Abstract
Birth defects are the leading cause of infant mortality, and they are caused by a combination of genetic and environmental factors. Environmental risk factors may contribute to birth defects in genetically susceptible infants by altering critical molecular pathways during embryogenesis, but experimental evidence for gene-environment interactions is limited. Fetal hyperglycemia associated with maternal diabetes results in a 5-fold increased risk of congenital heart disease (CHD), but the molecular basis for this correlation is unknown. Here, we show that the effects of maternal hyperglycemia on cardiac development are sensitized by haploinsufficiency of Notch1, a key transcriptional regulator known to cause CHD. Using ATAC-seq, we found that hyperglycemia decreased chromatin accessibility at the endothelial NO synthase (Nos3) locus, resulting in reduced NO synthesis. Transcription of Jarid2, a regulator of histone methyltransferase complexes, was increased in response to reduced NO, and this upregulation directly resulted in inhibition of Notch1 expression to levels below a threshold necessary for normal heart development. We extended these findings using a Drosophila maternal diabetic model that revealed the evolutionary conservation of this interaction and the Jarid2-mediated mechanism. These findings identify a gene-environment interaction between maternal hyperglycemia and Notch signaling and support a model in which environmental factors cause birth defects in genetically susceptible infants.
Keywords: Cardiology, Genetics
Keywords: Cardiovascular disease, Embryonic development, Epigenetics
Maternal hyperglycemia disrupts normal cardiac morphogenesis by epigenetically mediated dysregulation of a nitric oxide–Jarid2/Notch1 signaling pathway.
Introduction
Birth defects, defined as an abnormality affecting body structure and function that is present at birth, affect nearly 8 million children annually and are an important worldwide health problem (1). Among these, congenital heart disease (CHD) is the most common type of birth defect and is a leading noninfectious cause of infant morbidity and mortality (2, 3). While an increasing number of genetic contributors for CHD in humans have been identified, the etiology for majority of CHD remains unknown. The etiology for these cases is proposed to be multifactorial, as demonstrated in epidemiological studies, which show a significant role for both genetic and environmental determinants (4–6). Studies of genetic contributors have identified an increasing number of genes associated with CHD (7–9). The importance of gene dosage for the occurrence of cardiac malformations has been shown, along with the concept of an oligogenic etiology for CHD, supporting the concept that mild alterations in gene expression levels may lead disease phenotypes (10–13). Similarly, numerous environmental risk factors for CHD have been reported, each of which increases the risk of having a child with CHD (14). For the majority of these, exposure to a single teratogen raises the risk by only 2- to 10-fold, suggesting that additional factors, i.e., underlying genetic susceptibility, may be involved. While gene-environment interactions for birth defects have been proposed, there is limited molecular evidence demonstrating the mechanisms by which this occurs (15).
Among the noninherited risk factors, pregestational type 1 and type 2 maternal diabetes mellitus (matDM) is well-known, raising the risk of having an infant with CHD by 3- to 5-fold. The cardiac phenotypes include looping, conotruncal, and septation defects (14, 16, 17). MatDM constitutes an unfavorable environment for embryonic development, characterized by fetal exposure to elevated blood glucose levels, which is the proposed major teratogen for diabetic embryopathy (17, 18). However, the mechanisms by which alterations in maternal glucose homeostasis disrupt normal heart development remain poorly understood.
Maternal illnesses, which affect the fetal environment and cause birth defects, are proposed to be mediated by epigenetic mechanisms, including posttranslational modifications of histones, DNA methylation, and noncoding RNAs (19, 20). This tightly orchestrated spatiotemporal regulation is maintained by switching between a euchromatin (active) and a heterochromatin (silent) state, which is typically accomplished by the recruitment of chromatin-associated multisubunit protein complexes, which alter gene transcription by modulating chromatin accessibility (21, 22). With the tremendous advancements in epigenomics, these higher-order chromatin organization and accessible regions are able to be defined throughout the genome. Assay for transposase-accessible (Tn5) chromatin with high-throughput sequencing (ATAC-seq) is a robust method that has improved the ability to identify open chromatin regions, transcription factor (TF) occupancy, and regulatory elements, including promoters and enhancers (23, 24). These higher-order chromatin conformational studies allow us an opportunity to determine how environmental perturbations influence downstream gene expression required for normal embryonic development.
Hyperglycemia (HG) is known to result in the accumulation of ROS in the vasculature and developing embryo (25, 26). Increased ROS levels associated with reduced NO bioavailability are a hallmark of diabetes-mediated complications that lead to endothelial cell (EC) dysfunction in adult-onset cardiovascular disease (26–28). Since redox homeostasis plays an important role in regulating EC fate, it is important to understand how increased ROS and loss of NO affects the fetal cardiovascular system during development. It was recently shown that cellular exposure to NO can function as an epigenetic modulator by affecting global histone methylation status (29, 30). Targeted deletion of endothelial NO synthase (eNOS, also known as Nos3) in mice results in CHD, demonstrating the requirement of endothelial NO for proper heart development (31, 32). We previously demonstrated a genetic interaction between Nos3 and Notch1, as compound mutant mice exhibit a range of conotruncal and semilunar valve malformations, and suggested that loss of NO led to a further reduction of Notch1 signaling (33, 34). The Notch signaling pathway is a highly conserved intracellular signaling pathway critical in cell fate decisions and numerous developmental and disease processes; it includes the 4 Notch transmembrane receptors (Notch1–Notch4) that are activated by interactions with Jagged and Delta-like family of ligands on adjacent cells (35). Receptor-ligand recognition is followed by a series of proteolytic cleavages that releases Notch intracellular domain, allowing translocation to the nucleus, where it functions in a transcription complex [CBF1/Su(H)/Lag-1] to activate downstream target genes, including the HEY (Hes-related with YPRW motif) family of transcriptional repressors (36). Mutations in NOTCH1 have been linked to conotruncal and semilunar valve malformations in humans, but Notch1 heterozygote mice do not display cardiac malformations (33, 37–39).
Here, we describe a previously unrecognized gene-environment interaction between Notch1 and matDM in mice and investigate the epigenetic mechanisms by which maternal HG affects molecular pathways regulating cardiac development using ATAC-seq. These experiments reveal that HG alters the open chromatin status upstream of the Nos3 locus, contributing to decreased NO bioavailability. The resultant loss of NO is associated with enrichment of Jarid2, a transcriptional repressor, on the Notch1 locus and leads to further reduction in Notch1 levels below a critical threshold level required for normal cardiac development. The gene-environment interaction between matDM and Notch signaling is shown to be evolutionarily conserved using the Drosophila model system. These data reveal the epigenetic machinery by which maternal HG dysregulates the endothelial NO and Notch1 signaling pathways, leading to abnormal cardiac development.
Results
Gene-environment interaction between maternal HG and Notch1.
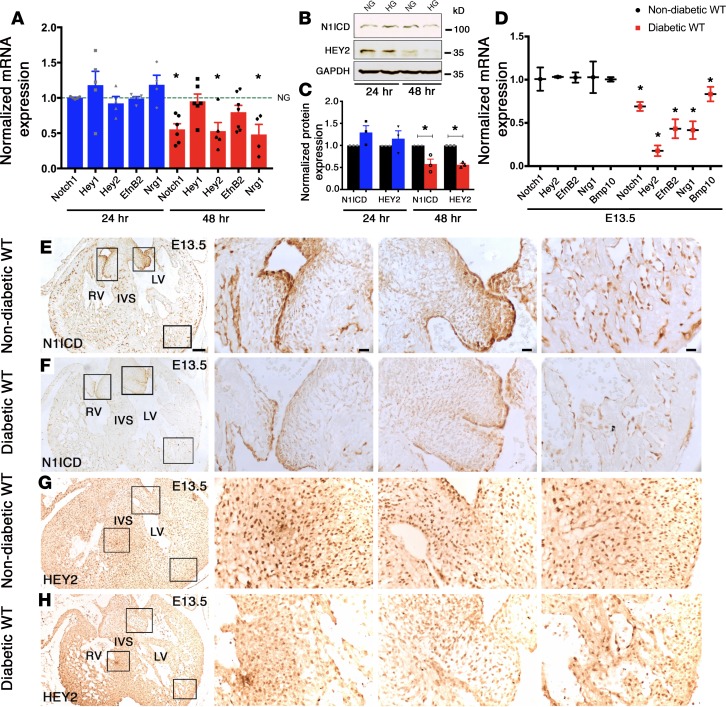
We previously reported that homozygous deletion of Nos3, a major source of NO production in ECs, in association with Notch1 haploinsufficiency, resulted in CHD in vivo and demonstrated that NO regulated the Notch1 signaling pathway (33). This led us to test whether expression of Notch1 and its downstream target genes is altered in HG, with its well-recognized association with decreased NO bioavailability. We examined the expression of Notch1 mRNA, along with several well-described downstream targets using atrioventricular cushion mesenchymal cells, an immortalized embryonic cardiac cell line (tsA58-AVM), cultured in normoglycemic (NG, 5.5 mM D-glucose) and HG (25 mM D-glucose) conditions by real-time qPCR (RT-qPCR) (40). There was significantly reduced expression of Notch1, Hey2, EfnB2, and Nrg1 with HG as compared with NG after 48 hours (Figure 1A). A similar reduction in expression was noted for NOTCH1 intracellular domain (N1ICD) and HEY2 by immunoblotting (Figure 1, B and C). To determine if HG had similar effects in utero, we generated a murine model of matDM by injection of streptozotocin (STZ), which is selectively toxic to pancreatic β cells (41). Using a STZ-induced matDM model, we examined the expression of the Notch1 signaling pathway in the embryonic heart exposed to maternal HG. Similar to our in vitro studies, there was downregulation of transcripts for Notch1, Hey2, EfnB2, Nrg1, and Bmp10 in E13.5 hearts exposed to maternal HG by RT-qPCR (Figure 1D). By immunohistochemistry, N1ICD and HEY2 expression was diminished in E13.5 hearts exposed to matDM as compared with nondiabetic controls (Figure 1, E–H). In combination, these studies demonstrated dysregulation of the Notch1 signaling pathway under HG stress in vitro and in vivo.
Figure 1. Hyperglycemia downregulates Notch1 pathway.
(A) Transcript levels of Notch1, Hey1, Hey2, EfnB2, and Nrg1 in AVM cells cultured in HG (25 mM) for 24 hours (blue) and 48 hours (red) by RT-qPCR (n ≥ 3). Comparisons to NG (5.5 mM), represented by the dotted line set to 1; data represent mean ± SEM. (B) N1ICD and HEY2 protein expression in AVM cells cultured in NG and HG for 24 hours and 48 hours. (C) Quantification of protein expression normalized to GAPDH (n = 3; mean ± SEM). (D) Examination of E13.5 WT murine hearts of nondiabetic and diabetic dams shows downregulation of Notch1, Hey2, EfnB2, Nrg1, and Bmp10 mRNA by RT-qPCR (n = 6 pooled hearts/group; mean ± SD). (E–H) Decreased expression of N1ICD and HEY2 in embryonic hearts exposed to maternal diabetes by immunohistochemistry (n = 3). Square boxes in left column in E–H are shown in higher magnifications from left to right. LV, left ventricle; RV, right ventricle; IVS, interventricular septum. *P < 0.05, 2-tailed Student’s t test in C, with Holm-Bonferroni correction in A and D. Scale bars: 100 μm (E–H, left column); 20 μm (E–H, center and right columns).
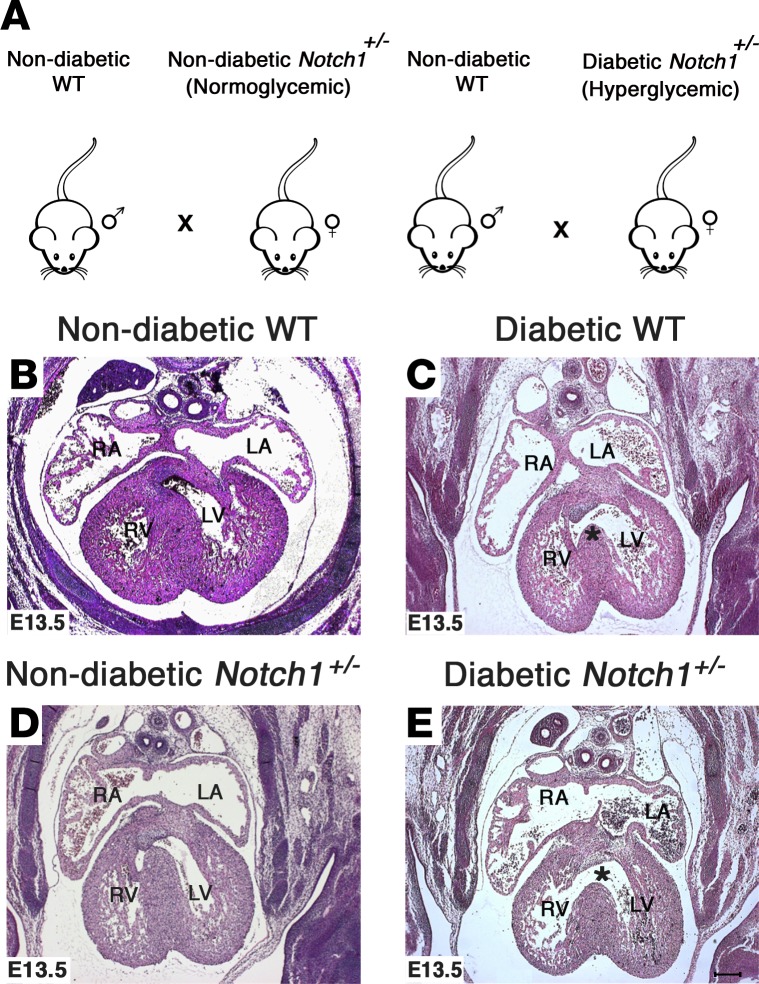
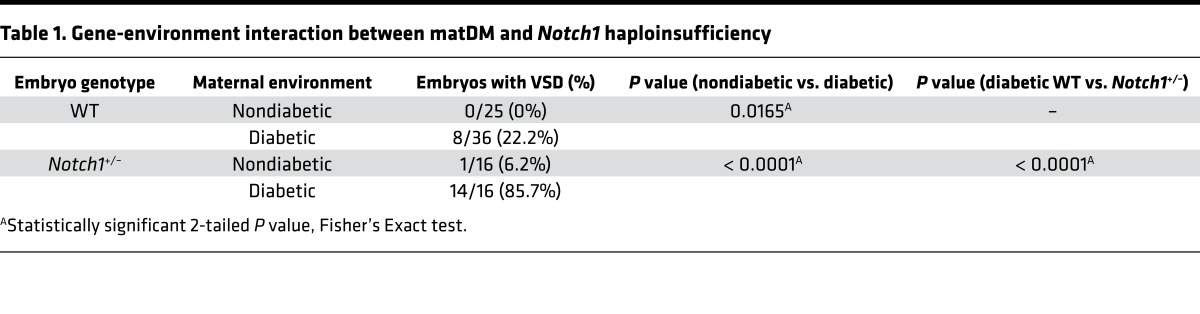
We next sought to determine if an in vivo gene-environment interaction exists between matDM and Notch1 heterozygosity (33, 34). For these studies, matDM with moderate HG was induced using STZ in WT and Notch1+/– females, which were subsequently bred with WT males (Figure 2A). We found an approximately 22% incidence of perimembranous ventricular septal defects (VSDs) (8 of 36 embryos; P = 0.0165) in WT E13.5 embryos exposed to matDM (maternal average blood glucose level ± SD = 259.59 ± 54.69 mg/dl) as compared with exposure to maternal NG (0 of 25 embryos) in the absence of growth retardation (Table 1; Figure 2, B and C; and Supplemental Figure 1, A–D; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.95085DS1). The incidence of VSDs was statistically higher in Notch1 heterozygote embryos (14 of 16 embryos; P < 0.0001) exposed to matDM (maternal average blood glucose level ± SD = 266.87 ± 62.58 mg/dl) when compared with nondiabetic Notch1+/– (1 of 16 embryos) and diabetic WT embryos (8 of 36 embryos) (Table 1 and Figure 2, D and E). 3D reconstruction of E13.5 hearts using AMIRA showed the perimembranous VSD location (Supplemental Figure 1, E and F). To determine if maternal Notch1 heterozygosity affected this interaction, we crossed WT females with matDM (maternal average blood glucose level ± SD = 293.67 ± 46.32 mg/dl) to nondiabetic Notch1+/– males (Supplemental Figure 2G) and observed a similar increased incidence of VSDs in Notch1+/– embryos (8 of 9 embryos) as compared with WT controls (2 of 7 embryos) (Supplemental Table 1 and Supplemental Figure 1, H–K). We also observed that embryos exposed to higher levels of maternal glycemic stress resulted in increased lethality and neural tube malformations, along with outflow tract defects, septal defects, and thin myocardium, demonstrating that fetal exposure to matDM impairs multiple aspects of development (Supplemental Figure 2, A–L). Taken together, we describe a gene-environment interaction between Notch1 haploinsufficiency and maternal HG, which affected the process of ventricular septation.
Figure 2. Exposure to maternal diabetes and Notch1 haploinsufficiency leads to increased incidence of ventricular septal defects.
(A) Breeding scheme demonstrating nondiabetic WT males crossed with nondiabetic (normoglycemic) and diabetic (hyperglycemic) Notch1+/– females. (B–E) Representative H&E-stained images showing the location of ventricular septal defects (*) in E13.5 WT and Notch1+/– hearts exposed to maternal diabetes as compared with nondiabetic WT and Notch1+/– controls. RA, right atrium, LA, left atrium; RV, right ventricle; LV, left ventricle. Scale bar: 200 μm.
Table 1. Gene-environment interaction between matDM and Notch1 haploinsufficiency.
HG alters chromatin accessibility and TF occupancy and reduces endothelial NO.
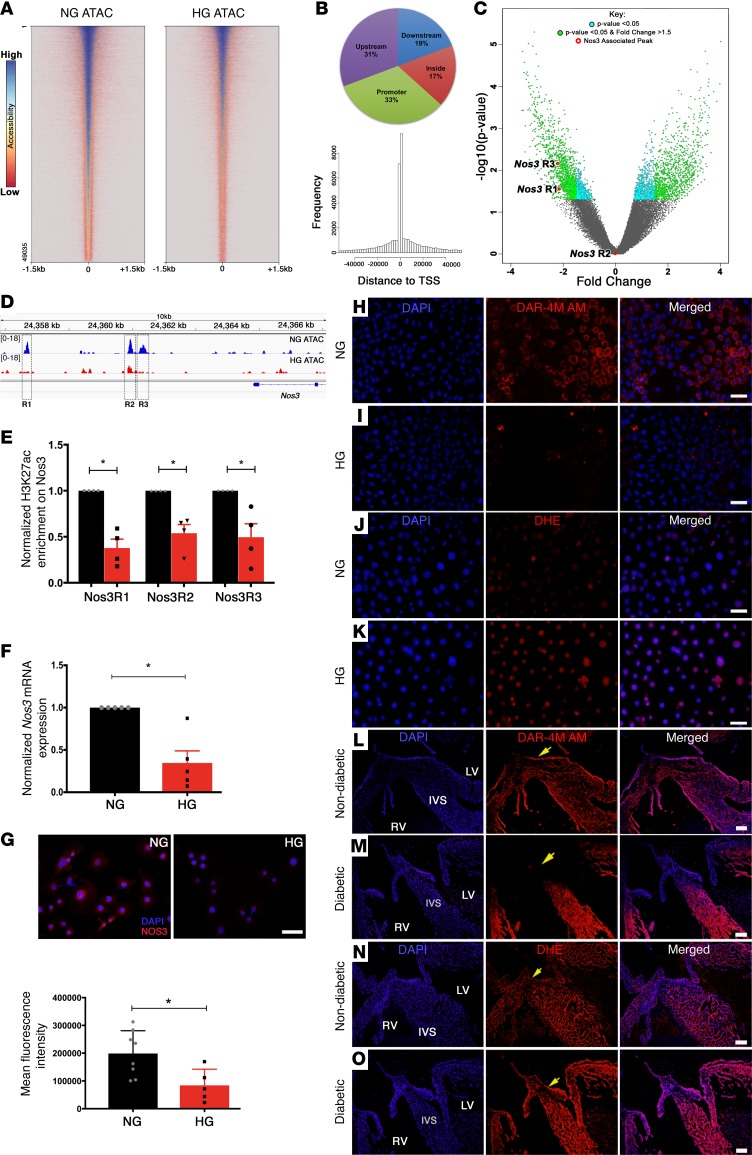
To investigate the epigenetic mechanisms by which the environmental stress of HG influences gene expression through dynamic changes in chromatin accessibility genome-wide, we performed high-throughput ATAC-seq in atrioventricular mesenchymal (AVM) cells in NG and HG conditions (23, 24). ATAC-seq analysis revealed 49,035 differential peaks, with an overall difference in signal intensity in HG compared with NG (Figure 3A and Supplemental Table 2). When aligned to mm10 genome, we found that these open chromatin regions were primarily distributed in the following four locations: (a) proximal promoters (±1.5 kb from transcriptional start site [TSS]; 33%); (b) upstream of genes (>1.5 kb upstream of TSS; 31%); (c) near intronic/exonic regions (>1.5 kb from TSS; 17%); and (d) downstream of nearest mapped genes (19%) (Figure 3B). Genome-wide data visualization of the peaks, using a volcano plot, highlights the dynamic changes in chromatin accessibility between the NG and HG AVM cells (Figure 3C). Next, we performed known and de novo TF-binding motif analysis on the NG data set, with HG as background and found that NG has enrichment of binding sites for TFs previously demonstrated to regulate gene expression in ECs (Supplemental Table 3). Among the 91 known and 29 de novo motifs (P > 1e–10) analyzed, we found multiple binding sites for Kruppel-like factors (KLFs, SP1) and the ETS (ELK1, EGR2, ETV1) family of TFs (42, 43) (highlighted in yellow, Supplemental Table 4). Not surprisingly, we also found sites for NRF1/2, which is essential for diabetes-dependent reduced expression of key genes in oxidative metabolism and mitochondrial function (44) (highlighted in yellow, Supplemental Table 4). Utilizing in vitro and in vivo model systems, it has previously been demonstrated that SP1 and KLF2/4 directly regulate Nos3 by binding to its promoter (45, 46). Closer examination of the Nos3 locus revealed a decrease in ATAC-seq signal with HG stress compared with NG (Figure 3D). Among three differentially open chromatin regions (R1, chr5:24357617–24357726; R2, chr5:24360864–24360991; R3, chr5:24361289–24361376), only Nos3R1 and Nos3R3 peaks exhibited statistically significant (P < 0.05) changes in chromatin accessibility (Figure 3C). These regions were further shown to be sites of transcription by the presence of an active histone mark (H3K27ac) using ChIP-qPCR in NG conditions, and there was a significant reduction (P < 0.05) in H3K27ac enrichment on these 3 loci (R1–R3) with HG (Figure 3E). Consistent with this relative chromatin closure in HG, Nos3 expression was significantly downregulated with HG using RT-qPCR and immunofluorescent staining in AVM cells (Figure 3, F and G). Common TFBS profiles for three open sites predicted using Genomatix software also identified putative binding sequences for SP1, ETS, and KLFs in these Nos3 enhancer regions (Supplemental Tables 5–7). The ATAC-seq signals revealed no statistically significant difference in chromatin openness between NG and HG at the proximal promoters of Notch1 and its downstream targets (Hey2, EfnB2, and Nrg1), although we did observe significant differences in the enhancer, intronic, and downstream regions of Hey2 (P = 0.011), EfnB2 (P = 0.031), and Nrg1 (P = 0.008) (Supplemental Figure 3, A–D).
Figure 3. Hyperglycemia alters chromatin accessibility, TF occupancy, and NO production.
(A) Loss of ATAC-seq peaks in HG- (25 mM) compared with NG-treated (5.5 mM) atrioventricular mesenchymal (AVM) cells. Heatmap generated from HG normalized to NG input, with peaks centered, including 1.5 kb upstream and downstream of the transcriptional start site. Red and blue represent low and high signal intensity, respectively. (B) Distribution of 49,035 differential peaks. Pie chart highlights distribution of peaks within promoter (33%), inside gene (17%), upstream (31%), and downstream (19%) regions. Histogram displays frequency of peaks at a given distance from TSS. (C) Volcano plot highlights fold change and significance of identified peaks as plotted by –log10(P value) versus fold change. (D) Integrative genome viewer tracks of normalized merged data sets, highlighting peak loss at Nos3 loci in HG. Square boxes highlight 3 regions (R1–R3) with relative chromatin closure in HG. (E) ChIP-qPCR with H3K27ac showed significant downregulation of activation marks at Nos3R1–R3 (n = 4). (F and G) Decreased expression of Nos3 in HG at mRNA and protein levels (n ≥ 5) by IF respectively. Measurement of NO (H and I) and superoxide (J and K) production in AVM cells maintained in NG and HG for 48 hours, probed with diaminorhodamine (DAR-4M AM) and dihydroethidium (DHE), respectively. (L and M) DAR-4M AM and (N and O) DHE staining of E13.5 murine hearts exposed to nondiabetic and diabetic environments demonstrates increased ROS and reduced NO with diabetes (n = 3 per group). Nuclear DAPI stain, blue; atrioventricular cushions (yellow arrows); LV, left ventricle; RV, right ventricle; IVS, interventricular septum. Mean ± SEM; *P < 0.05, 2-tailed Student’s t test in F and G, with Holm-Bonferroni correction in E. Scale bars: 50 μm (G–K); 100 μm (L–O).
We next examined levels of NO production using diaminorhodamine-4M AM (DAR-4M AM), a fluorescent probe that detects NO, and found reduced NO levels with HG in AVM cells (Figure 3, H and I, and Supplemental Figure 4, A–C). Using the STZ-induced matDM model, we examined E13.5 hearts exposed to matDM (>200 mg/dl blood glucose) and demonstrated reduced DAR-4M AM staining (Figure 3, L and M). As previously discussed, HG is associated with excessive production of ROS, including peroxide (O22–), superoxide (O2–), and hydroxyl (OH–) radicals, which in turn reacts with NO-producing reactive nitrogen species, including nitrates (NO3–), and peroxynitrite (ONOO–), leading to further reduction in NO availability (47). Consistent with this, we found elevated ROS with HG using dihydroethidium (DHE), a fluorescent probe that detects superoxide, in both AVM cells exposed to HG and in vivo embryonic hearts exposed to matDM (Figure 3, J, K, N, and O, and Supplemental Figure 4, D–F). The increase in superoxide corresponded to a marked reduction of NO, notably in the region of the atrioventricular cushions that are composed of endocardial-derived mesenchymal cells that have undergone endocardial-mesenchymal transformation (yellow arrows, Figure 3, L–O). These changes in NO production were seen predominantly in ECs of E13.5 hearts exposed to matDM (Supplemental Figure 4, G–L), consistent with the ATAC-seq data showing that HG alters chromatin accessibility to TFs known to regulate EC gene expression. To test if the cardiac phenotype could be recovered by quenching excess ROS generation, a subset of control and STZ-treated WT females were treated with an antioxidant, N-acetyl cysteine (NAC), in the drinking water from E0.5 until E13.5 (Supplemental Figure 5A). We found that addition of NAC treatment in vivo was unable to rescue the VSD phenotype in diabetic WT embryos (3 of 8 embryos [37%]; diabetic, maternal average blood glucose level ± SD = 297.33 ± 69.01 mg/dl versus 5 of 16 embryos [31%]; diabetic+NAC, maternal average blood glucose level ± SD = 303 ± 75.97 mg/dl; P = 1.000) compared with nondiabetic controls (0 of 14 embryos [0%]; P = 0.036 and 0.044, respectively) (Supplemental Figure 5, B–J, and Supplemental Table 8). The inability to rescue the phenotype by NAC could be explained by chromatin inaccessibility at Nos3 loci with HG, which does not allow for complete rescue of NO levels. Both in vitro and in vivo HG models demonstrated dual mechanisms for the association between HG and decreased NO production, relative chromatin closure at the upstream of Nos3 loci, and increased levels of ROS production.
Increased Jarid2 with HG.
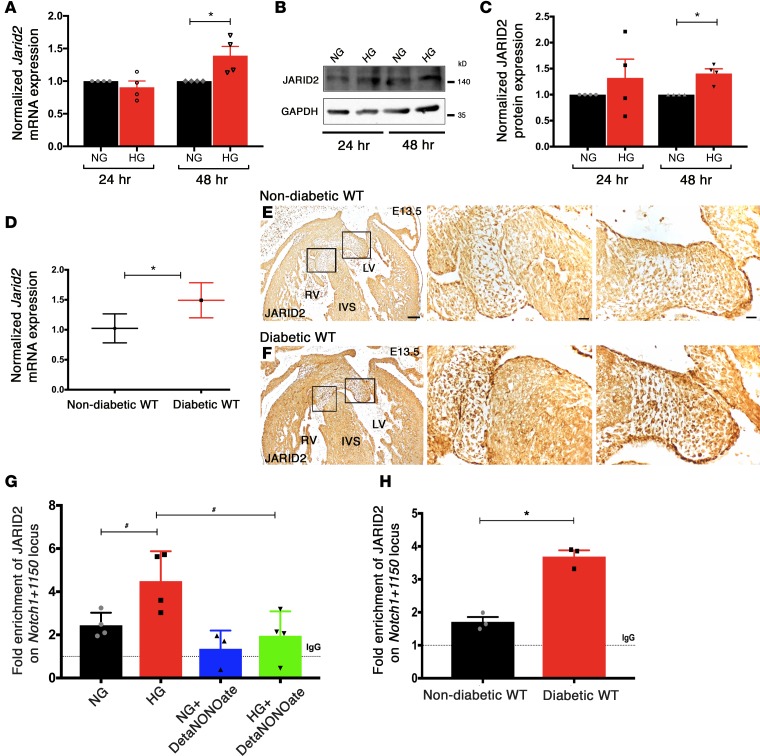
To elucidate the molecular mechanisms underlying matDM-mediated CHD in the setting of Notch1 haploinsufficiency, we examined the expression of Jarid2, which encodes a Jumonji-C and AT-rich interaction domain–containing protein that acts as a transcriptional repressor by recruiting silencing marks at histone H3 lysine 9 (H3K9me3) and lysine 27 (H3K27me3) in coordination with Setdb1 and polycomb repressive complex 2 (PRC2) (48, 49). Jarid2 had been previously linked to epigenetic regulation of endocardial Notch1, as the expression of Notch1 is upregulated in Jarid2 KO mice (50, 51). Examination of Jarid2 expression after 48 hours of HG in AVM cells demonstrated upregulation of Jarid2 mRNA and protein when compared with NG (Figure 4, A–C). To determine if similar changes occurred in vivo, we examined E13.5 hearts exposed to matDM and found increased Jarid2 expression as compared with controls by RT-qPCR and immunohistochemistry (Figure 4, D–F). Jarid2 has been also shown to negatively regulate Notch1 expression by directly binding to the Notch1+1150 locus with Setdb1 (50, 51). We examined if HG led to increased binding of JARID2 on the Notch1 locus to downregulate its expression by performing a ChIP-based PCR assay in AVM cells maintained in NG or HG conditions. By ChIP-qPCR, we demonstrated an enrichment of JARID2 binding on the Notch1+1150 locus in AVM cells with HG and a dramatic reduction in binding by ChIP-qPCR with addition of NO donor, DetaNONOate (Figure 4G). Experiments were also performed using immunoprecipitated DNA from E13.5 embryonic hearts exposed to matDM (maternal average blood glucose level ± SD = 296.58 ± 54.93 mg/dl) versus NG, and they demonstrated a similar enrichment of JARID2 (Figure 4H). Our in vitro and in vivo studies show that HG increases Jarid2 expression and its occupancy on the Notch1+1150 locus, leading to repression of Notch1 transcription, and suggested regulation by the level of NO.
Figure 4. Jarid2 mediated regulation of Notch1 with hyperglycemia.
(A) RT-qPCR and (B) representative immunoblot showing Jarid2 transcript and JARID2 protein levels in NG- (5.5 mM) and HG-treated (25 mM) AVM cells 24 and 48 hours after treatment (n = 4; mean ± SEM; *P < 0.05, 2-tailed Student’s t test). (C) Quantification of relative expression normalized to GAPDH (n = 4; mean ± SEM; *P < 0.05, 2-tailed Student’s t test). (D) Examination of E13.5 murine hearts (n = 6 hearts pooled together/group) exposed to nondiabetic and diabetic environments shows upregulation of Jarid2 mRNA by RT-qPCR (mean ± SD; *P < 0.05, 2-tailed Student’s t test). (E and F) Increased JARID2 protein in embryonic hearts exposed to maternal diabetes by immunohistochemistry (n = 3). Square boxes in E and F are shown in higher magnification from left to right. LV, left ventricle; RV, right ventricle; IVS, interventricular septum. Scale bars: 100 μm (E and F, left column); 20 μm (E and F, center and right columns). (G) ChIP-qPCR on AVM cells in NG or HG revealed enrichment of JARID2 on Notch1+1150 locus with HG and lack of enrichment with addition of 250 μM DetaNONOate (n = 4; mean ± SEM; #P < 0.05, 2-tailed Student’s t test, not significant with Holm-Bonferroni correction). (H) In vivo ChIP-qPCR on E13.5 hearts exposed to nondiabetic and diabetic environments (n = 3). Rabbit IgG served as mock control, shown as dotted line (set to 1) (mean ± SEM; *P < 0.05, 2-tailed Student’s t test).
Relationship between NO and Jarid2 and Notch1 expression.
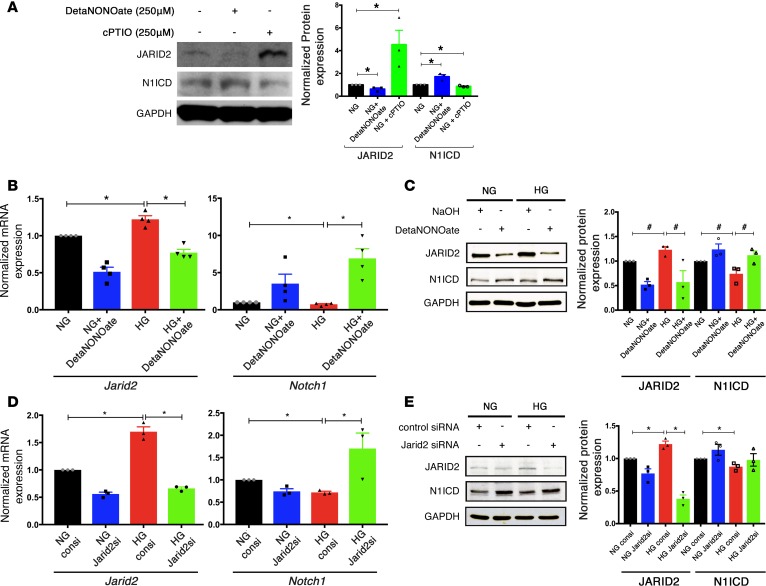
NO has been proposed to act as modulator of gene expression by affecting posttranslational modifications of histones, including the Jumonji C domain containing demethylases (29, 30). To determine if NO levels affected Jarid2 expression, AVM cells were cultured in NG for 48 hours, followed by treatment with 250 μM DetaNONOate (NO donor) or 250 μM cPTIO (NO scavenger). While NO donor decreased JARID2 and increased N1ICD protein expression, NO scavenger had the opposite effect, supporting a regulatory mechanism mediated by NO (Figure 5A). We next assessed whether the addition of NO donor under the setting of HG could prevent Jarid2 upregulation and rescue the reduction in Notch1 expression. Therefore, we cultured AVM cells in NG or HG, treated them with 250 μM DetaNONOate (NO donor), and found that the addition of exogenous NO decreases Jarid2 and increased Notch1 expression in NG and HG conditions (Figure 5, B and C). To determine if Jarid2 was necessary for downregulation of Notch1 expression in this system, we performed studies using Jarid2 siRNA. Knockdown of Jarid2 using siRNA resulted in an increased expression of Notch1 mRNA in AVM cells exposed to HG, as compared with treatment with a scrambled siRNA control, which led to the expected downregulation of Notch1 (Figure 5D). A similar rescue in N1ICD was found by immunoblot with JARID2 knockdown in AVM cells treated with HG (Figure 5E). To test whether addition of an antioxidant was able to restore JARID2 and N1ICD expression, we treated AVM cells cultured in NG and HG with 5 mM NAC. In cells exposed to HG, we noted a significant rescue in N1ICD expression back to basal levels with NAC treatment compared with HG alone (Supplemental Figure 6, M–P and T); while JARID2 expression was decreased with NAC, it remained significantly elevated when compared with basal NG levels (Supplemental Figure 6, I–L and S). We noted that NAC treatment was able to reduce total ROS generation in HG cells to baseline NG levels (Supplemental Figure 6, A–D and Q), but NO levels did not increase (Supplemental Figure 6, E–H and R), as shown by H2DCFDA (a marker for ROS) and DAR4M staining, respectively. The inability to rescue NO levels could possibly be due to decreased transcription of Nos3 mRNA due to alterations in chromatin dynamics. In aggregate, these in vitro rescue experiments demonstrate the role of NO in Jarid2-mediated regulation of Notch1 signaling.
Figure 5. NO mediated Jarid2 repression restores Notch1 expression.
(A) Immunoblot showing JARID2 and N1ICD levels in normoglycemic AVM cells treated with 250 μM DetaNONOate and 250 μM cPTIO as NO donor and quencher for 48 hours. The graph shows quantification (n = 3; mean ± SEM; *P < 0.05, 2-tailed Student’s t test). (B) Jarid2 and Notch1 mRNA expression and (C) JARID2 and N1ICD protein levels in AVM cells cultured in NG (5.5 mM) and HG (25 mM) with 250 μM DetaNONOate. 0.01 M NaOH served as vehicle control. Immunoblot quantification in C (n ≥ 3; mean ± SEM; #P < 0.05 by 2-tailed Student’s t test, not significant with Holm-Bonferroni correction). (D and E) RT-qPCR and representative immunoblot showing expression of Jarid2 and N1ICD in AVM cells after treatment with Jarid2 siRNA or control siRNA cultured in NG or HG. Immunoblot quantification in E (n ≥ 3; mean ± SEM; *P < 0.05, 2-tailed Student’s t test with Holm-Bonferroni correction in B, D, and E).
Evolutionary conservation of HG and Notch1 signaling interaction.
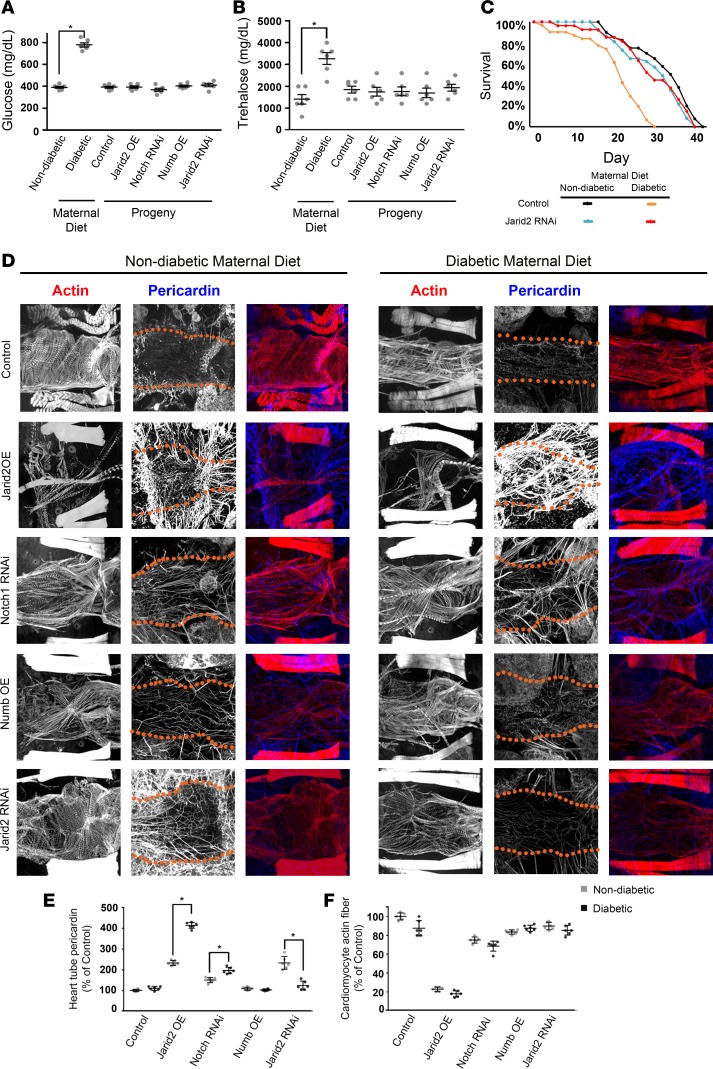
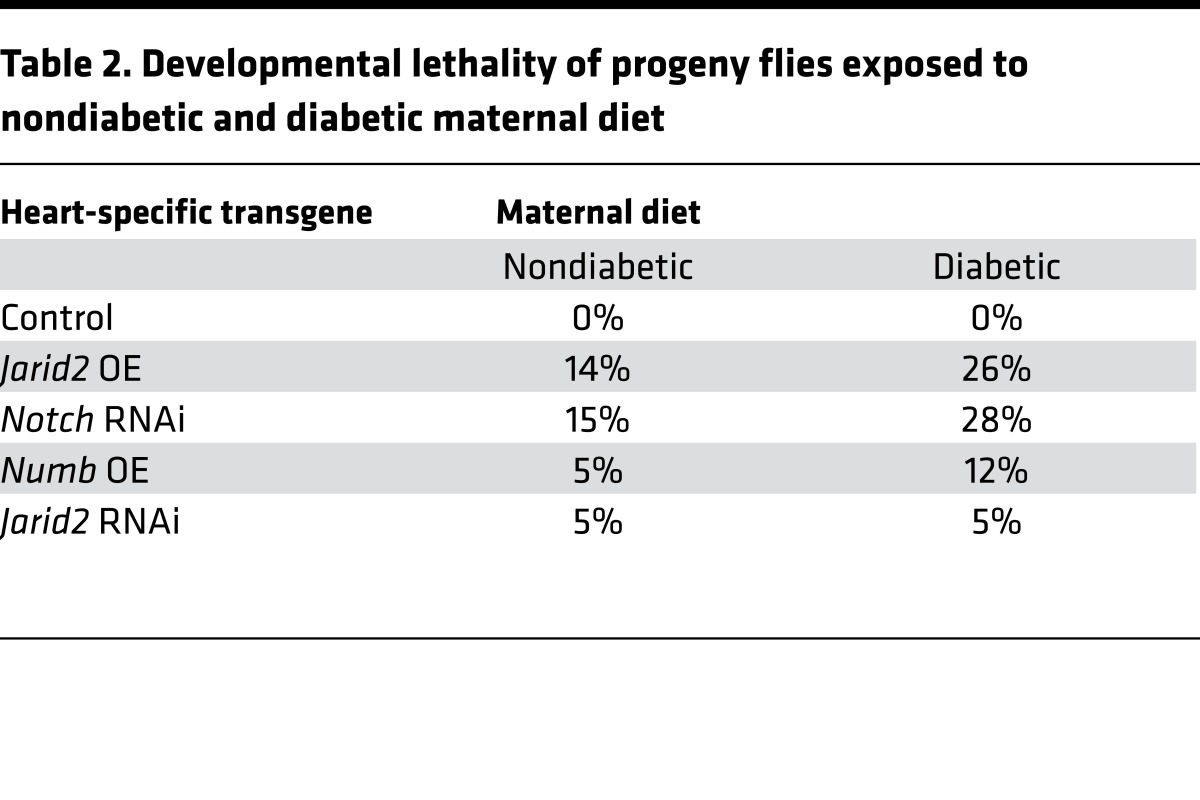
To determine if the gene-environment interaction between HG and Notch1 signaling is evolutionarily conserved, we utilized a Drosophila melanogaster model of HG by supplementing standard fly food with 0.2 g/ml sucrose (52). Expression of Notch signaling was manipulated by heart-specific overexpression (OE) of Notch1 RNAi-silencing gene, Numb or Jarid2 transgenes (known repressors of Notch signaling), or Jarid2 RNAi-silencing gene. Maternal WT flies maintained on a “diabetic” diet demonstrated elevated glucose and trehalose levels in hemolymph as compared with control females maintained on “nondiabetic” fly food (Figure 6, A and B). Progeny flies from both maternal “diabetic” or control diets maintained on standard food had normal glucose and trehalose levels similar to progeny flies expressing transgenes directing Notch1 RNAi, Numb OE, Jarid2 OE, and Jarid2 RNAi (Figure 6, A and B). When Notch signaling was decreased with Notch RNAi, Numb OE, or Jarid2 OE, there was notable lethality among the progeny of nondiabetic mothers, and this lethality was increased when combined with exposure to maternal HG (Table 2). We did not find a rescue of the lethality of progenies from nondiabetic and diabetic mothers with Jarid2 RNAi (Table 2), so we examined the maximum adult lifespan of the progenies expressing transgenes.
Figure 6. Overexpression of Jarid2 and Notch1 downregulation results in developmental mortality in the Drosophila maternal diabetes and gene-environment interaction model.
(A) Glucose and (B) trehalose levels measured in hemolymph of adult WT flies with standard and diabetic (0.2 g/ml) maternal diet, along with WT progeny flies and those overexpressing heart-specific Jarid2, Numb, and Notch1 RNAi and Jarid2 RNAi transgenes (n = 6 using 50 adult flies) (mean ± SEM; *P < 0.05 by 2-tailed Student’s t test). (C) Survival curves for adult progenies exposed to nondiabetic and diabetic maternal diet overexpressing heart-specific Jarid2 RNAi transgenes compared with controls. (D) Adult fly heart phenotypes in progeny of nondiabetic and diabetic mothers. Cardiomyocyte muscle fiber actin (phalloidin, red) and pericardin (blue) were stained. Red dotted lines delineate heart tube. Scale bar: 5 μm. Quantification of (E) heart tube pericardin deposition and (F) cardiomyocyte actin fiber density in adult progeny of nondiabetic and diabetic mothers (n = 6; mean ± SD; *P < 0.05, 2-tailed Student’s t test).
Table 2. Developmental lethality of progeny flies exposed to nondiabetic and diabetic maternal diet.
We found that the lifespan of WT progeny of diabetic mothers was approximately 25% shorter than that of progeny of nondiabetic mothers (Figure 6C). The lifespan was severely shortened by heart-specific Jarid2 or Numb OE in progeny of both diabetic and nondiabetic mothers, and this was true as well for progeny expressing Notch RNAi. In all cases, the effect was more severe in progeny of diabetic mothers (Supplemental Figure 7). Intriguingly, Jarid2 silencing by RNAi transgene expression rescued the reduced survival phenotype displayed by WT progeny of diabetic mothers, while having no adverse effect on lifespan of progeny of nondiabetic mothers (Figure 6C). We did note that Jarid2 knockdown alone did not cause a significant reduction in lifespan, while HG did decrease survival. This suggested that the lethality is not solely related to Jarid2.
Examination of the adult WT progeny exposed to the maternal HG diet demonstrated an apparent disorganization, lower density of cardiomyocyte actin fibers, and slightly elevated pericardin deposition (Figure 6, D–F). Jarid2 OE induced strikingly elevated pericardin in offspring of diabetic mothers and reduced cardiomyocyte actin fibers in progeny of diabetic and nondiabetic mothers. This phenotype was also induced to a lesser extent in progeny expressing Notch RNAi. Numb OE was not associated with significant changes in pericardin, but disorganization of cardiac actin was apparent. Silencing Jarid2 by RNAi transgene expression was associated with upregulated pericardin deposition in progeny of nondiabetic mothers, but not in progeny of diabetic mothers, suggesting a rescue effect of reducing Jarid2 level by RNAi that counteracts the increased Jarid2 caused by a maternal HG diet. Together, experiments with the Drosophila maternal HG model displayed an evolutionarily conserved gene-environment interaction model involving Jarid2 and Notch1.
Discussion
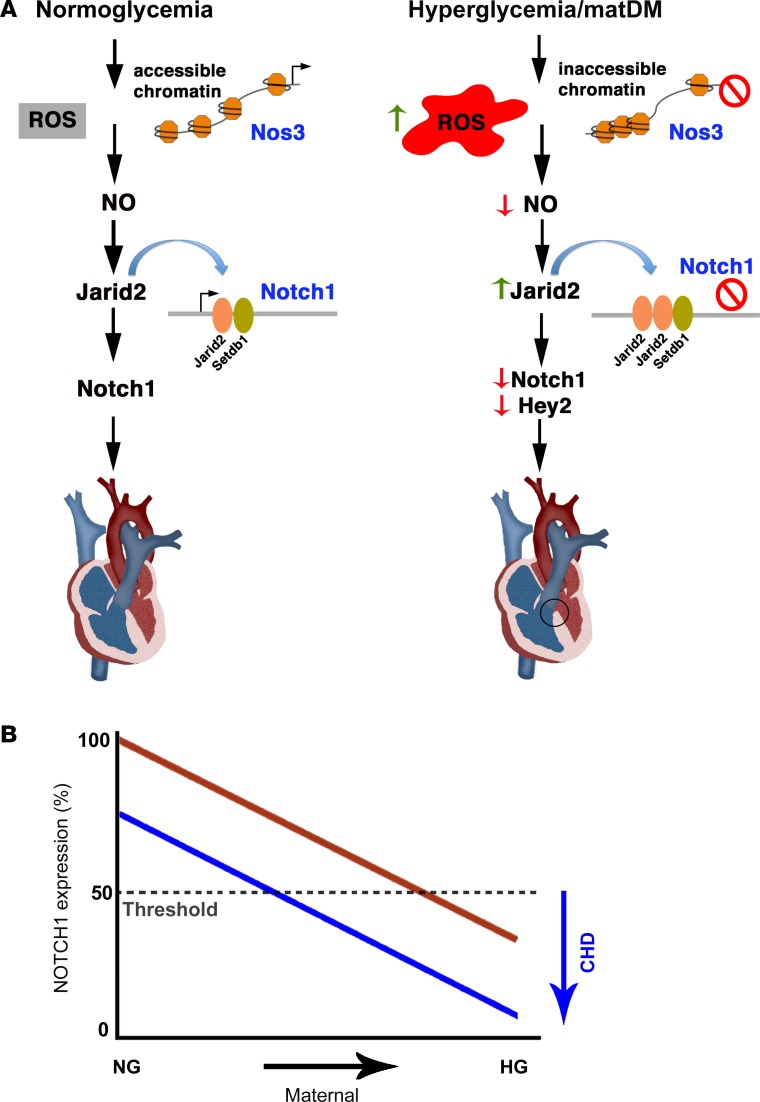
In this study, we demonstrate an epigenetic mechanism underlying the gene-environment interaction between Notch1 haploinsufficiency and matDM that results in CHD. We show using an in vitro ATAC-seq approach that HG stress alters overall chromatin accessibility and TF occupancy, predominantly for genes involved in EC gene regulation. Specifically, we noted reduced chromatin accessibility at the Nos3 locus and that this was associated with reduced NO along with increased oxidative stress in an embryonic cardiac cell line and the developing heart. Diminished NO production is associated with increased expression of Jarid2, a known epigenetic repressor of Notch1, and thereby results in a further reduction in Notch1 expression. The combination of in vitro and in vivo studies using both murine and fly model systems identifies an evolutionarily conserved molecular mechanism in which maternal HG raises the risk of CHD by reducing expression of Notch1 to a level below a threshold required for normal heart development and identifies potential mechanisms by which environmental factors cause CHD in genetically susceptible individuals (Figure 7A).
Figure 7. Gene-environment interaction model.
(A) Schematic diagram representing the normoglycemic condition and a proposed model in which HG results in increased ROS and reduced NO production by altering chromatin accessibility. Reduced NO is associated with increased expression of Jarid2 and its enrichment on the Notch1 locus with Setdb1. This results in repression of Notch1 and its downstream targets, culminating in congenital heart defects (highlighted by black circle). (B) Model showing gene-environment interaction between HG and Notch1 expression. In individuals without NOTCH1 mutations (red line), only severe HG crosses a threshold for congenital heart disease. Individuals with NOTCH1 mutations (blue) are a sensitized population that can cross the threshold with modest elevations in maternal blood glucose. HG, hyperglycemia; NG, normoglycemia.
Systems biology approaches have indicated that the maternal intrauterine environment modulates fetal metabolic programming, which in turn can have a profound effect on gene expression early in life and can be passed across generations through epigenetic modifications (53). Recent reports have shown that NO affects global histone methylation patterns, particularly by directly inhibiting the lysine-specific histone demethylase Kdm3a and likely other Jumonji C domain–containing demethylases (29, 30). Although the enzymatic activity of JARID2 has not yet been demonstrated, this DNA-binding protein functions as a transcriptional repressor by recruiting other repressive methyltransferases and has been shown to regulate the cardiac expression of Gata4, Nkx2.5, and Cyclin D1 (48–51, 54, 55). Here, we demonstrated that Jarid2 expression can be modulated by adding external NO donor and quencher, suggesting a link between NO and Jarid2; however, the precise mechanism remains to be elucidated. While we observed increased expression of Jarid2 in response to HG, the in vitro ATAC-seq data showed that there was no statistical difference in chromatin accessibility at the promoter of the Jarid2 locus between the two groups, suggesting alternative mechanisms for regulation of Jarid2 (Supplemental Figure 3E). TFBS profiling of 3 open chromatin regions (J2R1–J2R3) upstream of Jarid2 revealed a consensus binding sequence for SP1, KLFs, hypoxia response element, CREB, and NF-κB (Supplemental Tables 9–11), further suggesting that EC TFs may regulate the transcription of this gene under HG stress.
We also describe a mechanism of Notch1 regulation by Jarid2 in response to HG-induced changes in NO levels. This repression of Notch1 was restored with addition of a NO donor or by downregulation of Jarid2 in AVM cells (Figure 7A). Notch has been shown to alter NO production by activating a PI3 kinase/Akt pathway to phosphorylate Nos3, and this Notch-dependent autocrine loop is necessary to drive EMT in the developing atrioventricular canal (56). Given that NO serves as an important cardiovascular signaling molecule, Nos3 is regulated by multiple transcriptional, posttranscriptional, and posttranslational mechanisms (57–59). In parallel to this, our findings from in vitro ATAC-seq revealed that Nos3 open chromatin regions (R1–R3) also contain binding sites for KLFs, SP1, and the ETS-family of TFs, which can regulate Nos3’s expression through another feedback mechanism under HG stress (Supplemental Tables 5–7). This observation is supplemented by the fact that Klf2 and Klf4 ablation in embryos resulted in EC disruption by reducing Nos3 expression (60). Further characterization of these TFs on Nos3 enhancer regulation upon HG exposure is needed. ATAC-seq did not show a statistically significant difference between NG and HG at the Notch1 promoter (R1–R4, Supplemental Figure 3A) because of an inconsistent pattern in their chromatin accessible regions among replicates. Additionally, these regulatory regions may need to be opened in order for JARID2 and SETDB1 to bind and regulate its expression. Putative TFBS sites for GATA, KLFs, NRF1, PAX5/6/9, GLI3, PPARG, PRDM5, SMARCA3, and NF-κB (Supplemental Tables 12–15) were identified in these regions, some of which have been reported to regulate Notch1 expression.
In addition to our current gene-environment model involving Notch1, we examined 361 differential ATAC-seq peaks between NG and HG (Figure 3C) with statistical significance (P < 0.05) and a fold change of at least 1.5, which were located within ±1.5 kb of TSS (Supplemental Figure 8, A–C, and Supplemental Table 16). The downstream gene ontology (GO) analysis with this pruned data set identified terms related to chromatin regulation and embryo development and genes involved in maintenance of cellular architecture among the top 25 GO terms (Supplemental Figure 8A). Circle plot displayed the gene associated with peaks that were upregulated or downregulated within each GO term (Supplemental Figure 8, B and C). For example, peaks associated with the genes in the BMP, FoxP1, and Wnt cardiac morphogenetic signaling pathways were identified, and they have been linked to increased risk of fetal neurologic and cardiac developmental defects in matDM-exposed embryos (61–65). HG stress resulted in dysregulation of chromatin accessibility of genes important for reorganizing actin cytoskeleton, cell polarity, and alignment during tissue morphogenesis (66, 67). Interestingly, our data demonstrated that peaks associated with genes involved in chromatin regulation were also altered in the setting of HG (Supplemental Figure 8, A–C, and Supplemental Table 17). The classifications identified as most significant by GO terms matched the known HG biologic response to diabetes mellitus, indicating the usefulness of this technology to explain the epigenetic regulation of gene expression in the setting of environmental teratogen. Genome-wide analysis of chromatin conformational studies in vivo at different developmental time points will provide a better molecular framework to understand gene-environment interaction in the context of CHD.
Diabetes mellitus is associated with a multitude of metabolic disturbances, and elevated glucose levels appear to be the primary teratogen for diabetic embryopathy (12). Consistent with this, improved glucose control during pregnancy is associated with reduction in CHD risk and, alternatively, poor glycemic control is linked to a higher risk for CHD in offspring (10). We are unable to completely rule out the effect of lack of insulin or insulitis as a result of STZ toxicity on the murine cardiac phenotype. However, our studies in AVM cells and Drosophila model systems, along with the finding of Notch1 mRNA downregulation in an embryonic (HH24) chick model of HG exposure (data not shown), supports the teratogenicity of HG as opposed to the metabolic consequences related to insulin. Reduced cardiomyocyte proliferation and Cyclin D1 transcript expression was noticed in E13.5 embryos exposed to matDM (Supplemental Figure 9, A–J), which is either due to excess ROS production or the secondary effect of Notch1 downregulation in endocardial/endocardial-derived cells; however, the mechanism leading to VSD is beyond the scope of this study. It has been shown that overexpression of the superoxide dismutase (SOD) gene rescues CHD phenotypes in the offspring of mice with matDM but this may overexpression system may have numerous effects apart from decreased ROS (68). Similarly, NAC treatment of severely diabetic mice (average blood glucose >450 mg/dl) has been shown to partially rescue the CHD phenotypes (69). This finding is not inconsistent with our data, as complete rescue was not achieved with NAC with moderate maternal HG (average blood glucose >200 mg/dl), and this is likely due to changes in chromatin accessibility at the Nos3 locus. Of note, neither of these published studies examined NO levels or Nos3 expression. Our data suggest that increased incidence of CHD with matDM may be prevented by either exogenous NO donor supplementation or genetic upregulation of Nos3. Additionally, overlaying the ATAC-seq data with ChIP-seq using redox-sensitive TFs, such as NRF1/2 and NF-κB, will identify targets in HG-mediated oxidative stress that will help us understand the mechanism of how ROS alters chromatin accessibility and results in reduced Nos3 transcription.
While no study has directly linked levels of maternal HG in the first trimester when the heart develops with the risk of CHD, lower maternal glycosylated hemoglobin levels (hemoglobin A1c), which serve as an indicator of HG over the preceding 2–3 months, have been shown to be associated with a reduced risk of birth malformations in humans (70–72). It was recently reported that diabetes-induced elevation of maternal blood glucose levels during the 15th–18th week of gestation were correlated with CHD in a California-based case-control study (73). A mild but statistically significant increase in maternal serum glucose was associated with tetralogy of Fallot, a conotruncal heart defect, in offspring relative to unaffected controls, implying that subtle alterations in maternal glucose have the potential to serve as an environmental risk factor for CHD (73). These findings, in combination with our work, suggest a gene-environment interaction model where increasing maternal glucose levels interact to reduce Notch1 expression levels to cause CHD, and this interaction is sensitized by genetic susceptibility loci, i.e., Notch1 haploinsufficiency, to cross a threshold for CHD occurrence (Figure 7B). In addition to the importance of dosage on phenotypic variability, the variety of cardiac defects observed in diabetic pregnancies include defective endocardial cushions with perturbed endothelial-to-mesenchymal transformation to abnormal development and/or migration of cardiac neural crest cells (74), suggesting the potential involvement of different cell types. Therefore, it is equally important to understand what cell lineage is being modulated epigenetically in the setting of altered in utero environment. In summary, our work addresses the combinatorial role of genetic and environmental risk factors for normal cardiac morphogenesis in the developing embryo. We have shown how environmental contributors, such as matDM, can influence the expression of critical cardiac developmental genes via epigenetic mechanisms and suggest that environmental factors can act in a genetically sensitized population to further reduce expression levels below a threshold to cause CHD.
Methods
Cell culture experiments.
Immortalized AVM cells were cultured in DMEM (ThermoFisher Scientific) with 5.5 mM D-glucose (NG), L-Glutamine, and sodium pyruvate and supplemented with 10% FBS, 1% penicillin/streptomycin at 37°C in 5% CO2 chamber (40). To mimic HG conditions, cells were treated with D-glucose solution (A2494001, ThermoFisher Scientific) at a concentration of 25 mM (HG). Media was changed every 2–3 days, and cells were passaged at >90% confluence. All the experiments were performed between passages 4 and 7. Hydrogen peroxide (H2O2; AC302865000, Fisher Scientific) diluted in DMSO (D2650, Sigma-Aldrich) was added to culture media to serve as positive control for oxidative stress. NAC (O1049-25, Fisher Scientific), a thiol-containing antioxidant, was used to neutralize the effect of HG-mediated ROS generation in cells. 2, 2′-(Hydroxynitrosohydrazino) bis-ethanamine (DetaNONOate; AC328650250, Fisher Scientific) was used as NO donor and carboxy-PTIO, potassium salt (cPTIO; 81540, Cayman Chemical) was used as NO scavenger and dissolved in DMSO per manufacturer’s instructions. To test how NO regulates Notch1 expression via Jarid2 in HG conditions, 250 μM DetaNONOate was added to AVM cells in NG and HG for 48 hours. Media was refreshed daily with DetaNONOate, and 0.01 M NaOH served as negative control.
ATAC-seq.
A described ATAC-seq protocol was used to profile the pattern of accessible chromatin in the setting of NG and HG with minor modifications (23, 24). After 48 hours of culture in NG and HG, unfixed AVM cells were counted for nuclei preparation using a hemocytometer (Bright-Line, Hausser Scientific). In each experiment, 54,000 cells were centrifuged at 500 g for 5 minutes (4°C) and washed with 200 μl ice-cold 1× PBS. Cell pellets were gently resuspended in 200 μl cold lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3mM MgCl2, 0.1% IGEPAL CA-630; Sigma-Aldrich) supplemented with complete mini EDTA-free protease inhibitor cocktail (11836170001, Sigma-Aldrich) and spun at 500 g for 5 minutes (4°C). Nuclear pellets were transposed with 25.0 μl Tagmentation DNA buffer, 2.5 μl Transposase enzyme (Nextera DNA library preparation kit, FC-121-1030, Illumina), and 22.5 μl nuclease-free water for 30 minutes (37°C) followed by purification with the MinElute Reaction Cleanup kit (28204, Qiagen). Transposed DNA fragments were amplified using following PCR conditions: 72°C for 5 minutes; 98°C for 30 seconds; cycling at 98°C for 10 seconds, 63°C for 30 seconds, and 72°C for 1 minute in 50-μl reaction mix combining 10 μl Transposed DNA, 2.5 μl of 25 μM PCR primer 1 (common, Ad1_no mix), 2.5 μl of 25 μM PCR primer 2 (sample specific, Ad2.1–Ad2.6), 25 μl NEBNext High Fidelity 2X PCR master mix (M0541S, NEB), 0.3 μl 100× SYBR Green I (S7563, ThermoFisher Scientific), and 9.7 μl Nuclease-free water. Index primers were synthesized by Integrated DNA Technologies (Supplemental Table 18). qPCR side reaction was performed on 5 μl preamplified DNA for 19 cycles to reduce size bias and to determine appropriate number of PCR cycles for each experiment. Amplified fragments were purified with the Qiaquick PCR purification kit (28104, Qiagen) and run on 5% Mini-PROTEAN TBE gel (4565013, Bio-Rad Laboratories) and Agilent Bioanalyzer to check average fragment size. The remaining 45 μl of product was run for 10–13 cycles, and the amplified library was purified using the MinElute kit. Quality of libraries was assessed using the Agilent Bioanalyzer High Sensitivity DNA analysis kit by Biomedical Genomics Core at Nationwide Children’s Hospital (NCH). ATAC-seq libraries were sent for 50bp paired-end sequencing on Illumina HiSeq 2000 (Epigentek Group Inc.).
ATAC-seq and bioinformatics analysis.
Data analyses were performed in collaboration with Epigentek Group Inc. Raw reads were quality checked using FastQC version 0.10.1 (Babraham Bioinformatics). One NG sample was excluded from analysis due to low coverage. Nextera primers were trimmed from raw reads using cutadapt v1.0 with parameters –O 5 –m 30 –q 15, and trimmed reads were aligned to mm10 genome using Bowtie v1.0.0 with activation of “-m 1” so only unique reads were allowed. The mapping results in SAM were converted to BAM and sorted by coordinates using samtools v0.1.19. The mapping statistics were extracted using Picard version 1.90. Peak calling was performed using MACS2 v2.1.0 with default parameters. Affinity-based differential binding analysis and annotation were performed using DiffBind v1.12.3 (75) and ChIPpeakAnno v3.0.0, respectively (76). R package DiffBind employs edgeR for differential analysis, utilizing trimmed mean of M values (TMM) to account for sequence depth to normalize read counts between samples in differential peak calling analysis. The ATAC-seq data have been deposited in NCBI’s Gene Expression Omnibus (GEO GSE104520).
ATAC-seq and data visualization.
Data visualization was performed by normalizing the merged HG BAM file to the merged NG BAM file through random downsampling. Heatmaps were generated with Seqplots using 49,035 differential peaks called by DiffBind and comparing NG to HG, with features set as the midpoint and a plotting distance of 1.5 kb upstream and downstream of the feature. Signal was plotted as “feature centered” in decreasing fashion, utilizing the K-means clustering algorithm, and color scaling was set to 0.1. To visualize peaks in a genome browser, the ATAC-seq signal was normalized to NG input, converted to bigwig files, and visualized on Integrative Genomics Viewer (Broad Institute, ref. 77). Differential ATAC-seq peaks, identified by DiffBind, were visualized as a volcano plot [fold change versus log10(P value)] using R package ggplot2 (78).
Functional annotation.
Genes associated with “promoter” peaks, which were significantly different (P < 0.05) and had at least a 1.5-fold change, were subjected to functional annotation utilizing DAVID v6.8 (Database for Annotation, Visualization and Integrated Discover) and included GO direct analysis of biological processes, cellular components, and molecular functions (79, 80). The top 25 GO terms were plotted according to significance. Circle plot visualization was performed, using GOplot R package v1.0.2 (81) and focused on 12 of the top 25 GO terms: nucleosome assembly, regulatory region DNA binding, telomeric heterochromatin, chromatin binding, positive regulation of transcription DNA templated, negative regulation of BMP signaling pathway, embryo development, Wnt-activated receptor activity, actin filament, cytoskeleton, and microtubule.
Motif enrichment analysis.
To identify overrepresented motifs present in ATAC-seq data sets, motif enrichment analysis was performed using HOMER v4.4 (Hypergeometric Optimization of Motif EnRichment) (82). Enriched motifs were identified using the findMotifsGenome.pl command with default settings and the mm10 genome. To identify motifs that became inaccessible with HG, NG ATAC-seq was analyzed for enriched motifs with HG set as control. Both known and de novo–enriched motifs were identified.
TFBS prediction analysis.
Sequence-based TFBS prediction analysis was performed using Genomatix software suite v3.8 with default parameters, with Nos3 (R1, chr5:24357617–24357726; R2, chr5:24360864–24360991; R3, chr5:24361289–24361376), Notch1 (R1, chr2:26500935–26501063; R2, chr2:26503580–26503673; R3, chr2:26503787–26503942; R4, chr2:26507515–26507771), and Jarid2 (R1, chr13:44733879–44734017; R2, chr13:44733432–44733769; R3: chr13:44730497–44730770) loci identified to be open by ATAC-seq. The lists of predicted TFs include consensus sequences; matrix and core similarity scores are summarized (Supplemental Tables 5–7 and 9–15).
Experimental mouse models.
WT C57BL/6 male and female mice (4–8 weeks old) were used (Jackson Laboratory). Six- to eight-week-old female mice were used to induce type 1 diabetes mellitus by injecting STZ (NC0146241, Fisher Scientific) dissolved in 0.01 mol/l citrate buffer, pH 4.5, intraperitoneally at 75 mg/kg body weight for 3 consecutive days (74). Two weeks after STZ injection, mice were fasted for 8 hours during the light cycle, and glucose levels were measured using the AlphaTrak veterinary blood glucometer calibrated specifically for rodents from tail vein blood (Abbott Laboratories) before breeding and during embryo harvesting. Mice with fasting blood glucose ≥200 mg/dl (11 mmol/l) were defined as HG (83). A subset of nondiabetic and diabetic WT female mice received 4 mg/ml NAC (1 g/kg of body weight/d) in drinking water (69) from E0.5 until embryo harvest at E13.5. Notch1+/– mice were generated and genotyped as described previously (33).
Mouse embryo collection.
Timed mating was performed using nondiabetic and diabetic WT and Notch1+/– female mice with WT and Notch1+/– males. Mice were maintained on a 12-hour-light/dark cycle, with noon of the day of vaginal plug observation defined as E0.5. Pregnant mothers were sacrificed and embryos were collected at E13.5. WT littermates were used as controls for experiments.
Histologic and AMIRA analysis of embryos.
E13.5 embryos from multiple litters were harvested, fixed in 10% formalin (Fisher) overnight and washed with 1× PBS. Tissue sections (7 μm) were stained for H&E and imaged using Zeiss AxioImagerA2. For AMIRA 3D reconstruction, one embryo from the diabetic and control group was serially sectioned (7 μm), H&E stained, and imaged at ×5 magnification. Images were optimized with Adobe Photoshop Elements 9 and uploaded to AMIRA v5.5 imaging software. After manual alignment, material labels were added by tracing the outline of atrioventricular cushion and interventricular septum.
Measurement of ROS and NO production.
DHE (D-23107, ThermoFisher Scientific), a superoxide indicator; 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA; D-399, ThermoFisher Scientific), total ROS indicator; and DAR-4M AM (251765, Calbiochem, EMD Millipore), a NO indicator were used at 10 μM to determine endogenous ROS and NO production in AVM cells, respectively. DMSO served as negative control while 300 μM H2O2 and 250 μM DetaNONOate were positive controls for superoxide and NO staining. Cells were incubated with dye for 30 minutes (37°C) and washed 5 times with 1× PBS and nuclei were stained with DAPI (H-1500, Vector laboratories). Fluorescence was visualized using Olympus IX51 (×20 magnification) and captured using CellSens software. Mean fluorescence intensity of DHE- or DAR-4M AM–positive cells in 5 different fields was measured (ImageJ, NIH) and statistical comparisons between two groups were performed using Student’s t test (GraphPad Prism). Total ROS levels were measured in NG- and HG-treated AVM cells in presence/absence of 5 mM NAC using H2DCFDA staining. E13.5 embryos from nondiabetic and diabetic WT dams were fixed in 4% paraformaldehyde (PFA; 15710, Electron Microscopy Sciences, Fisher Scientific) for 20 minutes (4°C), subjected to 10%–30% sucrose gradient, embedded in Tissue-Tek OCT compound (50-363-579, Electron Microscopy Sciences, Fisher Scientific), and cryosectioned. Sections (8 μm) were permeabilized with 0.1% Triton X-100 in PBS (PBST) and incubated with 30 μM DHE and DAR-4M AM for 30 minutes (room temperature). Sections were washed with 1× PBST and mounted with Vectashield HardSet Antifade Mounting Medium with DAPI (H-1500, Vector Laboratories). Fluorescent images were visualized using Olympus BX51 microscope (×10 magnification). All staining experiments were performed in triplicate.
RNA purification and quantitative real-time PCR.
RNA was extracted from AVM cells cultured in NG and HG for 24 and 48 hours using TRIzol Reagent (15596018, ThermoFisher Scientific) per the manufacturer’s instructions. E13.5 hearts were dissected from nondiabetic and diabetic WT mice, harvested in TRIzol, and homogenized using TissueLyser II (Qiagen), followed by chloroform-isopropanol extraction and purification. RNA was quantified spectrophotometrically, and 500 ng to 1 μg of total RNA was used for reverse transcription using the SuperScript VILO cDNA Synthesis Kit (11754-050, ThermoFisher Scientific). SYBR Green–based real-time qRT-PCR was performed for following genes: mouse Nos3, Notch1, Hey1, Hey2, Nrg1, EphrinB2, Bmp10, Jarid2, and Cyclin D1 using Applied Biosystems 7500 real-time PCR (ABI) machine. Mean relative gene expression was calculated after normalizing Ct values to Gapdh using the ΔΔCt method. Three independent experiments were performed in triplicate in vitro with 6 embryos per group. Cell lysates from AVM cells treated with 250 μM DetaNONOate versus 0.01 M NaOH (control) and 10 nM control siRNA versus Jarid2 siRNA were also extracted 48 hours after treatment following above protocol, and qRT-PCR was performed for Jarid2 and Notch1. Oligonucleotide sequences of these genes are provided in Supplemental Table 19.
Immunofluorescence and immunohistochemistry.
For immunofluorescence staining, AVM cells were cultured in NG and HG in a 12-well plate for 48 hours and fixed in 2.5% PFA. Cells were permeabilized with 0.1% PBST at room temperature for 5 minutes, and nonspecific immunoreactions were blocked using IF block buffer containing 1% BSA (A2153, Sigma-Aldrich) in 1× PBST for 1 hour at room temperature. Nonspecific immunoreactions were washed 3 times with PBST. Cells were incubated overnight with mouse anti-NOS3 (sc-376751; 1:100, Santa Cruz Biotechnology), rabbit anti-JARID2 (1:250, NB100-2214, Novus Biologicals; 13594S, Cell Signaling Technology), and rabbit anti-N1ICD (1:250, ab8925, Abcam) primary antibodies. Following day, after PBST wash, cells were incubated with Alexa Fluor 594/488 anti-mouse and anti-rabbit highly cross-adsorbed secondary antibodies (ThermoFisher Scientific) for 1 hour at dark and washed 3 times with 1× PBST. Nuclei were stained using either propidium iodide (11348639001, Sigma-Aldrich) or Vectashield Mounting Medium with DAPI. Images were taken using Olympus IX51 (×20 magnification). A similar protocol was performed to stain E13.5 heart sections after deparaffinization using xylene and grades of ethanol, followed by antigen retrieval using citrate-based Antigen Unmasking solution (H-3300, Vector laboratories) for 30 minutes. After permeabilization and blocking with 1% BSA in PBST for 1 hour, tissue sections were incubated with endothelial marker, rabbit anti-PECAM-1 (M20, 1:250, Santa Cruz Biotechnology); mitosis marker, rabbit anti-phospho-Histone H3 (PHH3; 1:250, 06-570, EMD Millipore); and cardiomyocyte marker, mouse anti-cardiac Troponin T (cTnT; 1:250, ab8295; Abcam) primary antibodies overnight at 4°C. Following 1× PBST wash, sections were incubated with goat anti-rabbit and anti-mouse secondary antibodies that were conjugated to Alexa Fluor 594/488 for 1 hour at room temperature in the dark. Sections were washed 3 times with 1× PBST, and sections were counterstained with Vectashield HardSet Antifade Mounting Medium with DAPI. Fluorescent images were visualized using Olympus BX51 (×40 magnification). All staining experiments were performed in triplicate.
For immunohistochemical staining, embryonic heart sections were deparaffinized in xylene and rehydrated in grades of ethanol and 1× PBS. Briefly, after antigen retrieval tissue, sections were incubated with 3% H2O2 diluted in water at room temperature for 10 minutes to quench endogenous peroxidase activity and blocked by 5% normal goat serum in 1× TBS containing 0.1% Tween-20 (Sigma-Aldrich) for 1 hour at room temperature to avoid nonspecific binding per manufacturer’s protocol. Following this, sections were incubated with rabbit anti-N1ICD (1:250, ab8925, Abcam) and anti-JARID2 (1:250, NB100-2214, Novus Biologicals) and anti-HEY2 (1:250, AB5716, EMD Millipore) primary antibodies diluted in SignalStain Antibody Diluent (8112, Cell Signaling Technology) overnight at 4°C. Normal IgG served as negative control. Next, sections were washed with 1× TBST and incubated with SignalStain Boost IHC Detection Reagent (HRP, rabbit, 8114, Cell Signaling Technology) for 30 minutes at room temperature. Sections were washed 3 times with 1× TBST and visualized using the SignalStain DAB Substrate Kit (8059, Cell Signaling Technology) and imaged using Zeiss AxioImagerA2 (×10 and ×40 magnification).
Immunoblotting.
Cell lysate was extracted from AVM cells cultured in NG and HG conditions. Briefly, cells were rinsed with ice-cold 1× PBS and lysed with RIPA Lysis and Extraction Buffer (89900, ThermoFisher Scientific) supplemented with Halt Protease Inhibitor Cocktail (EDTA free, 100×) (87785, ThermoFisher Scientific). Cell lysates were centrifuged at 15,871 g for 15 minutes (4°C) and supernatant were collected. Protein concentrations were measured using the Pierce BCA Protein Assay Kit (23227, ThermoFisher Scientific). 10–20 μg total protein was boiled for 10 minutes with 5× sample loading buffer containing 10% SDS and β-mercaptoethanol and separated in 8% polyacrylamide gel electrophoresis. After transferring the proteins into polyvinylidene difluoride membrane, blocking was performed with 5% nonfat milk (in 1× TBST). Blots were probed using primary antibodies against rabbit anti-N1ICD (1:1,000; ab8925, Abcam), rabbit anti-HEY2 (1:1,000; AB5716, EMD Millipore), rabbit anti-JARID2 (1:1,000; NB100-2214, Novus Biologicals, 13594, Cell Signaling), and mouse anti-GAPDH (1:1,000; NB300-221, Novus Biologicals) and HRP-conjugated anti-rabbit and anti-mouse secondary antibodies (1:5000, PI-1000, PI-2000, Vector Laboratories). Membranes were developed using either Pierce ECL Western Blotting Substrate (32106, ThermoFisher Scientific) or SuperSignal West Dura Extended Duration Substrate (34075, ThermoFisher Scientific). Lysates prepared from NG AVM cells after treatment with 250 μM DetaNONOate and 250 μM cPTIO for 48 hours were run on SDS-PAGE and probed against JARID2 and N1ICD. Similarly, cell lysates from AVM cells were collected 48 hours after DetaNONOate treatment and 72 hours after Jarid2 siRNA treatment in presence of HG and NG, and Western blotting was performed for N1ICD and JARID2, as described above. Immunoblotting was performed in three independent experiments. Densitometric analysis was performed to normalize the protein levels with respect to GAPDH as loading control using ImageJ software (NIH; http://imagej.nih.gov/ij/).
ChIP-qPCR.
ChIP was performed with digested chromatin from AVM cells maintained in NG and HG, with and without addition of 250 μM DetaNONOate using the SimpleChIP Plus Enzymatic Chromatin IP Kit (9004, Cell Signaling Technology). Briefly, cells were fixed by cross-linking with 1% formaldehyde solution (F8775, Sigma-Aldrich) and chromatin fragmented using both micrococcal nuclease (MNase) digestion and sonication. Chromatin was immunoprecipitated with rabbit anti-Histone H3K27ac (ab4729, Abcam), rabbit anti-JARID2 (NB100-2214, Novus Biologicals), Histone H3 (D2B12) XP rabbit monoclonal antibody (positive control), and normal rabbit IgG (mock/no antibody control) at 4°C overnight following the manufacturer’s instructions. Histone-DNA complexes were captured by Protein G agarose beads followed by reversal of cross-linking and DNA purification. Immunoprecipitated DNA from H3K27ac pull-down was amplified for 3 Nos3 (R1–R3) regions found to be differentially accessible in response to HG by ATAC-seq. Similarly, oligonucleotides were designed to amplify the Notch1+1150 locus to detect JARID2 enrichment in NG and HG.
For in vivo ChIP, at least 15 nondiabetic WT and 20 diabetic WT E13.5 hearts were harvested, minced, and homogenized. Chromatin from tissues was cross-linked with 1.5% formaldehyde, followed by MNase digestion at 37°C for 20 minutes, and fragmented with 3 sets of-20 second pulses using the Virsonic 100 Ultrasonic sonicator. The rest of the immunoprecipitation steps was performed following the protocol described above. JARID2-immunoenriched DNA from both in vitro and in vivo ChIP was amplified for the Notch1+1150 locus by SYBR Green–based quantitative PCR. Fold enrichment was calculated by 2–ΔΔCt methods, with comparison between IgG mock control and either H3K27ac or JARID2. Supplemental Table 20 contains oligonucleotide sequences encompassing Notch1 and Nos3 loci.
Jarid2 siRNA.
Predesigned siRNA targeting the coding region of murine Jumonji (sc-60873, Santa Cruz Biotechnology) was used to knockdown Jarid2. This siRNA consists of pools of 3–5 target-specific 19- to 25-nt siRNAs designed to silence gene expression. A control siRNA-A (sc-37007, Santa Cruz Biotechnology) was used as scramble siRNA, targeting no known gene. AVM cells maintained in NG and HG at 60%–70% confluency were transfected with 10 nM Jarid2 or control siRNA using Lipofectamine 3000 Transfection Reagent (L3000015, ThermoFisher Scientific). Cell lysates were collected and examined for JARID2 and N1ICD protein expression 72 hours after transfection.
Drosophila Studies.
Maternal WT flies were maintained on nondiabetic (standard fly food) or diabetic (standard fly food with 0.2 g/ml sucrose) diets. Notch1 was downregulated either by overexpressing Jarid2 (Jarid2 OE) and Numb (Numb OE) or by silencing Notch with RNAi. Jarid2 was also knocked down using a silencing transgene (Jarid2 RNAi). Progeny flies expressing indicated upstream activator sequence transgenes under the control of cardiac cell–specific 4XHand-Gal4 driver were compared with WT control flies. Progeny flies of indicated genotypes were reared and maintained on nondiabetic standard fly food, as a function of mothers reared and maintained on nondiabetic versus diabetic diet. Glucose and trehalose levels in hemolymph of adult flies were measured. Developmental lethality was presented as the percentage of progeny flies expressing the indicated transgene that fail to emerge as adults, relative to sibling flies not expressing transgene. Survival of adult progenies of diabetic versus nondiabetic mothers with WT and transgenes expressing Jarid2 OE, Numb OE, Notch RNAi, and Jarid2 RNAi were compared. 50 male flies of each genotype were transferred within one day of emergence to vials containing nondiabetic standard fly food. Surviving flies were counted every 2 days. Histological analysis was performed on adult fly hearts in progeny of nondiabetic and diabetic mothers. Cardiomyocyte muscle fiber actin was visualized with phalloidin. Somatic muscle fibers on either side of the heart tube were stained. Cardiomyocytes and pericardial cells were visualized by nuclear-localized GFP expression driven by Hand gene promoter (Hand-GFP). Pericardin was stained and visualized by immunofluorescence. Pericardin produced by cells of the fat body on either side of the heart tube was antibody labeled.
Statistics.
Data are presented as either average ± SD or mean ± SEM. Two-tailed Student’s t test and Fisher’s exact test (for categorical data) were performed to determine statistical significance using the GraphPad Prism 7 software package. Holm-Bonferroni correction was applied to t test P values for Figures 1, 3, 4, and 5 to determine correction for multiplicity of testing using SAS v.9.4. P values of less than 0.05 were considered statistically significant.
Study approval.
This study was approved by the Institutional Animal Care and Use Committee at NCH (protocol AR13-00056) and conducted in accordance with the NIH’s Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011).
Author contributions
VG and MB conceived the study and experimental design. MB performed and analyzed the experiments. KJ provided the AVM cell line. JYZ and ZH designed and performed experiments in Drosophila, with input from MB and VG. ATAC-seq bioinformatic analyses, with input from Epigentek Inc., was performed by SL. UM performed NO donor and quencher experiments in AVM cells. MB and VG wrote the manuscript with input from coauthors.
Supplementary Material
Acknowledgments
The authors thank members of the Biomorphology Core at NCH for histology support. We are grateful to N. Huang, D. Bauer, and J. Feller for technical assistance; E. Alexy for biostatistical analysis; and T. Vondriska, B. Lilly, and D. Srivastava for helpful comments and critical review of the manuscript. MB was supported by the NCH Postdoctoral Idea Award and an American Heart Association Postdoctoral Fellowship (15POST25510006). ZH is supported by grants from NIH (R01-HL090801; R01-NK098410), and VG is supported by funding from NIH (R01-HL121797; R01-HL109758) and NCH.
Version 1. 10/19/2017
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(20):e95085. https://doi.org/10.1172/jci.insight.95085.
Contributor Information
Madhumita Basu, Email: madhumita.basu@nationwidechildrens.org.
Stephanie LaHaye, Email: Stephanie.LaHaye@nationwidechildrens.org.
Uddalak Majumdar, Email: uddalak.majumdar@nationwidechildrens.org.
Kai Jiao, Email: kjiao@uab.edu.
Zhe Han, Email: ZHan@childrensnational.org.
Vidu Garg, Email: vidu.garg@nationwidechildrens.org.
References
- 1. Christianson A, Howson CP, Modell CB. March of Dimes Global Report on Birth Defects: The Hidden Toll of Dying and Disabled Children. March of Dimes. http://www.marchofdimes.org/global-report-on-birth-defects-the-hidden-toll-of-dying-and-disabled-children-full-report.pdf Accessed September 22, 2017.
- 2.Mozaffarian D, et al. Heart disease and stroke statistics--2015 update: a report from the American Heart Association. Circulation. 2015;131(4):e29–322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39(12):1890–1900. doi: 10.1016/S0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 4.Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126(6):1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Gelb BD, Chung WK. Complex genetics and the etiology of human congenital heart disease. Cold Spring Harb Perspect Med. 2014;4(7):a013953. doi: 10.1101/cshperspect.a013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nora JJ. Multifactorial inheritance hypothesis for the etiology of congenital heart diseases. The genetic-environmental interaction. Circulation. 1968;38(3):604–617. doi: 10.1161/01.CIR.38.3.604. [DOI] [PubMed] [Google Scholar]
- 7.Zaidi S, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498(7453):220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Homsy J, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350(6265):1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sifrim A, et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet. 2016;48(9):1060–1065. doi: 10.1038/ng.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fahed AC, Gelb BD, Seidman JG, Seidman CE. Genetics of congenital heart disease: the glass half empty. Circ Res. 2013;112(4):707–720. doi: 10.1161/CIRCRESAHA.112.300853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azhar M, Ware SM. Genetic and developmental basis of cardiovascular malformations. Clin Perinatol. 2016;43(1):39–53. doi: 10.1016/j.clp.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Priest JR, et al. De novo and rare variants at multiple loci support the oligogenic origins of atrioventricular septal heart defects. PLoS Genet. 2016;12(4):e1005963. doi: 10.1371/journal.pgen.1005963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, et al. The complex genetics of hypoplastic left heart syndrome. Nat Genet. 2017;49(7):1152–1159. doi: 10.1038/ng.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel SS, Burns TL. Nongenetic risk factors and congenital heart defects. Pediatr Cardiol. 2013;34(7):1535–1555. doi: 10.1007/s00246-013-0775-4. [DOI] [PubMed] [Google Scholar]
- 15.Zhu H, Kartiko S, Finnell RH. Importance of gene-environment interactions in the etiology of selected birth defects. Clin Genet. 2009;75(5):409–423. doi: 10.1111/j.1399-0004.2009.01174.x. [DOI] [PubMed] [Google Scholar]
- 16.Ferencz C, Loffredo CA, Correa-Villasenor, Wilson PD. Genetic and Environmental Risk Factors of Major Cardiovascular Malformations, The Baltimore Washington Infant Study, (1981-1989) Perspectives in Pediatric Cardiology. 1993:5 [Google Scholar]
- 17.Øyen N, et al. Prepregnancy Diabetes and Offspring Risk of Congenital Heart Disease: A Nationwide Cohort Study. Circulation. 2016;133(23):2243–2253. doi: 10.1161/CIRCULATIONAHA.115.017465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ornoy A, Reece EA, Pavlinkova G, Kappen C, Miller RK. Effect of maternal diabetes on the embryo, fetus, and children: congenital anomalies, genetic and epigenetic changes and developmental outcomes. Birth Defects Res C Embryo Today. 2015;105(1):53–72. doi: 10.1002/bdrc.21090. [DOI] [PubMed] [Google Scholar]
- 19.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 Suppl:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 20.van Weerd JH, Koshiba-Takeuchi K, Kwon C, Takeuchi JK. Epigenetic factors and cardiac development. Cardiovasc Res. 2011;91(2):203–211. doi: 10.1093/cvr/cvr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meier K, Brehm A. Chromatin regulation: how complex does it get? Epigenetics. 2014;9(11):1485–1495. doi: 10.4161/15592294.2014.971580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper ME, El-Osta A. Epigenetics: mechanisms and implications for diabetic complications. Circ Res. 2010;107(12):1403–1413. doi: 10.1161/CIRCRESAHA.110.223552. [DOI] [PubMed] [Google Scholar]
- 23.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creager MA, Lüscher TF, Cosentino F, Beckman JA. Diabetes and vascular disease: pathophysiology, clinical consequences, and medical therapy: Part I. Circulation. 2003;108(12):1527–1532. doi: 10.1161/01.CIR.0000091257.27563.32. [DOI] [PubMed] [Google Scholar]
- 26.Cai H, Harrison DG. Endothelial dysfunction in cardiovascular diseases: the role of oxidant stress. Circ Res. 2000;87(10):840–844. doi: 10.1161/01.RES.87.10.840. [DOI] [PubMed] [Google Scholar]
- 27.Giacco F, Brownlee M. Oxidative stress and diabetic complications. Circ Res. 2010;107(9):1058–1070. doi: 10.1161/CIRCRESAHA.110.223545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paneni F, Beckman JA, Creager MA, Cosentino F. Diabetes and vascular disease: pathophysiology, clinical consequences, and medical therapy: part I. Eur Heart J. 2013;34(31):2436–2443. doi: 10.1093/eurheartj/eht149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hickok JR, Vasudevan D, Antholine WE, Thomas DD. Nitric oxide modifies global histone methylation by inhibiting Jumonji C domain-containing demethylases. J Biol Chem. 2013;288(22):16004–16015. doi: 10.1074/jbc.M112.432294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vasudevan D, et al. Nitric oxide regulates gene expression in cancers by controlling histone posttranslational modifications. Cancer Res. 2015;75(24):5299–5308. doi: 10.1158/0008-5472.CAN-15-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng Q, et al. Development of heart failure and congenital septal defects in mice lacking endothelial nitric oxide synthase. Circulation. 2002;106(7):873–879. doi: 10.1161/01.CIR.0000024114.82981.EA. [DOI] [PubMed] [Google Scholar]
- 32.Lee TC, Zhao YD, Courtman DW, Stewart DJ. Abnormal aortic valve development in mice lacking endothelial nitric oxide synthase. Circulation. 2000;101(20):2345–2348. doi: 10.1161/01.CIR.101.20.2345. [DOI] [PubMed] [Google Scholar]
- 33.Bosse K, et al. Endothelial nitric oxide signaling regulates Notch1 in aortic valve disease. J Mol Cell Cardiol. 2013;60:27–35. doi: 10.1016/j.yjmcc.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koenig SN, Bosse K, Majumdar U, Bonachea EM, Radtke F, Garg V. Endothelial Notch1 Is Required for Proper Development of the Semilunar Valves and Cardiac Outflow Tract. J Am Heart Assoc. 2016;5(4):4. doi: 10.1161/JAHA.115.003075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de la Pompa JL, Epstein JA. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell. 2012;22(2):244–254. doi: 10.1016/j.devcel.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garg V, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437(7056):270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 38.Kerstjens-Frederikse WS, et al. Cardiovascular malformations caused by NOTCH1 mutations do not keep left: data on 428 probands with left-sided CHD and their families. Genet Med. 2016;18(9):914–923. doi: 10.1038/gim.2015.193. [DOI] [PubMed] [Google Scholar]
- 39.Theodoris CV, et al. Long telomeres protect against age-dependent cardiac disease caused by NOTCH1 haploinsufficiency. J Clin Invest. 2017;127(5):1683–1688. doi: 10.1172/JCI90338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng Y, et al. Sema6D acts downstream of bone morphogenetic protein signalling to promote atrioventricular cushion development in mice. Cardiovasc Res. 2016;112(2):532–542. doi: 10.1093/cvr/cvw200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leiter EH, Schile A. Genetic and pharmacologic models for type 1 diabetes. Curr Protoc Mouse Biol. 2013;3(1):9–19. doi: 10.1002/9780470942390.mo120154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Val S, Black BL. Transcriptional control of endothelial cell development. Dev Cell. 2009;16(2):180–195. doi: 10.1016/j.devcel.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atkins GB, Jain MK. Role of Krüppel-like transcription factors in endothelial biology. Circ Res. 2007;100(12):1686–1695. doi: 10.1161/01.RES.0000267856.00713.0a. [DOI] [PubMed] [Google Scholar]
- 44.Patti ME, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci USA. 2003;100(14):8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar S, et al. Hydrogen peroxide decreases endothelial nitric oxide synthase promoter activity through the inhibition of Sp1 activity. DNA Cell Biol. 2009;28(3):119–129. doi: 10.1089/dna.2008.0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sangwung P, et al. KLF2 and KLF4 control endothelial identity and vascular integrity. JCI Insight. 2017;2(4):e91700. doi: 10.1172/jci.insight.91700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afanas’ev I. Signaling of reactive oxygen and nitrogen species in Diabetes mellitus. Oxid Med Cell Longev. 2010;3(6):361–373. doi: 10.4161/oxim.3.6.14415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pasini D, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464(7286):306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 49.Kim TG, Chen J, Sadoshima J, Lee Y. Jumonji represses atrial natriuretic factor gene expression by inhibiting transcriptional activities of cardiac transcription factors. Mol Cell Biol. 2004;24(23):10151–10160. doi: 10.1128/MCB.24.23.10151-10160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mysliwiec MR, Bresnick EH, Lee Y. Endothelial Jarid2/Jumonji is required for normal cardiac development and proper Notch1 expression. J Biol Chem. 2011;286(19):17193–17204. doi: 10.1074/jbc.M110.205146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mysliwiec MR, Carlson CD, Tietjen J, Hung H, Ansari AZ, Lee Y. Jarid2 (Jumonji, AT rich interactive domain 2) regulates NOTCH1 expression via histone modification in the developing heart. J Biol Chem. 2012;287(2):1235–1241. doi: 10.1074/jbc.M111.315945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Na J, et al. A Drosophila model of high sugar diet-induced cardiomyopathy. PLoS Genet. 2013;9(1):e1003175. doi: 10.1371/journal.pgen.1003175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sookoian S, Gianotti TF, Burgueño AL, Pirola CJ. Fetal metabolic programming and epigenetic modifications: a systems biology approach. Pediatr Res. 2013;73(4 Pt 2):531–542. doi: 10.1038/pr.2013.2. [DOI] [PubMed] [Google Scholar]
- 54.Nakajima K, et al. Coordinated regulation of differentiation and proliferation of embryonic cardiomyocytes by a jumonji (Jarid2)-cyclin D1 pathway. Development. 2011;138(9):1771–1782. doi: 10.1242/dev.059295. [DOI] [PubMed] [Google Scholar]
- 55.Barth JL, et al. Jarid2 is among a set of genes differentially regulated by Nkx2.5 during outflow tract morphogenesis. Dev Dyn. 2010;239(7):2024–2033. doi: 10.1002/dvdy.22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang AC, et al. Notch initiates the endothelial-to-mesenchymal transition in the atrioventricular canal through autocrine activation of soluble guanylyl cyclase. Dev Cell. 2011;21(2):288–300. doi: 10.1016/j.devcel.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 57.Tai SC, Robb GB, Marsden PA. Endothelial nitric oxide synthase: a new paradigm for gene regulation in the injured blood vessel. Arterioscler Thromb Vasc Biol. 2004;24(3):405–412. doi: 10.1161/01.ATV.0000109171.50229.33. [DOI] [PubMed] [Google Scholar]
- 58.Matouk CC, Marsden PA. Epigenetic regulation of vascular endothelial gene expression. Circ Res. 2008;102(8):873–887. doi: 10.1161/CIRCRESAHA.107.171025. [DOI] [PubMed] [Google Scholar]
- 59.Krause BJ, Costello PM, Muñoz-Urrutia E, Lillycrop KA, Hanson MA, Casanello P. Role of DNA methyltransferase 1 on the altered eNOS expression in human umbilical endothelium from intrauterine growth restricted fetuses. Epigenetics. 2013;8(9):944–952. doi: 10.4161/epi.25579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiplunkar AR, et al. The Krüppel-like factor 2 and Krüppel-like factor 4 genes interact to maintain endothelial integrity in mouse embryonic vasculogenesis. BMC Dev Biol. 2013;13:40. doi: 10.1186/1471-213X-13-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bai Y, Wang J, Morikawa Y, Bonilla-Claudio M, Klysik E, Martin JF. Bmp signaling represses Vegfa to promote outflow tract cushion development. Development. 2013;140(16):3395–3402. doi: 10.1242/dev.097360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang B, et al. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development. 2004;131(18):4477–4487. doi: 10.1242/dev.01287. [DOI] [PubMed] [Google Scholar]
- 63.Jin Y, et al. Effects of oxidative stress on hyperglycaemia-induced brain malformations in a diabetes mouse model. Exp Cell Res. 2016;347(1):201–211. doi: 10.1016/j.yexcr.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 64.Pavlinkova G, Salbaum JM, Kappen C. Maternal diabetes alters transcriptional programs in the developing embryo. BMC Genomics. 2009;10:274. doi: 10.1186/1471-2164-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang SW, et al. Genetic abnormalities in FOXP1 are associated with congenital heart defects. Hum Mutat. 2013;34(9):1226–1230. doi: 10.1002/humu.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.López-Escobar B, et al. The effect of maternal diabetes on the Wnt-PCP pathway during embryogenesis as reflected in the developing mouse eye. Dis Model Mech. 2015;8(2):157–168. doi: 10.1242/dmm.017723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hien TT, et al. Elevated glucose levels promote contractile and cytoskeletal gene expression in vascular smooth muscle via Rho/protein kinase c and actin polymerization. J Biol Chem. 2016;291(7):3552–3568. doi: 10.1074/jbc.M115.654384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang F, Fisher SA, Zhong J, Wu Y, Yang P. Superoxide dismutase 1 in vivo ameliorates maternal diabetes mellitus-induced apoptosis and heart defects through restoration of impaired Wnt signaling. Circ Cardiovasc Genet. 2015;8(5):665–676. doi: 10.1161/CIRCGENETICS.115.001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moazzen H, et al. N-Acetylcysteine prevents congenital heart defects induced by pregestational diabetes. Cardiovasc Diabetol. 2014;13:46. doi: 10.1186/1475-2840-13-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kitzmiller JL, Wallerstein R, Correa A, Kwan S. Preconception care for women with diabetes and prevention of major congenital malformations. Birth Defects Res Part A Clin Mol Teratol. 2010;88(10):791–803. doi: 10.1002/bdra.20734. [DOI] [PubMed] [Google Scholar]
- 71. Leslie RDG, John PN, Pyke DA, White JM. Hemoglobin A1 in diabetic pregnancy. Lancet. 1978;2(8097):958–959. https://www.sciencedirect.com/science/article/pii/S0140673678925254.
- 72.Miller E, et al. Elevated maternal hemoglobin A1c in early pregnancy and major congenital anomalies in infants of diabetic mothers. N Engl J Med. 1981;304(22):1331–1334. doi: 10.1056/NEJM198105283042204. [DOI] [PubMed] [Google Scholar]
- 73.Priest JR, Yang W, Reaven G, Knowles JW, Shaw GM. Maternal midpregnancy glucose levels and risk of congenital heart disease in offspring. JAMA Pediatr. 2015;169(12):1112–1116. doi: 10.1001/jamapediatrics.2015.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumar SD, Dheen ST, Tay SS. Maternal diabetes induces congenital heart defects in mice by altering the expression of genes involved in cardiovascular development. Cardiovasc Diabetol. 2007;6:34. doi: 10.1186/1475-2840-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ross-Innes CS, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481(7381):389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu LJ, et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag New York:2009. [Google Scholar]
- 79.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 80.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Walter W, Sánchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31(17):2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
- 82.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kennedy AJ, Ellacott KL, King VL, Hasty AH. Mouse models of the metabolic syndrome. Dis Model Mech. 2010;3(3-4):156–166. doi: 10.1242/dmm.003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.