Genetic diversity on the Comoros Islands shows early seafaring as major determinant of human biocultural evolution in the Western Indian Ocean. Eur J Hum Genet. 2010;19:89–94. doi: 10.1038/ejhg.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neparáczki E, Juhász Z, Pamjav H, Fehér T, Csányi B, Zink A, Maixner F, Pálfi G, Molnár E, Pap I, Kustár Á, Révész L, Raskó I, Török T. Genetic structure of the early Hungarian conquerors inferred from mtDNA haplotypes and Y-chromosome haplogroups in a small cemetery. Mol Genet Genom. 2016 doi: 10.1007/s00438-016-1267-z. [DOI] [PubMed] [Google Scholar]
- Nogueiro I, Manco L, Gomes V, Amorim A, Gusmão L. Phylogeographic analysis of paternal lineages in NE Portuguese Jewish communities. Am J Phys Anthropol. 2010;141:373–381. doi: 10.1002/ajpa.21154. [DOI] [PubMed] [Google Scholar]
- Noguera MC, Schwegler A, Gomes V, Briceño I, Alvarez L, Uricoechea D, Amorim A, Benavides E, Silvera C, Charris M, Bernal JE, Gusmão L. Colombia’s racial crucible: Y chromosome evidence from six admixed communities in the Department of Bolivar. Ann Hum Biol. 2014;41:453–459. doi: 10.3109/03014460.2013.852244. [DOI] [PubMed] [Google Scholar]
- Nonaka I, Minaguchi K, Takezaki N. Y-chromosomal binary haplogroups in the Japanese population and their relationship to 16 Y-STR polymorphisms. Ann Hum Genet. 2007;71:480–495. doi: 10.1111/j.1469-1809.2006.00343.x. [DOI] [PubMed] [Google Scholar]
- O’Rourke DH, Raff JA. The Human Genetic History of the Americas: the Final Frontier. Curr Biol. 2010;20:R202–R207. doi: 10.1016/j.cub.2009.11.051. [DOI] [PubMed] [Google Scholar]
- Pakendorf B, Novgorodov IN, Osakovskij VL, Stoneking M. Mating patterns amongst Siberian reindeer herders: inferences from mtDNA and Y-chromosomal analyses. Am J Phys Anthropol. 2007;133:1013–1027. doi: 10.1002/ajpa.20590. [DOI] [PubMed] [Google Scholar]
- Quinque D, Kittler R, Kayser M, Stoneking M, Nasidze I. Evaluation of saliva as a source of human DNA for population and association studies. Anal Biochem. 2006;353:272–277. doi: 10.1016/j.ab.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Raff JA, Bolnick DA. Palaeogenomics: genetic roots of the first Americans. Nature. 2014;506:162–163. doi: 10.1038/506162a. [DOI] [PubMed] [Google Scholar]
- Raghavan M, Steinrücken M, Harris K, Schiffels S, Rasmussen S, DeGiorgio M, Albrechtsen A, Valdiosera C, Ávila-Arcos MC, Malaspinas A-S, Eriksson A, Moltke I, Metspalu M, Homburger JR, Wall J, Cornejo OE, Moreno-Mayar JV, Korneliussen TS, Pierre T, Rasmussen M, Campos PF, Damgaard PB, Allentoft ME, Lindo J, Metspalu E, Rodriguez-Varela R, Mansilla J, Henrickson C, Seguin-Orlando A, Malmstrom H, Stafford T, Shringarpure SS, Moreno-Estrada A, Karmin M, Tambets K, Bergstrom A, Xue Y, Warmuth V, Friend AD, Singarayer J, Valdes P, Balloux F, Leboreiro I, Vera JL, Rangel-Villalobos H, Pettener D, Luiselli D, Davis LG, Heyer E, Zollikofer CPE, Ponce de Leon MS, Smith CI, Grimes V, Pike K-A, Deal M, Fuller BT, Arriaza B, Standen V, Luz MF, Ricaut F, Guidon N, Osipova L, Voevoda MI, Posukh OL, Balanovsky O, Lavryashina M, Bogunov Y, Khusnutdinova E, Gubina M, Balanovska E, Fedorova S, Litvinov S, Malyarchuk B, Derenko M, Mosher MJ, Archer D, Cybulski J, Petzelt B, Mitchell J, Worl R, Norman PJ, Parham P, Kemp BM, Kivisild T, Tyler-Smith C, Sandhu MS, Crawford M, Villems R, Smith DG, Waters MR, Goebel T, Johnson JR, Malhi RS, Jakobsson M, Meltzer DJ, Manica A, Durbin R, Bustamante CD, Song YS, Nielsen R, Willerslev E. Genomic evidence for the Pleistocene and recent population history of native Americans. Science. 2015;349:aab3884. doi: 10.1126/science.aab3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M, Li Y, Lindgreen S, Pedersen JS, Albrechtsen A, Moltke I, Metspalu M, Metspalu E, Kivisild T, Gupta R, Bertalan M, Nielsen K, Gilbert MTP, Wang Y, Raghavan M, Campos PF, Kamp HM, Wilson AS, Gledhill A, Tridico S, Bunce M, Lorenzen ED, Binladen J, Guo X, Zhao J, Zhang X, Zhang H, Li Z, Chen M, Orlando L, Kristiansen K, Bak M, Tommerup N, Bendixen C, Pierre TL, Grønnow B, Meldgaard M, Andreasen C, Fedorova SA, Osipova LP, Higham TFG, Ramsey CB, Hansen TO, Nielsen FC, Crawford MH, Brunak S, Sicheritz-Pontén T, Villems R, Nielsen R, Krogh A, Wang J, Willerslev E. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–762. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M, Anzick SL, Waters MR, Skoglund P, DeGiorgio M, Stafford TW, Rasmussen S, Moltke I, Albrechtsen A, Doyle SM, Poznik GD, Gudmundsdottir V, Yadav R, Malaspinas AS, White SS 5th, Allentoft ME, Cornejo OE, Tambets K, Eriksson A, Heintzman PD, Karmin M, Korneliussen TS, Meltzer DJ, Pierre TL, Stenderup J, Saag L, Warmuth VM, Lopes MC, Malhi RS, Brunak S, Sicheritz-Ponten T, Barnes I, Collins M, Orlando L, Balloux F, Manica A, Gupta R, Metspalu M, Bustamante CD, Jakobsson M, Nielsen R, Willerslev E (2014) The genome of a Late Pleistocene human from a Clovis burial site in western Montana. Nature 506:225–229. doi:10.1038/nature13025 [DOI] [PMC free article] [PubMed]
- Regueiro M, Alvarez J, Rowold D, Herrera RJ. On the origins, rapid expansion and genetic diversity of native Americans from hunting-gatherers to agriculturalists. Am J Phys Anthropol. 2013;150:333–348. doi: 10.1002/ajpa.22207. [DOI] [PubMed] [Google Scholar]
- Sala A, Corach D. Analysis of admixture and genetic structure of two Native American groups of Southern Argentinean Patagonia. Mol Biol Rep. 2014;41:1533–1543. doi: 10.1007/s11033-013-2999-z. [DOI] [PubMed] [Google Scholar]
- Sandoval J, Lacerda D, Jota M, Salazar-Granara A, Vieira P, Acosta O, Cuellar C, Revollo S, Fujita R, Santos F. The genetic history of indigenous populations of the Peruvian and Bolivian Altiplano: the legacy of the Uros. PLoS One. 2013;8:e73006. doi: 10.1371/journal.pone.0073006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seielstad M, Yuldasheva N, Singh N, Underhill P, Oefner P, Shen P, Wells RS. A novel Y-chromosome variant puts an upper limit on the timing of first entry into the Americas. Am J Hum Genet. 2003;73:700–705. doi: 10.1086/377589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Zhivotovsky LA, King R, Mehdi SQ, Edmonds CA, Chow C-ET, Lin AA, Mitra M, Sil SK, Ramesh A, Rani MVU, Thakur CM, Cavalli-Sforza LL, Majumder PP, Underhill PA. Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet. 2006;78:202–221. doi: 10.1086/499411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Rai E, Bhat AK, Bhanwer AS, Bamezai RNK. A novel subgroup Q5 of human Y-chromosomal haplogroup Q in India. BMC Evol Biol. 2007;7:232. doi: 10.1186/1471-2148-7-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Ayub Q, Vermeulen M, Shao RG, Zuniga S, Van Der Gaag K, De Knijff P, Kayser M, Xue Y, Tyler-Smith C. A worldwide survey of human male demographic history based on Y-SNP and Y-STR data from the HGDP-CEPH populations. Mol Biol Evol. 2010;27:385–393. doi: 10.1093/molbev/msp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B, Xiao C, Deka R, Seielstad MT, Kangwanpong D, Xiao J, Lu D, Underhill P, Cavalli-Sforza L, Chakraborty R, Jin L. Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum Genet. 2000;107:582–590. doi: 10.1007/s004390000406. [DOI] [PubMed] [Google Scholar]
- Tambets K, Rootsi S, Kivisild T, Help H, Serk P, Loogväli E-L, Tolk H-V, Reidla M, Metspalu E, Pliss L, Balanovsky O, Pshenichnov A, Balanovska E, Gubina M, Zhadanov S, Osipova L, Damba L, Voevoda M, Kutuev I, Bermisheva M, Khusnutdinova E, Gusar V, Grechanina E, Parik J, Pennarun E, Richard C, Chaventre A, Moisan J-P, Barac L, Pericic M, Rudan P, Terzic R, Mikerezi I, Krumina A, Baumanis V, Koziel S, Rickards O, Stefano GFD, Anagnou N, Pappa KI, Michalodimitrakis E, Ferak V, Furedi S, Komel R, Beckman L, Villems R. The western and eastern roots of the Saami—the story of genetic “outliers” told by mitochondrial DNA and Y chromosomes. Am J Hum Genet. 2004;74:661–682. doi: 10.1086/383203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona-Santos E, Carvalho-Silva DR, Pettener D, Luiselli D, De Stefano GF, Labarga CM, Rickards O, Tyler-Smith C, Pena SDJ, Santos FR. Genetic differentiation in South Amerindians is related to environmental and cultural diversity: evidence from the Y chromosome. Am J Hum Genet. 2001;68:1485–1496. doi: 10.1086/320601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres JB, Vilar MG, Torres GA, Gaieski JB, Hernandez RB, Browne ZE, Stevenson M, Walters W, Schurr TG. Genetic diversity in the Lesser Antilles and its implications for the settlement of the Caribbean basin. PLoS One. 2015;10:1–27. doi: 10.1371/journal.pone.0139192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toscanini U, Gusmão L, Berardi G, Gomes V, Amorim A, Salas A, Raimondi E. Male lineages in South American native groups: evidence of M19 traveling south. Am J Phys Anthropol. 2011;146:188–196. doi: 10.1002/ajpa.21562. [DOI] [PubMed] [Google Scholar]
- Toscanini U, Vullo C, Berardi G, Llull C, Borosky A, Gómez A, Pardo-Seco J, Salas A. A comprehensive Y-STR portrait of Argentinean populations. Forensic Sci Int Genet. 2016;20:1–5. doi: 10.1016/j.fsigen.2015.09.002. [DOI] [PubMed] [Google Scholar]
- Tremblay M, Vézina H. New estimates of intergenerational time intervals for the calculation of age and origins of mutations. Am J Hum Genet. 2000;66:651–658. doi: 10.1086/302770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonné-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ. Y chromosome sequence variation and the history of human populations. Nat Genet. 2000;26:358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- Varzari A, Kharkov V, Nikitin AG, Raicu F, Simonova K, Stephan W, Weiss EH, Stepanov V. Paleo-Balkan and Slavic contributions to the genetic pool of Moldavians: insights from the Y chromosome. PLoS One. 2013 doi: 10.1371/journal.pone.0053731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-C, Li H. Evaluating the Y chromosomal STR dating in deep-rooting pedigrees. Investig Genet. 2015;6:8. doi: 10.1186/s13323-015-0025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-C, Gilbert MTP, Jin L, Li H. Evaluating the Y chromosomal timescale in human demographic and lineage dating. Investig Genet. 2014;5:12. doi: 10.1186/2041-2223-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ayub Q, Xue Y, Tyler-Smith C. A comparison of Y-chromosomal lineage dating using either resequencing or Y-SNP plus Y-STR genotyping. Forensic Sci Int Genet. 2013;7:568–572. doi: 10.1016/j.fsigen.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, Su B, Pitchappan R, Shanmugalakshmi S, Balakrishnan K, Read M, Pearson NM, Zerjal T, Webster MT, Zholoshvili I, Jamarjashvili E, Gambarov S, Nikbin B, Dostiev A, Aknazarov O, Zalloua P, Tsoy I, Kitaev M, Mirrakhimov M, Chariev A, Bodmer WF. The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA. 2001;98:10244–10249. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Li H, Lu D, Song X, Zhang F, He Y, Li F, Gao Y, Mao X, Zhang L, Qian J, Tan J, Jin J, Huang W, Deka R, Su B, Chakraborty R, Jin L. Genetic evidence supports demic diffusion of Han culture. Nature. 2004;431:302–305. doi: 10.1038/nature02878. [DOI] [PubMed] [Google Scholar]
- Wilson IJ, Weale ME, Balding DJ, Wilson LJ. Inferences from DNA data: population histories, evolutionary processes and forensic match inferences from DNA data: population histories, evolutionary processes and forensic match probabilities. Source J R Stat Soc Ser A (Statistics Soc J R Stat Soc A) 2003;166:155–201. doi: 10.1111/1467-985X.00264. [DOI] [Google Scholar]
- Xue Y, Zerjal T, Bao W, Zhu S, Shu Q, Xu J, Du R, Fu S, Li P, Hurles ME, Yang H, Tyler-Smith C. Male demography in East Asia: a north–south contrast in human population expansion times. Genetics. 2006;172:2431–2439. doi: 10.1534/genetics.105.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Wang C-C, Zheng H-X, Wang W, Qin Z-D, Wei L-H, Wang Y, Pan X-D, Fu W-Q, He Y-G, Xiong L-J, Jin W-F, Li S-L, An Y, Li H, Jin L. Y chromosomes of 40% Chinese descend from three Neolithic super-grandfathers. PLoS One. 2014;9:e105691. doi: 10.1371/journal.pone.0105691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunusbayev B, Metspalu M, Metspalu E, Valeev A, Litvinov S, Valiev R, Akhmetova V, Balanovska E, Balanovsky O, Turdikulova S, Dalimova D, Nymadawa P, Bahmanimehr A, Sahakyan H, Tambets K, Fedorova S, Barashkov N, Khidiyatova I, Mihailov E, Khusainova R, Damba L, Derenko M, Malyarchuk B, Osipova L, Voevoda M, Yepiskoposyan L, Kivisild T, Khusnutdinova E, Villems R. The genetic legacy of the expansion of Turkic-speaking nomads across Eurasia. PLoS Genet. 2015;11:1–24. doi: 10.1371/journal.pgen.1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal T, Wells RS, Yuldasheva N, Ruzibakiev R, Tyler-Smith C. A genetic landscape reshaped by recent events: Y-chromosomal insights into central Asia. Am J Hum Genet. 2002;71:466–482. doi: 10.1086/342096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y-B, Li H-J, Cai D-W, Li C-X, Zhang Q-C, Zhu H, Zhou H. Ancient DNA from nomads in 2500-year-old archeological sites of Pengyang, China. J Hum Genet. 2010;55:215–218. doi: 10.1038/jhg.2010.8. [DOI] [PubMed] [Google Scholar]
- Zhao YB, Zhang Y, Li HJ, Cui YQ, Zhu H, Zhou H. Ancient DNA evidence reveals that the Y chromosome haplogroup Q1a1 admixed into the Han Chinese 3,000 years ago. Am J Hum Biol. 2014;821:813–821. doi: 10.1002/ajhb.22604. [DOI] [PubMed] [Google Scholar]
- Zhao Y-B, Zhang Y, Zhang Q-C, Li H-J, Cui Y-Q, Xu Z, Jin L, Zhou H, Zhu H. Ancient DNA reveals that the genetic structure of the northern han chinese was shaped prior to 3,000 years ago. PLoS One. 2015;10:e0125676. doi: 10.1371/journal.pone.0125676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, Cinnioğlu C, Kayser M, Morar B, Kivisild T, Scozzari R, Cruciani F, Destro-Bisol G, Spedini G, Chambers GK, Herrera RJ, Yong KK, Gresham D, Tournev I, Feldman MW, Kalaydjieva L. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Shi H, Bin Qi X, Duan ZY, Tan PP, Jin L, Su B, Ma RZ. Extended Y chromosome investigation suggests postglacial migrations of modern humans into East Asia via the northern route. Mol Biol Evol. 2011;28:717–727. doi: 10.1093/molbev/msq247. [DOI] [PubMed] [Google Scholar]
- Zoossmann-Diskin A. The origin of Eastern European Jews revealed by autosomal, sex chromosomal and mtDNA polymorphisms. Biol Direct. 2010;5:57. doi: 10.1186/1745-6150-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Y-chromosomal STR data haplotypes used within haplogroup Q (XLSX 59 kb)
Frequencies of Haplogroup Q in the world (XLSX 47 kb)
National composition of population and citizenship in Russia (XLSX 25 kb)