Abstract
Eosinophilic esophagitis (EoE) is an allergic inflammatory disease of the esophagus mediated by an IL-13–driven epithelial cell transcriptional program. Herein, we show that the cytoskeletal protein synaptopodin (SYNPO), previously associated with podocytes, is constitutively expressed in esophageal epithelium and induced during allergic inflammation. In addition, we show that the SYNPO gene is transcriptionally and epigenetically regulated by IL-13 in esophageal epithelial cells. SYNPO was expressed in the basal layer of homeostatic esophageal epithelium, colocalized with actin filaments, and expanded into the suprabasal epithelium in EoE patients, where expression was elevated 25-fold compared with control individuals. The expression level of SYNPO in esophageal biopsies correlated with esophageal eosinophil density and was improved following anti–IL-13 treatment in EoE patients. In esophageal epithelial cells, SYNPO gene silencing reduced epithelial motility in a wound healing model, whereas SYNPO overexpression impaired epithelial barrier integrity and reduced esophageal differentiation. Taken together, we demonstrate that SYNPO is induced by IL-13 in vitro and in vivo, is a nonredundant regulator of epithelial cell barrier function and motility, and is likely involved in EoE pathogenesis.
Keywords: Gastroenterology, Inflammation
Actin-associated protein synaptopodin is a nonredundant regulator of epithelial cell barrier function and motility,and is likely involved in the pathogenesis of eosinophilic esophagitis.
Introduction
The hallmark of allergic inflammation is a robust epithelial response of the inflamed tissue to the Th2 cell–derived cytokines IL-4 and IL-13 (1–3). Binding of these proatopy cytokines with their common receptor IL-4 receptor α (IL-4Rα) on epithelial cells of multiple origin, including skin, nasal, lung, esophageal, and intestinal epithelium, causes phosphorylation of the STAT6 and initiates transcriptional programs that lead to hallmark features of allergic inflammation (4, 5).
Eosinophilic esophagitis (EoE) is a chronic inflammatory disorder characterized by allergic hypersensitivity to food and eosinophilic infiltration in the esophageal mucosa (6). EoE pathogenesis is driven by IL-13–mediated transcriptional responses of the esophageal epithelium (7, 8), which are collectively improved by steroids (9). Structural and transcriptional changes observed in the inflamed epithelium in biopsies from patients with EoE can be recapitulated in vitro by treating esophageal epithelial cells with IL-13 (10–13). IL-13 is thought to mediate these responses through epigenetic changes, including elevated levels of histone acetylation and decreased DNA methylation in the promoters of critical mediators, such as eotaxin 3 (also known as CCL26), CAPN14, and NTRK1 (14–16). Co-occurrence of epigenetic changes with transcriptional response suggests that integrating transcriptional and epigenetic analyses may serve as a useful approach to discover molecules with functional roles in EoE pathogenesis, especially since EoE is influenced by gene-environment interactions (17).
Herein, we report that synaptopodin (SYNPO), a proline-rich, actin-associated protein, primarily studied in the context of podocytes (18–20), is an esophageal epithelial protein associated with the actin cytoskeleton and regulated by IL-13. SYNPO is induced by IL-13 in esophageal cells in a STAT6-dependent manner and epigenetically modified in the promoter region in response to IL-13, and its increased expression in EoE biopsies is normalized following treatment with humanized anti–IL-13 therapy. Experimental modulation of SYNPO expression in esophageal epithelial cells leads to decreased epithelial motility in a wound healing assay and decreased transepithelial resistance, increased FITC-dextran flux, and loss of esophageal differentiation. We propose that the actin-associated protein SYNPO is an esophageal epithelial gene product involved in Th2-associated allergic responses, including epithelial cell motility and barrier integrity.
Results
SYNPO is transcriptionally and epigenetically regulated by IL-13 in esophageal epithelial cells.
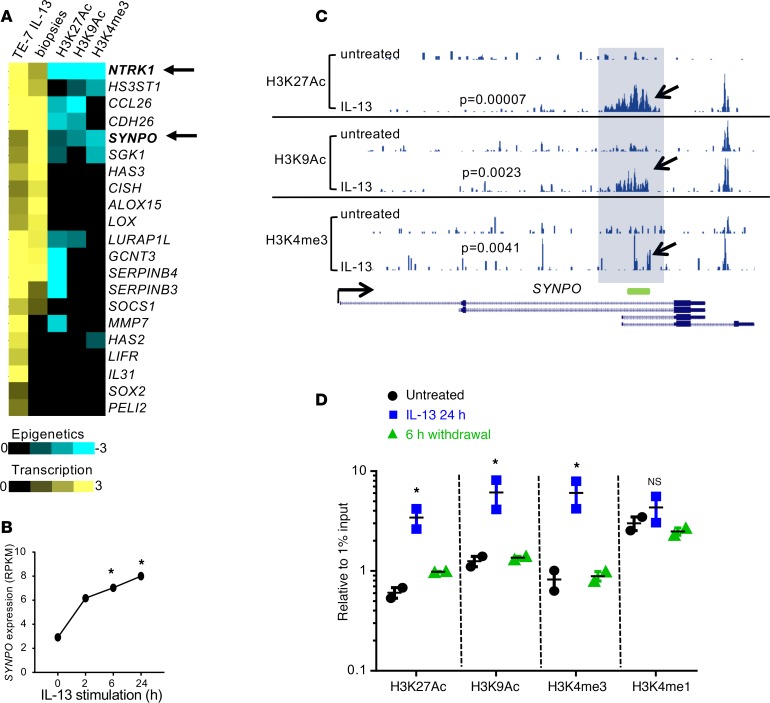
By combining transcriptional and epigenetic analyses of esophageal epithelial cells stimulated by IL-13 (Gene Expression Omnibus database [GEO] GSE57637), we aimed to identify molecules with potential functional roles in EoE pathogenesis. We focused on the genes that were induced by IL-13 (at 2, 6, and 24 hours; differential expression determined by DESeq analysis, P < 0.05) and had significantly elevated levels of activating epigenetic marks in their promoters (histone H3 lysine 9 acetylation [H3K9Ac], histone H3 lysine 27 acetylation [H3K27Ac], histone H3 lysine 4 trimethylation [H3K4me3]; MAnorm, P < 0.01). With these criteria, we initially identified 21 genes that were induced by IL-13 at least 2-fold at all tested time points (Figure 1A, TE-7 IL-13 column); of these 21 genes, 15 genes were significantly upregulated in EoE biopsies compared with healthy controls (Figure 1A, biopsies column). Ten of the fifteen genes were also significantly epigenetically modified, as was evident from elevated levels of activating epigenetic marks in their promoters (Figure 1A, last 3 columns). Several known IL-13 target genes were identified, including NTRK1, CCL26, CDH26, SERPINB3, and SERPINB4 (7, 15, 21). Notably, epigenetically modified IL-13 target genes commonly had one or two activating epigenetic marks elevated in their promoter regions. For example, SERPINB3 and SERPINB4 had elevated H3K27Ac, whereas CCL26 and CDH26 showed increases in both H3K27Ac and H3K9Ac. The only two genes that were transcriptionally upregulated by IL-13 and had three epigenetic marks elevated in their promoters were NTRK1 (consistent with our previous report, ref. 15) and SYNPO (arrows, Figure 1A). Kinetic analysis showed that SYNPO expression was increased after treatment with IL-13 over a 24-hour period (Figure 1B) and the SYNPO promoter region was epigenetically modified (Figure 1C, compare untreated tracks with IL-13–treated tracks). Accordingly, increased levels of the activating epigenetic marks H3K27Ac, H3K9Ac, and H3K4me3 were detected by ChIP-PCR analysis, but histone H3 lysine 4 monomethylation (H3K4mel), which marks inactive enhancers (22), was not increased. The epigenetic changes were highly dynamic, as they were fully normalized after 6 hours of IL-13 withdrawal (Figure 1D).
Figure 1. Transcriptional and epigenetic regulation of SYNPO in TE-7 cells.
(A) The heatmap shows transcriptional and epigenetic changes in TE-7 cells in response to IL-13 stimulation for 6 hours or eosinophilic esophagitis (EoE) biopsies compared with controls. The yellow color represents log2 fold change in gene expression compared with controls, and the blue color represents M values for the indicated histone mark (note that negative M value indicates increased level of epigenetic marks; see Methods for details). (B) The expression of SYNPO in TE-7 cells stimulated with IL-13 at 100 ng/ml is shown (data are from ref. 15). *P < 0.05, DESeq analysis. RPKM, reads per kilobase per million reads. (C) An image capture from the USCS browser shows the epigenetic landscape for H3K27Ac, H3K9Ac, and H3K4me3 marks in TE-7 cells in the SYNPO gene. The promoter area of SYNPO isoforms 1 and 2 (see Figure 2A for details) is shaded. Arrows point to the histone mark peaks that are significantly increased after 6 hours of IL-13 stimulation. P values comparing IL-13–stimulated cells with untreated cells were calculated by using a MAnorm algorithm. The green rectangle shows the position of the probe used for ChIP-PCR experiments. The read depths of the aligned sequences are of the same scale. (D) The level of the indicated epigenetic marks was assessed in the promoter of SYNPO isoforms 1 and 2 by ChIP-PCR after a 24-hour IL-13 stimulation and subsequent IL-13 withdrawal for 6 hours. One of two independent experiments is shown; the dots represent the mean ± SD for technical replicates. *P < 0.05, compared with untreated cells by ANOVA.
SYNPO expression in an esophageal cell line and primary esophageal cells.
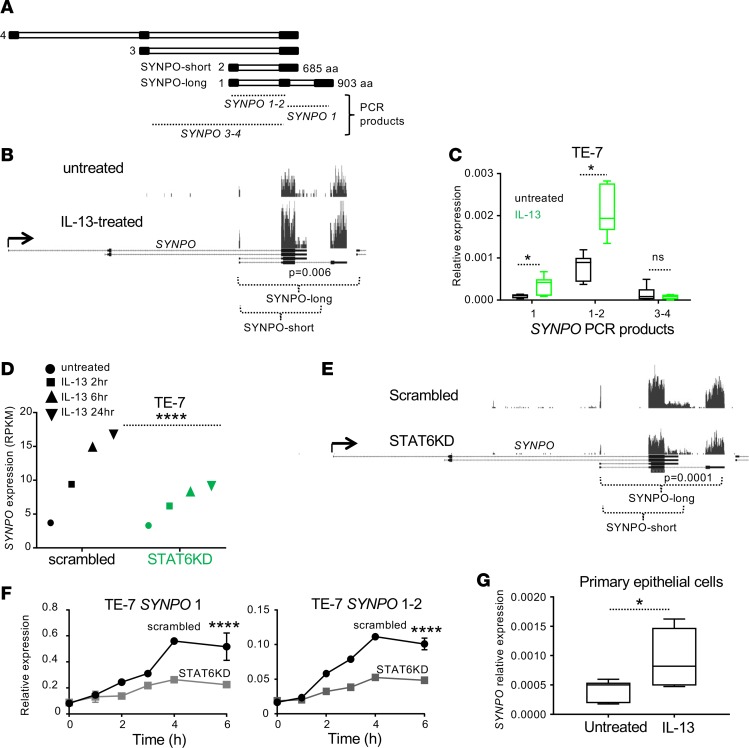
SYNPO is expressed as 4 isoforms (Figure 2A), with isoforms 1 and 2 (SYNPO-long and SYNPO-short, respectively) being most commonly expressed (20, 23). Epigenetic analysis suggested that the long and short isoforms were expressed in esophageal TE-7 cells and induced by IL-13 (Figure 1C, epigenetic mark locations). Distribution of the aligned reads from RNA sequencing (Figure 2B), as well as assessment of the expression of SYNPO isoforms by RT-PCR in TE-7 cells (Figure 2C), validated this hypothesis. Indeed, SYNPO was expressed as long and short isoforms (1 and 2, respectively) that were upregulated by IL-13, whereas isoforms 3 and 4 were expressed at low levels and did not respond to IL-13 (Figure 2C). IL-13 is a primary activator of the STAT6 signaling pathway (24). To test whether SYNPO induction by IL-13 is mediated through STAT6, we assessed kinetics of SYNPO expression in esophageal epithelial cells after IL-13 stimulation following STAT6 knockdown (STAT6KD) (15). Expression of both the long and short SYNPO isoforms was substantially downregulated in STAT6KD cells, as evident from the decreased expression of SYNPO in RNA-sequencing and RT-PCR analyses (Figure 2, D–F). We further tested SYNPO expression in primary epithelial cells isolated from esophageal biopsies and demonstrated that SYNPO was also increased in these cells in response to IL-13 stimulation (Figure 2G).
Figure 2. SYNPO expression in esophageal epithelial cells.
(A) Schematic structure of the SYNPO isoforms; black and white rectangles represent exons and introns, respectively. SYNPO-long and SYNPO-short represent the common protein isoforms 1 and 2, respectively. The SYNPO regions detected by RT-PCR are indicated by dotted lines; SYNPO 1–2 and SYNPO 3–4 represent common PCR products for indicated isoforms. (B) An image capture from the USCS browser shows aligned RNA-sequencing reads on the SYNPO gene in TE-7 cells either untreated or treated with IL-13 for 24 hours. Black rectangles represent exons. The indicated P value comparing expression under these conditions was calculated by DESeq analysis (59). Note that reads are detected only on the exons corresponding to SYNPO-long and SYNPO-short isoforms. (C) Expression of SYNPO isoforms by TE-7 cells induced by IL-13 was assessed by RT-PCR. *P < 0.05, t test. (D) Expression of SYNPO was assessed by RNA sequencing in TE-7 cells subjected to gene silencing control (scrambled) and STAT6 knockdown (STAT6KD) pool. SYNPO expression in RPKM is shown. ****P < 0.0001 for 24-hour time point, DESeq analysis. (E) An image capture from the USCS browser shows aligned RNA-sequencing reads on the SYNPO gene in control TE-7 cells and STAT6KD pool from D. Note that both isoforms are affected by the knockdown. The indicated P value comparing expression under these conditions was calculated by DESeq analysis (59). (F) Expression of SYNPO isoforms (mean ± SD) was assessed by RT-PCR. ****P < 0.001, Holm-Sidak method. (G) Expression of SYNPO was analyzed in primary esophageal epithelial cells stimulated with IL-13. Combined data from cells isolated from 3 independent biopsies are shown. *P < 0.05, t test. For C, F, and G, expression was normalized to expression of GAPDH. For box-and-whisker plots, the box represents 50th percentile of the data, whiskers show minimum and maximum values, and the line in the box represents the median. For B and E, the read depth is of the same scale within each panel.
SYNPO is an actin-associated protein in esophageal epithelial cells.
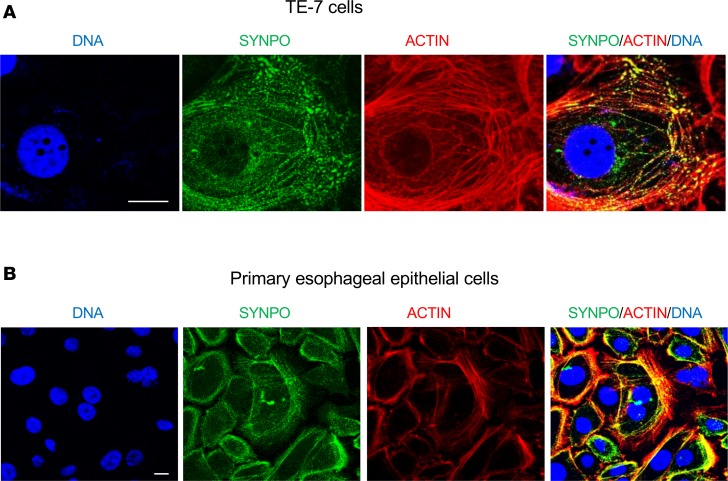
In podocytes, SYNPO is an actin-associated protein localized to the cytoskeleton (25). We tested whether SYNPO in esophageal epithelial cells possesses similar properties. By immunofluorescence staining, we showed that SYNPO was partially colocalized with actin fibers in both TE-7 (Figure 3A) and primary esophageal epithelial cells (Figure 3B). Collectively, our data establish that SYNPO is an actin-associated protein and that it is a direct transcriptional and epigenetic target of IL-13 in esophageal epithelial cells.
Figure 3. SYNPO is an actin-associated protein in esophageal epithelial cells.
(A) Representative immunofluorescence images of SYNPO (green) in the esophageal epithelial TE-7 cell line. (B) Primary esophageal epithelial cells isolated from biopsies. DNA was labeled by Hoechst (blue), and actin was stained with phalloidin (red). The merged image shows partial colocalization of SYNPO and actin (yellow). Original magnification: ×60 (A); ×40 (B); scale bars: 10 μM.
SYNPO is elevated in EoE, correlates with disease severity, and is normalized following humanized anti–IL-13 treatment.
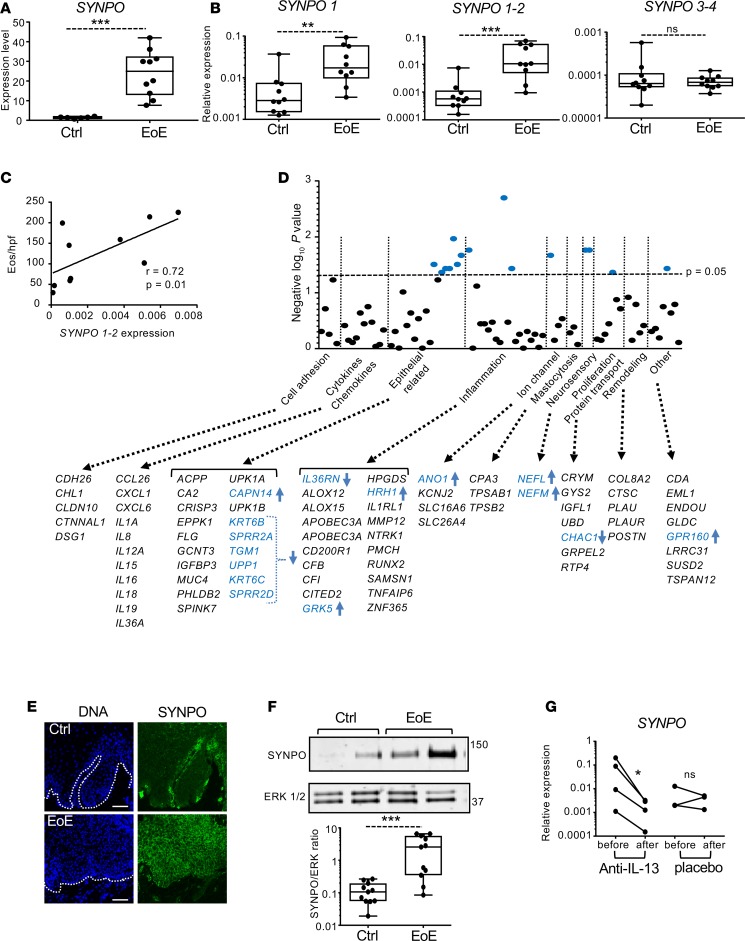
We aimed to characterize SYNPO expression in the esophageal epithelium. RNA-sequencing data from esophageal biopsies demonstrated significantly elevated SYNPO expression in patients with EoE compared with subjects without EoE (Figure 4A and data from ref. 8), and its expression was fully normalized following successful treatment (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.96789DS1 and data from ref 26). Using independent biopsies, we further demonstrated that esophageal SYNPO was expressed as long and short isoforms and that both isoforms were significantly upregulated in EoE (Figure 4B). Isoforms 3 and 4 were detected at low levels, and their expression was not modified in EoE (Figure 4B). Moreover, expression of the long and short SYNPO isoforms positively correlated with disease severity, as defined by the number of eosinophils per high-power field in the esophageal epithelium (r = 0.72, P = 0.01, Figure 4C). We further correlated SYNPO expression with expression of other genes from the RNA-sequencing analysis of EoE patients (8). Focusing on gene categories previously used for molecular diagnosis of the disease (27), SYNPO expression correlated primarily with epithelial, inflammatory, and neurosensory molecular markers of EoE, showing mostly negative correlation with epithelial-related genes (Figure 4D and Supplemental Table 1). The gene with the strongest correlation was G protein–coupled receptor kinase 5 (GRK5) (r = 0.9, P = 0.002), a serine/threonine kinase involved in cellular signaling, including NF-κB transcriptional activity (28). Among epithelial-related genes, CAPN14 uniquely positively correlated with SYNPO (r = 0.73, P = 0.03). Immunofluorescence analysis of human esophageal biopsies from patients with and without EoE showed that SYNPO was expressed in the basal epithelial layer in patients without EoE, whereas its expression increased throughout the suprabasal layers of the esophageal epithelium, mirroring basal zone hyperplasia, in patients with EoE (Figure 4E). Additionally, independent analysis by Western blot confirmed that SYNPO protein levels are significantly increased in EoE biopsies compared with controls (Figure 4F; see complete unedited blots in Supplemental Figure 2A). We further assessed the potential role of IL-13 in regulating SYNPO expression in the esophageal biopsies in EoE by comparing SYNPO expression in EoE patients treated with anti–IL-13 antibody QAX576 to that of untreated controls (29). In this study, intravenous injection of QAX576 led to significant improvement of molecular markers of EoE, including CCL26 (29). We found that QAX576 treatment, but not placebo treatment, reduced SYNPO expression in EoE patients (Figure 4G). Collectively, these results provide evidence that SYNPO is substantially upregulated in the inflamed esophageal epithelium in EoE patients as a part of a dysregulated epithelial differentiation program and that this pathway is IL-13 dependent in humans, at least in part.
Figure 4. Expression of SYNPO in esophageal biopsies and correlation with EoE transcriptome.
(A) The normalized expression level for SYNPO in esophageal biopsy tissue from patients with active EoE (n = 10) and control patients (n = 6) (8). ***P < 0.001, t test. (B) The relative expression level of SYNPO isoforms (see Figure 2A) in esophageal biopsies from patients with active EoE and without EoE (Ctrl) was quantified by measuring expression of SYNPO PCR products. For A–C, each dot represents an esophageal biopsy from independent patients. (A and B) **P < 0.01, ***P < 0.001, Holm-Sidak method. (C) The correlation between the expression of SYNPO (PCR product 1–2) and the number of eosinophils per high-power field (Eos/hpf) in an EoE esophageal biopsies. The Spearman correlation (r) and P value are indicated. (D) The graph shows correlation of SYNPO expression with EoE molecular signature genes from RNA-sequencing analysis of EoE patients (8). Gene categories were based on those used for the molecular diagnostics panel for EoE (27). The significance of the correlation is plotted as a negative log(10) P value. The horizontal dashed line represents P = 0.05. The genes probed in each category are indicated; significantly correlated genes are in blue font. Arrows indicate positive (up) and negative (down) correlation. (E) Representative immunofluorescence of SYNPO in esophageal biopsies from independent patients. The white dotted line demarcates the basement membrane. Scale bar: 100 μM. (F) A representative Western blot for SYNPO in control and EoE biopsies; ERK 1/2 is used as a loading control. Quantification of SYNPO expression was performed in control and EoE biopsies from independent patients (n = 11 per each group). ***P < 0.001, Holm-Sidak method. (G) Relative expression level of SYNPO was assessed in the EoE patients either treated with anti–IL-13 antibody or placebo, as described in ref. 29. “Before” corresponds to the beginning of the study, and “after” corresponds to day 85 of the study. *P < 0.05, t test. For A, B, and G expression level was normalized to GAPDH. For box-and-whisker plots, the box represents 50th percentile of the data, whiskers show minimum and maximum values, and the line in the box represents the median.
Effects of altered SYNPO expression in esophageal epithelial cells.
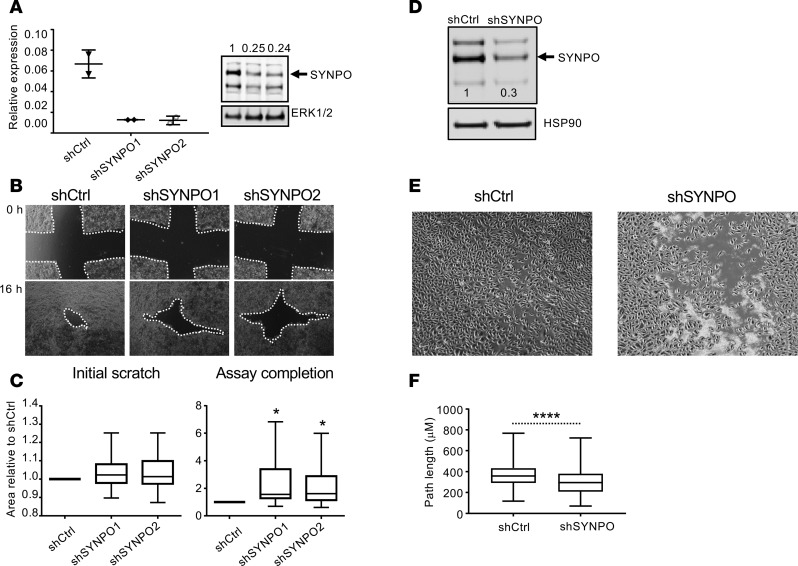
In the light of the significant correlation of SYNPO expression with genes related to epithelial differentiation, we aimed to explore the functional role of SYNPO in esophageal epithelium. SYNPO has a role in cellular migration in normal kidney podocytes (18), and its downregulation reduces endothelial cell motility (30). We therefore tested whether downregulation of SYNPO alters migration properties of esophageal epithelial cells. We first gene-silenced SYNPO with two independent shRNAs in TE-7 cells; this stably downregulated SYNPO to approximately 25% of its baseline protein expression level (Figure 5A; see complete unedited blots in Supplemental Figure 2B). We then assessed epithelial cell reconstitution using a wound healing assay (18). We found that TE-7 cells with gene-silenced SYNPO closed the wound slower than control cells (P < 0.05; Figure 5, B and C). Using an independent esophageal cell line, EPC2 cells, the wound closed slower in cells with SYNPO downregulated than in control cells (Figure 5, D and E; see complete unedited blots in Supplemental Figure 2C). By quantifying the overall distance that EPC2 cells traveled after wound induction, we found that SYNPO downregulation significantly decreased the length of the path traveled by the cells comparing SYNPO-downregulated cells and control EPC2 cells (P < 0.0001, Figure 5F).
Figure 5. Effect of SYNPO downregulation on esophageal epithelial cell mobility.
(A) Representative RT-PCR and Western blot of SYNPO after stable SYNPO downregulation by two shRNAs in TE-7 cells. The arrow points to SYNPO-long isoform. Expression of SYNPO normalized to ERK 1/2 and then to control shRNA is indicated; the dots represent the mean ± SD for technical duplicates. (B) A wound healing assay was performed in TE-7 cells. Representative images show the scratch area at the beginning of the experiment (0 hours, initial scratch) and at the endpoint (16 hours after initial scratch, assay completion). The white dotted line indicates the scratch area that was quantified in C. (C) Quantification is shown for 5 independent experiments performed in quadruplicates and normalized to the scratch area in control shRNA (shCtrl) samples at the endpoint (shCtrl assay completion); *P < 0.05, ANOVA. (D) A Western blot for SYNPO after stable SYNPO downregulation in EPC2 cells by shRNA (shSYNPO) is shown; the arrow points to SYNPO-long isoform. The relative level of SYNPO expression normalized to HSP90 and then to shCtrl SYNPO expression is indicated by the numbers on the image. (E) A representative image of the wound healing assay simultaneously performed for control cells and cells with downregulated SYNPO expression (shCtrl and shSYNPO, respectively) is shown for the assay completion (8 hours) time point. Note that the control cells are fully confluent, whereas the shSYNPO cells still have visible gaps. (F) The overall distance traveled by 300 cells in the wound over the course of the wound healing assay for three independent experiments combined are shown; ****P < 0.0001, t test. For box-and-whisker plots, graphs the box represents 50th percentile of the data, whiskers show minimum and maximum values, and the line in the box represents the median. Original magnification, ×10 (B and E).
We subsequently subjected EPC2 cells to growth at the air-liquid interface (ALI), which models esophageal differentiation under normal and pathologic conditions. This approach allowed us to assess the epithelial integrity by measuring transepithelial electrical resistance (TEER), FITC-dextran flux, and expression of key epithelial differentiation markers (10, 11). We experimentally overexpressed SYNPO in EPC2 cells by lentiviral transduction; these cells had approximately 5-fold higher SYNPO-long expression compared with control cells (Figure 6A). Following cellular differentiation by ALI (Figure 6B), there was a loss of the superficial keratinized layer in SYNPO-overexpressing cells (Figure 6C, arrow in control). Measuring TEER revealed a significant decrease in resistance in SYNPO-overexpressing cells (Figure 6D) and increased epithelial permeability to low-molecular-weight dextran compared with control cells (Figure 6E). Moreover, a number of genes linked to esophageal epithelial differentiation — CRNN, KRTs, serine protease inhibitors SPINK5 and SPINK7, and genes from the epidermal differentiation complex (SPRR2E, S100A9) but not involucrin (IVL) — had significantly decreased expression with SYNPO overexpression (Figure 6F). In summary, overexpression of SYNPO in esophageal epithelial cells modifies motility, barrier integrity, and the epithelial cell differentiation program.
Figure 6. Effect of SYNPO overexpression on epithelial differentiation and barrier function.
(A) The SYNPO-long level in overexpressed pools (SYNPO OE a and OE b, arrow) was quantified by Western blot and normalized to HSP90. (B) A schematic outline of the differentiation protocol for EPC2 cells grown at ALI. (C) Representative H&E staining of control (Ctrl) and SYNPO-overexpressing (OE) EPC2 cells grown at the ALI. Arrow points to the keratinized layer of differentiated epithelium. Scale bar: 50 μM. (D and E) The transepithelial resistance (TEER) and FITC-dextran flux measurements are shown for EPC2 cells grown at the ALI. (D) Data were normalized to TEER of control samples at day 4 of culture and represent an average of 3–4 independent experiments performed in triplicate. (E) Data are representative of 2 independent experiments performed with 3 independent ALI cultures. ****P < 0.0001, 1-way ANOVA. (F) The expression level of the indicated genes was assessed by RT-PCR at day 14 of the ALI culture. Expression was normalized to GAPDH and to control samples. The combined data for SYNPO OE pools from 3–4 independent experiments are shown (n = 7 for control cultures, n = 13 for SYNPO OE cultures). ***P < 0.001, ****P < 0.0001, t test. For D–F, the data are presented as mean ± SEM.
Discussion
Herein, we report that the actin-associated protein SYNPO, previously studied in the context of kidney epithelium in renal pathologies, is an esophageal epithelial gene product involved in Th2-associated allergic responses, including esophageal epithelial cell motility and differentiation. By utilizing genome-wide transcriptional and epigenetic analyses of IL-13–stimulated esophageal epithelial cells, we identify SYNPO as a transcriptional and epigenetic target of IL-13. Following stimulation with IL-13, SYNPO expression is induced in esophageal epithelial cells in a STAT6-dependent manner and is accompanied by a broad spectrum of elevated activating epigenetic marks in the promoter of the gene. In the homeostatic esophagus, SYNPO is expressed in the basal layer of the epithelium, and its expression expands into suprabasal epithelial layers in the esophagi of EoE patients compared with controls. In EoE, expression of SYNPO correlates with severity of the disease, as defined by esophageal eosinophil counts, and is normalized following anti–IL-13 treatment of EoE patients or STAT6 knockdown in esophageal epithelial cells. Functionally, we demonstrate that SYNPO gene silencing decreased esophageal epithelial cell motility in a wound healing assay, whereas SYNPO overexpression altered the differentiation and epithelial integrity of esophageal epithelial cells. Significant correlation of SYNPO expression with a subset of epithelial-related genes supports involvement of SYNPO in epithelial differentiation and EoE pathogenesis. Figure 7 summarizes the results reported in this study.
Figure 7. Schematic representation of SYNPO expression and regulation in the esophageal epithelium.
Under normal conditions (NL) esophageal epithelium is characterized by strong intercellular contacts, and barrier integrity is preserved. In the homeostatic esophagus, SYNPO is expressed at the basal layer of the esophageal epithelium and interacts with actin filaments. SYNPO is transcribed at low levels, and its promoter is decorated by low levels of activating epigenetic marks. In EoE, IL-13 is secreted by tissue-resident lymphocytes, causing STAT-6–dependent transcriptional activation of SYNPO in esophageal epithelium accompanied by elevated levels of activating epigenetic marks in the promoter of SYNPO gene. Basal zone hyperplasia and expansion of SYNPO expression into the suprabasal layers of the esophagus are evident. Subsequently, epithelial barrier integrity is altered and eosinophils penetrate esophageal mucosa. Treatment of EoE patients with anti–IL-13 antibody or knocking down STAT6 in esophageal epithelial cells counteracts IL-13–dependent transcriptional and epigenetic changes of SYNPO. Note that cell nuclei are omitted for clarity.
The actin-associated protein SYNPO is localized in the foot processes of kidney podocytes and dendritic spines of telencephalic synapses in brain, where it is associated with the postsynaptic density (25). While regulation of cytoskeletal dynamics by SYNPO is likely critical for function of both organs, SYNPO has been primarily studied in the context of nephropathies (31–33). Idiopathic nephrotic syndrome is a clinical syndrome associated with a variety of kidney pathologies, including minimal change disease (MCD) (34), in which there is effacement of the foot processes of podocytes, leading to loss of epithelial integrity and increased permeability associated with proteinuria (35, 36). Importantly, MCD can be successfully treated by immunosuppressive drugs, suggesting that immunologic responses are critical (23, 37, 38). Numerous reports associate MCD with atopy, as evidenced by elevated expression of IgE and increased prevalence of atopy in patients with MCD compared with control subjects (39–42). In line with these studies, T cells from patients with MCD produce elevated amounts of IL-13 compared with those from control subjects (43–45), which may contribute to the observed IgE in these patients (46, 47). Importantly, podocytes have been shown to respond to IL-13 stimulation via the IL-13 receptor, leading to decreased TEER and increased epithelial permeability (48), which suggests that exposure to IL-13 can cause MCD. Indeed, IL-13 overexpression in a rat model of MCD yields a podocyte gene expression profile that evidences a major loss of expression of podocyte-specific structural and functional genes (49). Notably, exposure to milk is a common trigger for both EoE and nephrotic syndrome, and milk elimination significantly improves symptoms of both diseases, including steroid-resistant idiopathic nephrotic syndrome (50–52). It is interesting to speculate that these findings might unify the pathogeneses of nephrotic syndrome and atopy through cellular and molecular responses to food allergens and IL-13. Additionally, these findings suggest that SYNPO may function as a common IL-13–driven mediator of both disease processes.
Most findings that underscore the critical role of IBF in pathogenesis of EoE have focused on structural epithelial proteins, such as DSG1, or proteases, such as CAPN14 (11, 14, 53). Our findings — that actin-associated proteins are significantly upregulated in EoE, correlate with EoE severity, and alter esophageal epithelial integrity in cultured cells — signify the potential involvement of SYNPO and other actin-binding proteins in the pathogenesis of EoE. Dysregulation of intercellular junctions suggests a mechanism whereby inflamed and hyperproliferative esophageal epithelial tissue can be penetrated by allergens, thereby amplifying allergic inflammation. Accordingly, SYNPO expression is highly correlated with a subset of esophageal-enriched genes involved in epithelial differentiation. Notably, the majority of these genes are downregulated in EoE, except CAPN14, which is considered to be a genetic driver of the disease and is linked to epithelial differentiation (14, 53, 54). These data further support that SYNPO dysregulation is a part of a broader alteration in esophageal epithelial differentiation program in response to IL-13.
In summary, herein, we have shown that SYNPO is upregulated in the esophagi of patients with EoE, that it is normalized by humanized anti–IL-13 treatment and is directly induced by IL-13 by an epigenetic mechanism involving histone acetylation and methylation, and that it regulates epithelial repair responses, including wound healing and barrier function, at least in part. These findings demonstrate a role of the actin-related protein SYNPO in allergic inflammatory responses and specifically in EoE pathogenesis and provide a common pathogenic step, involving IL-13–induced SYNPO-mediated epithelial cell dysfunction, that may be involved in allergic responses and nephrotic syndrome, two diseases processes whose association has remained enigmatic. Mechanistically, SYNPO overexpression impairs epithelial cell differentiation with loss of expression of structural esophageal-specific genes and protease inhibitors associated with differentiated esophageal epithelium. Of interest, IL-13 can mediate its signaling through an NADPH oxidase–dependent pathway (55), and an NADPH oxidase inhibitor has been shown to have activity against atopic dermatitis in humans (56). It would be interesting to assess the level of SYNPO expression in atopic dermatitis and its potential contribution to the pathogenesis of this disease. Collectively, our findings show that SYNPO is upregulated by IL-13 in EoE and has the capacity to regulate multiple esophageal epithelial cell responses, including cellular motility, barrier integrity, and differentiation; as such, it represents a pathogenic regulator of EoE and highlights the potential utility of anti–IL-13 treatment focused on normalizing SYNPO-driven responses.
Methods
Cell culture and treatment.
The squamous esophageal epithelial cell line TE-7 (a gift from Pierre Hainaut, Institute for Advanced Biosciences, INSERM 1209, UMR CNRS 5309, Université Grenoble-Alpes, Grenoble, France), which was originally selected from human esophageal tumors (57), was maintained in RPMI-1640 media (Invitrogen) supplemented with 5% FCS. The esophageal epithelial cell line (hTERT-immortalized EPC2 line) was a gift from Anil Rustgi (University of Pennsylvania, Philadelphia, Pennsylvania, USA). The EPC2 cell monolayers were grown on 0.4-μm pore–sized polyester permeable supports (Corning Incorporated, Corning, NY) in keratinocyte serum–free media (KSFM) (Life Technologies). Once the cells were confluent, the media were switched to high-calcium ([Ca2+] = 1.8 mM) KSFM for an additional 3–5 days. Epithelial differentiation was then induced by removing culture media from the inner chamber of the permeable support and maintaining the esophageal epithelial cells for 5–7 days at the ALI. STAT6KD clones were generated by a shRNA approach as previously described (15). The cells were treated with IL-13 (Peprotech) at a final concentration of 100 ng/ml for the indicated periods of time.
TEER and FITC-dextran measurements.
EPC2 cells were subjected to ALI on 12-well and 24-well filter inserts as previously described (53). TEER and 3- to 5-kDa FITC-dextran (Sigma-Aldrich) paracellular flux were measured using an EVOM (World Precision Instruments) and a fluorescence plate reader (BioTek), respectively.
SYNPO gene silencing and overexpression.
SYNPO gene silencing in EPC2 and TE-7 cells was generated by using an shRNA approach. Lentiviral shRNA vectors against SYNPO (MISSION shRNA, Sigma-Aldrich, TRCN0000008655 [targets all 4 isoforms in CDS, CCGCAAATCCATGTTTACTTT, designated as shSYNPO1] and TRCN0000008656 [targets isoform 1 in CDS, CTGGCGCGAAACATCATCAAT, designated as shSYNPO2]) were used to produce lentiviral particles by the Cincinnati Children’s Hospital Medical Center Lenti-shRNA Library Core in pLKO.1-puro format. A vector that targets no known mammalian genes was used as a control (SHC002 SIGMA MISSION pLKO.1-puro Non-Mammalian shRNA Control). The viruses were generated by the Lenti-X HTX Packaging System (Takara Bio USA). The TE-7 cells were kept and transduced in RPMI 5% FCS media as described in ref. 15. For SYNPO overexpression, the coding sequence of the long SYNPO isoform (NM_007286) from the SYNPO Human cDNA clone (OriGene) was cloned into the pLVX-IRES-Puro lentiviral vector by the In-Fusion method (Takara). The EPC2 cells were grown and transduced in KSFM. The viruses, obtained from a 60-mm dish of 293T cells, were precipitated using Lenti-X concentrator solution and resuspended in 300 μl KSFM. KSFM (150 μl) was mixed with polybrene to a final concentration of 5 μg/ml and used to transduce EPC2 cells grown to 60%–70% confluency on a 6-well plate by centrifugation at 2,000 g at room temperature for 1 hour. All cells underwent puromycin selection (1 μg/ml) for 1–2 weeks and were later kept in puromycin at 0.5 μg/ml. The puromycin was removed 24 hours prior to the beginning of the experiment. Expression levels were validated by RT-PCR and Western blot analyses.
Immunofluorescence of cells and biopsies.
For immunofluorescence, the cells were grown on μ-Slide 8 Well, ibiTreat (Ibidi USA). The cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 for 10 minutes. All washes were performed with PBS for 5 minutes. Blocking was performed in 10% goat serum for 30 minutes at room temperature, and the cells were incubated with primary antibodies against SYNPO (sc-21537, Santa Cruz) at 1:250 dilution for 2 hours at room temperature. The secondary antibody donkey anti-goat Alexa Fluor 488 (Invitrogen) was added for 1 hour at room temperature at 1:500 dilution. The DNA was counterstained with Hoechst 33342 added to the first wash at 5 μg/ml for 20 minutes at room temperature, and actin was stained with phalloidin Alexa Fluor 568 (Thermo Fisher Scientific Inc.) for 1 hour. The immunofluorescence staining of the biopsies was performed as previously described (11) using SYNPO antibodies on at least 4 distal esophageal biopsies from control individuals and from patients with active EoE. The nuclei were stained with DAPI. The slides were blocked with PBS with 10% donkey serum. The secondary antibodies (1:500 dilution) used were donkey anti-goat Alexa Fluor 488 (Life Technologies). Imaging was performed with a Nikon A1 inverted confocal microscope.
mRNA extraction, heatmap generation, quantitative RT-PCR, ChIP-PCR, and Western blotting.
RNA isolation, RT-PCR, and ChIP-PCR were performed as described previously (15). For ChIP-PCR, the H3K4me3 (17-614, Millipore), H3K9Ac (ab10812, Abcam), and H3K27Ac (pAb-196-050, Diagenode Inc.) antibodies were used. The RNA-sequencing and ChIP-sequencing data for TE-7 cells were previously uploaded to GEO (GSE57637). For heatmap generation, genes that were upregulated by IL-13 at 2, 6, and 24 hours in TE-7 cells were chosen that intersected with RNA-sequencing data from the EoE biopsies (8). For assessment of SYNPO expression in EoE patients treated with anti–IL-13 antibody (QAX56), RNA was isolated from the esophageal biopsies of the patients from our previous study (29). Patients received QAX56 (n = 4) or placebo (n = 3) treatment (day 85 of the study), and the expression level of SYNPO was determined by RT-PCR using primers that recognized both the long and short isoforms of the gene. RT-PCR was performed in quadruplicates for each sample. Epigenetic changes were assessed after stimulation with IL-13 for 6 hours, and the MAnorm algorithm was applied to identify significantly different epigenetic changes (58). M values of less than –1 (increased histone mark) or more than 1 (decreased histone mark) were considered significant; a value of 0 was assigned to M values that were not significant following the analysis. For the heatmap generation, the log2 fold change for RNA-sequencing data and M values were used. Clustering was performed in the Cluster 3 program using Euclidian distance and average linkage parameters, and Java Tree was used for visualizing the results (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm). The levels of histone modifications on the promoters were assessed by the Prime Time TaqMan qPCR assay (Integrated DNA Technologies Inc.) and normalized to the levels of modifications in the input sample. The sequences GGCCATTACTCAGGCCTCTCCATTTT for the probe, TCCTTTCAGTGGGAGGAATG for the forward primer, and AGGTCAGGAAGGAACCAATG for the reverse primer were used to quantify epigenetic marks in the SYNPO promoter. The procuring and processing of esophageal biopsies was performed as described previously (15). Western blotting of cell lysates was performed as described previously (15). Mouse monoclonal antibody against p44/42 MAPK (ERK1/2) (3A7, 9107, Cell Signaling Technology) and mouse monoclonal anti-HSP90 antibody, clone 4C10 (OriGene) were used as loading controls.
Wound healing assay.
For the wound healing assay, the control and SYNPO knockdown TE-7 cells were grown on a 12-well plate until confluency. The wound was made with a 200-l culture tip, the cells were washed twice with RPMI media containing 5% FCS, and images were taken immediately and 16 hours after the wound was made. The surface area was quantified by ImageJ software (NIH). For the EPC2 cells, control and SYNPO knockdown cells were simultaneously seeded on a Culture-Insert 4 Well in μ-Dish 35 mm, high ibiTreat (Ibidi USA) in KSFM (2 fields per cell type) and grown until confluency. Two hours prior to the assay, the cells were labeled with 1 mM SiR-DNA dye (CY-SC007, Cytoskeleton) without verapamil. The insert was removed, cells were washed 3 times with KSFM and mounted on the humidified chamber on a Nikon Ti-E SpectraX Widefield Microscope, and images were taken every minute for 8 hours. Both far-red and bright-field images were collected. Quantification of cell parameters, including distance traveled over the length of the experiment, was performed by Nikon software at the Cincinnati Children’s Hospital Medical Center Confocal Imaging Core. The region of interest was set in the gap between cells, and SiR-DNA was used to track cells by creating binary masks.
Statistics.
Statistical analysis was performed using Prism 6.0 software. Comparisons among groups were performed by the ANOVA tests; the 2-tailed t test was applied for comparisons of two groups of data. Spearman correlation analysis was performed to correlate SYNPO expression with the genes from the EoE transcriptome. A statistical probability of P < 0.05 was considered significant; Holm-Sidak correction was applied when appropriate to correct for multiple samples. For box-and-whisker plots, the box represents the 50th percentile of the data, whiskers show minimum and maximum values, and the line in the box represents the median. RNA sequencing was analyzed by DESeq (59).
Study approval.
This study was performed with the approval of the Cincinnati Children’s Hospital Medical Center IRB (protocol 2008-0090). Informed consent was obtained from patients or their legal guardians for donation of tissue samples for research and to have clinical information entered into the Cincinnati Center for Eosinophilic Disorders Eosinophilic Gastrointestinal Disorder database.
Author contributions
MR, JT, and JMC performed experiments and data analysis; MR wrote the paper; MER supervised the study; and JPA contributed conceptual content.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R37 AI045898, R01 AI124355, and U19 AI070235 (to MER) and F30 DK109573 and T32 GM063483 (to JT); the Campaign Urging Research for Eosinophilic Disease (to MER); the Buckeye Foundation (to MER); and the Sunshine Charitable Foundation and its supporters, Denise A. Bunning and David G. Bunning (to MER). We thank Shawna Hottinger for editorial assistance and the Cincinnati Children’s Hospital Medical Center Confocal Imaging Core, especially Matthew Kofron for confocal and wound healing assay imaging.
Version 1. 10/19/2017
Electronic publication
Footnotes
Conflict of interest: MER is a consultant for Immune Pharmaceuticals, NKT Therapeutics, PulmOne, Celgene, Spoon Guru, Shire, and Novartis; has an equity interest in Immune Pharmaceuticals, NKT Therapeutics, PulmOne, and Spoon Guru; and receives royalties for reslizumab (Teva Pharmaceuticals). MER and MR are inventors of several patents, owned by Cincinnati Children’s Hospital Medical Center.
Reference information:JCI Insight. 2017;2(20):e96789. https://doi.org/10.1172/jci.insight.96789.
Contributor Information
Mark Rochman, Email: Mark.Rochman@cchmc.org.
Jared Travers, Email: jared.travers@cchmc.org.
J. Pablo Abonia, Email: pablo.abonia@cchmc.org.
Marc E. Rothenberg, Email: rothenberg@cchmc.org.
References
- 1.Gour N, Wills-Karp M. IL-4 and IL-13 signaling in allergic airway disease. Cytokine. 2015;75(1):68–78. doi: 10.1016/j.cyto.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wills-Karp M. Interleukin-13 in asthma pathogenesis. Immunol Rev. 2004;202:175–190. doi: 10.1111/j.0105-2896.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- 3.Barnes PJ. Pathophysiology of allergic inflammation. Immunol Rev. 2011;242(1):31–50. doi: 10.1111/j.1600-065X.2011.01020.x. [DOI] [PubMed] [Google Scholar]
- 4.Zimmermann N, et al. Transcript signatures in experimental asthma: identification of STAT6-dependent and -independent pathways. J Immunol. 2004;172(3):1815–1824. doi: 10.4049/jimmunol.172.3.1815. [DOI] [PubMed] [Google Scholar]
- 5.Krishnamurthy P, Kaplan MH. STAT6 and PARP family members in the development of t cell-dependent allergic inflammation. Immune Netw. 2016;16(4):201–210. doi: 10.4110/in.2016.16.4.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis BP, Rothenberg ME. Mechanisms of disease of eosinophilic esophagitis. Annu Rev Pathol. 2016;11:365–393. doi: 10.1146/annurev-pathol-012615-044241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanchard C, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120(6):1292–1300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 8.Sherrill JD, et al. Analysis and expansion of the eosinophilic esophagitis transcriptome by RNA sequencing. Genes Immun. 2014;15(6):361–369. doi: 10.1038/gene.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caldwell JM, et al. Glucocorticoid-regulated genes in eosinophilic esophagitis: a role for FKBP51. J Allergy Clin Immunol. 2010;125(4):879–888.e8. doi: 10.1016/j.jaci.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kc K, Rothenberg ME, Sherrill JD. In vitro model for studying esophageal epithelial differentiation and allergic inflammatory responses identifies keratin involvement in eosinophilic esophagitis. PLoS ONE. 2015;10(6):e0127755. doi: 10.1371/journal.pone.0127755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherrill JD, et al. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. 2014;7(3):718–729. doi: 10.1038/mi.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanchard C, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 2010;184(7):4033–4041. doi: 10.4049/jimmunol.0903069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rochman M, et al. Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2017;140(3):738–749.e3. doi: 10.1016/j.jaci.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kottyan LC, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014;46(8):895–900. doi: 10.1038/ng.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rochman M, et al. Neurotrophic tyrosine kinase receptor 1 is a direct transcriptional and epigenetic target of IL-13 involved in allergic inflammation. Mucosal Immunol. 2015;8(4):785–798. doi: 10.1038/mi.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim E, Rothenberg ME. Demethylation of the human eotaxin-3 gene promoter leads to the elevated expression of eotaxin-3. J Immunol. 2014;192(1):466–474. doi: 10.4049/jimmunol.1302454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexander ES, et al. Twin and family studies reveal strong environmental and weaker genetic cues explaining heritability of eosinophilic esophagitis. J Allergy Clin Immunol. 2014;134(5):1084–1092.e1. doi: 10.1016/j.jaci.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asanuma K, Yanagida-Asanuma E, Faul C, Tomino Y, Kim K, Mundel P. Synaptopodin orchestrates actin organization and cell motility via regulation of RhoA signalling. Nat Cell Biol. 2006;8(5):485–491. doi: 10.1038/ncb1400. [DOI] [PubMed] [Google Scholar]
- 19.Yanagida-Asanuma E, et al. Synaptopodin protects against proteinuria by disrupting Cdc42:IRSp53:Mena signaling complexes in kidney podocytes. Am J Pathol. 2007;171(2):415–427. doi: 10.2353/ajpath.2007.070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asanuma K, et al. Synaptopodin regulates the actin-bundling activity of alpha-actinin in an isoform-specific manner. J Clin Invest. 2005;115(5):1188–1198. doi: 10.1172/JCI23371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caldwell JM, et al. Cadherin 26 is an alpha integrin-binding epithelial receptor regulated during allergic inflammation. Mucosal Immunol. 2017;10(5):1190–1201. doi: 10.1038/mi.2016.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hon GC, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Hum Mol Genet. 2009;18(R2):R195–R201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faul C, et al. The actin cytoskeleton of kidney podocytes is a direct target of the antiproteinuric effect of cyclosporine A. Nat Med. 2008;14(9):931–938. doi: 10.1038/nm.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goenka S, Kaplan MH. Transcriptional regulation by STAT6. Immunol Res. 2011;50(1):87–96. doi: 10.1007/s12026-011-8205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mundel P, Heid HW, Mundel TM, Krüger M, Reiser J, Kriz W. Synaptopodin: an actin-associated protein in telencephalic dendrites and renal podocytes. J Cell Biol. 1997;139(1):193–204. doi: 10.1083/jcb.139.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanchard C, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116(2):536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen T, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology. 2013;145(6):1289–1299. doi: 10.1053/j.gastro.2013.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorriento D, et al. The G-protein-coupled receptor kinase 5 inhibits NFkappaB transcriptional activity by inducing nuclear accumulation of IkappaB alpha. Proc Natl Acad Sci USA. 2008;105(46):17818–17823. doi: 10.1073/pnas.0804446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothenberg ME, et al. Intravenous anti-IL-13 mAb QAX576 for the treatment of eosinophilic esophagitis. J Allergy Clin Immunol. 2015;135(2):500–507. doi: 10.1016/j.jaci.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 30.Mun GI, Park S, Kremerskothen J, Boo YC. Expression of synaptopodin in endothelial cells exposed to laminar shear stress and its role in endothelial wound healing. FEBS Lett. 2014;588(6):1024–1030. doi: 10.1016/j.febslet.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 31.Deller T, et al. Synaptopodin-deficient mice lack a spine apparatus and show deficits in synaptic plasticity. Proc Natl Acad Sci USA. 2003;100(18):10494–10499. doi: 10.1073/pnas.1832384100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barisoni L, Kriz W, Mundel P, D’Agati V. The dysregulated podocyte phenotype: a novel concept in the pathogenesis of collapsing idiopathic focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 1999;10(1):51–61. doi: 10.1681/ASN.V10151. [DOI] [PubMed] [Google Scholar]
- 33.Srivastava T, Garola RE, Whiting JM, Alon US. Synaptopodin expression in idiopathic nephrotic syndrome of childhood. Kidney Int. 2001;59(1):118–125. doi: 10.1046/j.1523-1755.2001.00472.x. [DOI] [PubMed] [Google Scholar]
- 34.Maas RJ, Deegens JK, Smeets B, Moeller MJ, Wetzels JF. Minimal change disease and idiopathic FSGS: manifestations of the same disease. Nat Rev Nephrol. 2016;12(12):768–776. doi: 10.1038/nrneph.2016.147. [DOI] [PubMed] [Google Scholar]
- 35.Chugh SS, Clement LC, Macé C. New insights into human minimal change disease: lessons from animal models. Am J Kidney Dis. 2012;59(2):284–292. doi: 10.1053/j.ajkd.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho MH, Hong EH, Lee TH, Ko CW. Pathophysiology of minimal change nephrotic syndrome and focal segmental glomerulosclerosis. Nephrology (Carlton) 2007;12 Suppl 3:S11–S14. doi: 10.1111/j.1440-1797.2007.00875.x. [DOI] [PubMed] [Google Scholar]
- 37.Baris HE, et al. The effect of systemic corticosteroids on the innate and adaptive immune system in children with steroid responsive nephrotic syndrome. Eur J Pediatr. 2016;175(5):685–693. doi: 10.1007/s00431-016-2694-x. [DOI] [PubMed] [Google Scholar]
- 38.Kronbichler A, et al. Rituximab treatment for relapsing minimal change disease and focal segmental glomerulosclerosis: a systematic review. Am J Nephrol. 2014;39(4):322–330. doi: 10.1159/000360908. [DOI] [PubMed] [Google Scholar]
- 39.Abdel-Hafez M, Shimada M, Lee PY, Johnson RJ, Garin EH. Idiopathic nephrotic syndrome and atopy: is there a common link? Am J Kidney Dis. 2009;54(5):945–953. doi: 10.1053/j.ajkd.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wittig HJ, Goldman AS. Nephrotic syndrome associated with inhaled allergens. Lancet. 1970;1(7646):542–543. doi: 10.1016/s0140-6736(70)90770-1. [DOI] [PubMed] [Google Scholar]
- 41.Wei CC, Lin CL, Shen TC, Li YF. Risk of idiopathic nephrotic syndrome among children with asthma: a nationwide, population-based cohort study. Pediatr Res. 2015;78(2):212–217. doi: 10.1038/pr.2015.80. [DOI] [PubMed] [Google Scholar]
- 42.Wei CC, Tsai JD, Lin CL, Shen TC, Li TC, Chung CJ. Increased risk of idiopathic nephrotic syndrome in children with atopic dermatitis. Pediatr Nephrol. 2014;29(11):2157–2163. doi: 10.1007/s00467-014-2835-2. [DOI] [PubMed] [Google Scholar]
- 43.Yap HK, Cheung W, Murugasu B, Sim SK, Seah CC, Jordan SC. Th1 and Th2 cytokine mRNA profiles in childhood nephrotic syndrome: evidence for increased IL-13 mRNA expression in relapse. J Am Soc Nephrol. 1999;10(3):529–537. doi: 10.1681/ASN.V103529. [DOI] [PubMed] [Google Scholar]
- 44.Kimata H, Fujimoto M, Furusho K. Involvement of interleukin (IL)-13, but not IL-4, in spontaneous IgE and IgG4 production in nephrotic syndrome. Eur J Immunol. 1995;25(6):1497–1501. doi: 10.1002/eji.1830250604. [DOI] [PubMed] [Google Scholar]
- 45.Cheung W, Wei CL, Seah CC, Jordan SC, Yap HK. Atopy, serum IgE, and interleukin-13 in steroid-responsive nephrotic syndrome. Pediatr Nephrol. 2004;19(6):627–632. doi: 10.1007/s00467-004-1438-8. [DOI] [PubMed] [Google Scholar]
- 46.Munitz A, Brandt EB, Mingler M, Finkelman FD, Rothenberg ME. Distinct roles for IL-13 and IL-4 via IL-13 receptor alpha1 and the type II IL-4 receptor in asthma pathogenesis. Proc Natl Acad Sci USA. 2008;105(20):7240–7245. doi: 10.1073/pnas.0802465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van der Pouw Kraan TC, Van der Zee JS, Boeije LC, De Groot ER, Stapel SO, Aarden LA. The role of IL-13 in IgE synthesis by allergic asthma patients. Clin Exp Immunol. 1998;111(1):129–135. doi: 10.1046/j.1365-2249.1998.00471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Den Berg JG, et al. Interleukin-4 and interleukin-13 act on glomerular visceral epithelial cells. J Am Soc Nephrol. 2000;11(3):413–422. doi: 10.1681/ASN.V113413. [DOI] [PubMed] [Google Scholar]
- 49.Chan CY, et al. Novel role of Vav1-Rac1 pathway in actin cytoskeleton regulation in Interleukin-13-induced minimal change-like nephropathy. Clin Sci (Lond) 2016;120(24):2317–2327. doi: 10.1042/CS20160312. [DOI] [PubMed] [Google Scholar]
- 50.Gonsalves N, Yang GY, Doerfler B, Ritz S, Ditto AM, Hirano I. Elimination diet effectively treats eosinophilic esophagitis in adults; food reintroduction identifies causative factors. Gastroenterology. 2012;142(7):1451–1459. doi: 10.1053/j.gastro.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 51.Laurent J, Lagrue G. Dietary manipulation for idiopathic nephrotic syndrome. A new approach to therapy. Allergy. 1989;44(8):599–603. doi: 10.1111/j.1398-9995.1989.tb04207.x. [DOI] [PubMed] [Google Scholar]
- 52.Sieniawska M, Szymanik-Grzelak H, Kowalewska M, Wasik M, Koleska D. The role of cow’s milk protein intolerance in steroid-resistant nephrotic syndrome. Acta Paediatr. 1992;81(12):1007–1012. doi: 10.1111/j.1651-2227.1992.tb12164.x. [DOI] [PubMed] [Google Scholar]
- 53.Davis BP, et al. Eosinophilic esophagitis-linked calpain 14 is an IL-13-induced protease that mediates esophageal epithelial barrier impairment. JCI Insight. 2016;1(4):e86355. doi: 10.1172/jci.insight.86355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sleiman PM, et al. GWAS identifies four novel eosinophilic esophagitis loci. Nat Commun. 2014;5:5593. doi: 10.1038/ncomms6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu H, et al. Interleukin-4 and interleukin-13 increase NADPH oxidase 1-related proliferation of human colon cancer cells. Oncotarget. 2017;8(24):38113–38135. doi: 10.18632/oncotarget.17494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stoff B, MacKelfresh J, Fried L, Cohen C, Arbiser JL. A nonsteroidal alternative to impetiginized eczema in the emergency room. J Am Acad Dermatol. 2010;63(3):537–539. doi: 10.1016/j.jaad.2009.05.027. [DOI] [PubMed] [Google Scholar]
- 57.Nishihira T, Hashimoto Y, Katayama M, Mori S, Kuroki T. Molecular and cellular features of esophageal cancer cells. J Cancer Res Clin Oncol. 1993;119(8):441–449. doi: 10.1007/BF01215923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 2012;13(3):R16. doi: 10.1186/gb-2012-13-3-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.