Abstract
Klotho is a renal protein involved in phosphate homeostasis, which is downregulated in renal disease. It has long been considered an antiaging factor. Two Klotho gene transcripts are thought to encode membrane-bound and secreted Klotho. Indeed, soluble Klotho is detectable in bodily fluids, but the relative contributions of Klotho secretion and of membrane-bound Klotho shedding are unknown. Recent advances in RNA surveillance reveal that premature termination codons, as present in alternative Klotho mRNA (for secreted Klotho), prime mRNAs for degradation by nonsense-mediated mRNA decay (NMD). Disruption of NMD led to accumulation of alternative Klotho mRNA, indicative of normally continuous degradation. RNA IP for NMD core factor UPF1 resulted in enrichment for alternative Klotho mRNA, which was also not associated with polysomes, indicating no active protein translation. Alternative Klotho mRNA transcripts colocalized with some P bodies, where NMD transcripts are degraded. Moreover, we could not detect secreted Klotho in vitro. These results suggest that soluble Klotho is likely cleaved membrane-bound Klotho only. Furthermore, we found that, especially in acute kidney injury, splicing of the 2 mRNA transcripts is dysregulated, which was recapitulated by various noxious stimuli in vitro. This likely constitutes a novel mechanism resulting in the downregulation of membrane-bound Klotho.
Keywords: Aging, Nephrology
Keywords: Chronic kidney disease, RNA processing
The human secreted Klotho alternative mRNA is a nonsense-mediated mRNA decay target, precluding its translation, and its splicing is dysregulated in renal disease.
Introduction
Deficiency of Klotho (sometimes denoted as αKlotho) shortens lifespan and leads to a phenotype resembling human aging (1), whereas overexpression of Klotho extends lifespan by 20%–30% (2) and protects against the development of many pathological phenotypes, particularly renal disease (3–7). Klotho is therefore considered a potential target for treatment of renal and aging-related diseases. The Klotho gene contains 5 exons, and the corresponding membrane-bound Klotho protein is predominantly expressed in the kidney (8). This membrane-bound protein functions as a coreceptor to FGF receptor 1c, enabling FGF23 signaling and promoting phosphaturia (9–11). Membrane-bound Klotho expression is dramatically reduced in patients with chronic kidney disease (CKD), which — due to end-organ resistance — fosters an early rise in FGF23 levels and a later rise in serum phosphate (12). Membrane-bound Klotho is proteolytically cleaved into soluble Klotho proteins, which may be the Klotho proteins chiefly responsible for exerting systemic beneficial effects after shedding to blood, urine, or cerebrospinal fluid (13, 14). However, alternative splicing also generates an alternative mRNA transcript, assumed since its discovery in 1998 to code for a secreted Klotho protein (8). In this transcript, 50 nucleotides of intronic sequence after exon 3 are followed by a premature termination codon (PTC), precluding translation of (downstream) exon 5 encoding the transmembrane domain (8), leading to the long-standing assumption that the resultant protein is secreted. While the existence of this alternative transcript is well documented (15–18), the existence of this putative secreted Klotho protein has never been demonstrated, nor has the process of Klotho secretion. The only study to date to specifically address this issue found that any putative secreted Klotho protein smaller than the full-length 130 kDa soluble Klotho could not be detected in human serum or CSF (13). Mice also express an alternative Klotho mRNA (15, 16, 19–22), a protein of which is also yet to be found in blood (13, 23). Rats, though, have only been found to express membrane-bound Klotho mRNA (24), which intuitively undercuts a supposed conserved role for secreted Klotho in physiology.
Recent advances in RNA surveillance mechanisms indicate that mRNA transcripts with PTCs are targets for nonsense-mediated mRNA decay (NMD). Although different models for NMD triggers are yet to be unified, the presence of a PTC > 50 nucleotides upstream of an exon-exon junction and with a sufficiently large distance to the poly(A) tail constitutes a well-characterized, potent initiator for NMD. Up-frameshift protein 1 (UPF1), an NMD core factor, binds to the 3’ UTR of all mRNA molecules and is displaced by the ribosome during the pioneer round of translation (25). If not displaced, as occurs when a PTC is encountered, UPF1 is phosphorylated, subsequently recruiting factors that initiate degradation of the aberrant mRNA (26). The human alternative Klotho mRNA transcript has been coined as a possible NMD candidate (27), although this has not yet been substantiated by evidence. Murine klotho mRNAs were shown to be susceptible to NMD after introduction of a PTC in exon 1, adding credence to the possibility that a human PTC-containing Klotho mRNA may also be an NMD target (28).
Not only is Klotho expression especially decreased in renal disease (29, 30), maintained or elevated Klotho expression levels have been demonstrated to have substantial renoprotective effects (3, 7, 31). The kidney is also the principal source of Klotho (32). Given the pleiotropic and profound effects of soluble Klotho proteins, it is of great importance to determine the structures of soluble Klotho proteins and the structure-function relationships between soluble Klotho and the molecules with which Klotho interacts. Furthermore, of particular importance are the mechanisms of shedding and/or secretion of Klotho, which could be targeted as a treatment strategy to upregulate Klotho levels. In this study, we aimed to determine whether the alternative Klotho mRNA transcript is an NMD target mRNA, and whether this has any relevance for renal disease.
Results
Detection of membrane-bound and alternative Klotho mRNA transcripts.
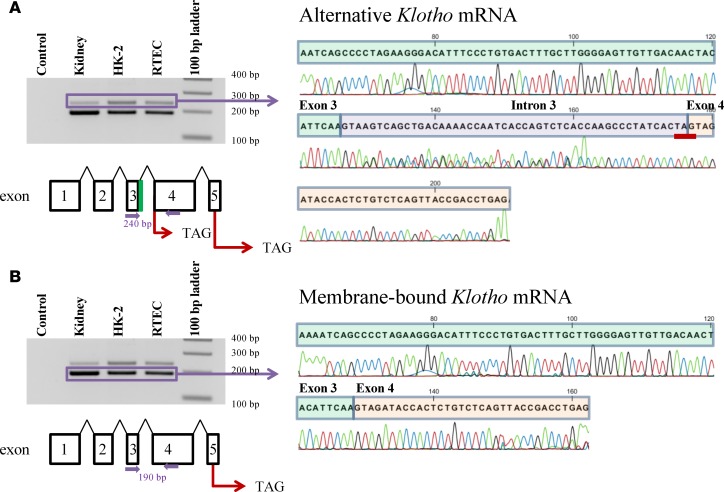
We confirmed the expression of both membrane-bound and alternative Klotho mRNA transcripts in human kidney, primary human renal tubular epithelial cells (RTECs), and a human renal epithelial cell line (HK-2) using reverse transcription PCR (RT-PCR). Using primers that amplify both alternative Klotho mRNA (Figure 1A) and membrane-bound Klotho mRNA (Figure 1B), we detected both at predicted amplicon lengths of 240 and 190 bp, respectively. DNA sequencing confirmed the transcripts to be normal Klotho mRNA coding for membrane-bound Klotho, with exon 3 and exon 4 being contiguous (corresponding to the lower band), and to be alternative Klotho mRNA putatively coding for secreted Klotho protein, with exon 3 extended by 50 nucleotides of intronic sequence, creating an in-frame PTC (TAG) (corresponding to the upper band).
Figure 1. Identification of human Klotho mRNA transcripts by RT-PCR and DNA sequencing.
(A) The alternative Klotho mRNA transcript is detected in human kidney, primary renal tubular epithelial cells, and in the human HK-2 renal cell line. PCR using primers spanning exons 3 and 4 (indicated by closed purple arrows) yields 2 PCR products. DNA sequencing confirms that the 240 bp PCR product corresponds to the alternative Klotho mRNA, which contains a 50 bp insertion (purple) between exon 3 (green) and 4 (orange), yielding a premature TAG stop codon (underlined in red). The alternative Klotho exons are depicted schematically, including the intronic sequence between exons 3 and 4 (green). (Note: nucleotide 145 in this sequence has similarly low signals for G and C, but is a G according to published sequences.) (B) The membrane-bound Klotho mRNA transcript is detected in human kidney, in primary renal tubular epithelial cells, and in the human HK-2 renal cell line. PCR using primers spanning exons 3 and 4 (indicated by closed purple arrows) yields 2 PCR products. DNA sequencing confirms that the 190 bp PCR product corresponds to the membrane-bound Klotho mRNA, which does not contain the 50 bp insertion between exon 3 and 4. The membrane-bound Klotho exons are depicted schematically.
Inhibition of NMD prevents degradation of alternative Klotho mRNA.
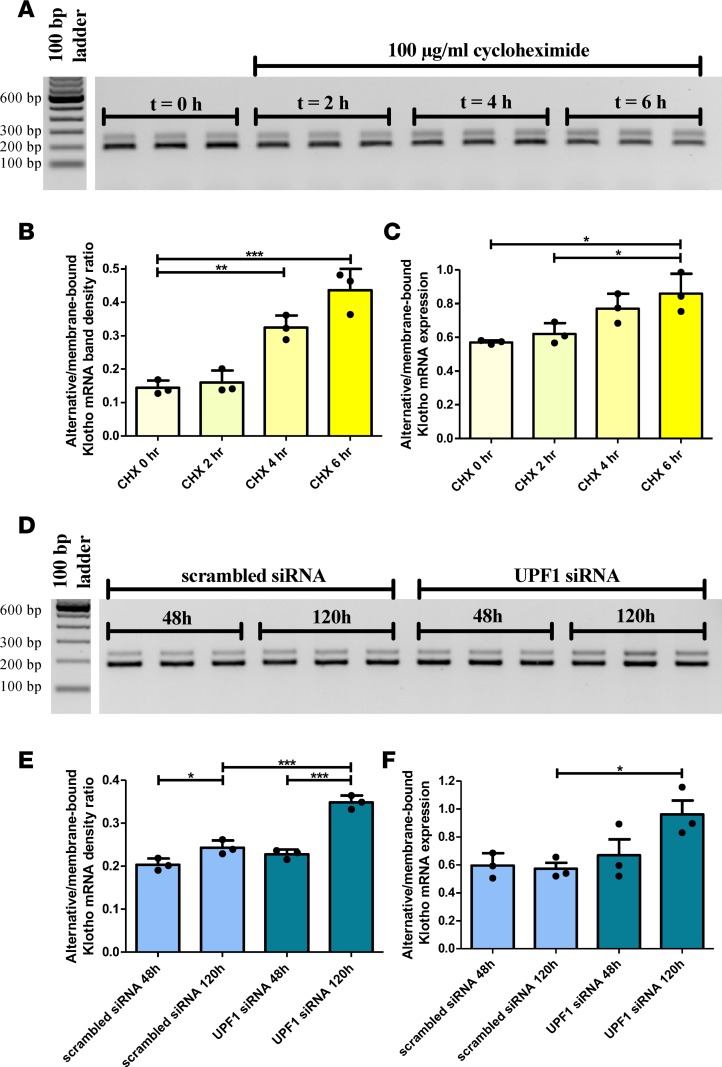
To assess whether alternative Klotho mRNA is subject to degradation by NMD, we inhibited protein translation in HK-2 cells using cycloheximide (CHX), since NMD is a translation-dependent process, inhibited by protein synthesis inhibitors (33). In 6 hours, we observed a time-dependent accumulation of alternative Klotho mRNA using RT-PCR/densitometry (alternative/membrane-bound Klotho mRNA ratio increase from 0.14 ± 0.02 to 0.44 ± 0.06, P < 0.001) (Figure 2, A and B, and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.94375DS1) and quantitative PCR (qPCR) (alternative/membrane-bound Klotho mRNA expression level increase from 0.57 ± 0.01 to 0.86 ± 0.12, P = 0.013) (Figure 2C and Supplemental Table 1). This CHX-induced increase suggests that alternative Klotho mRNA is an NMD target. To confirm these findings, we silenced the Upf1 gene (Supplemental Figure 1), which codes for NMD core factor UPF1, in HK-2 cells. After 120 hours, we observed an increase in alternative Klotho mRNA abundance as compared with scrambled siRNA–transfected controls, using RT-PCR/densitometry (ratios 0.24 ± 0.02 versus 0.35 ± 0.02 after 120 hours, P < 0.001) (Figure 2, D and E, and Supplemental Table 2) and qPCR (relative expression level 0.57 ± 0.07 versus 0.96 ± 0.17, P = 0.027) (Figure 2F and Supplemental Table 2). The lack of alternative Klotho mRNA accumulation at 48 hours may indicate that UPF1 was not yet depleted. In short, inhibition of NMD induces accumulation of alternative Klotho mRNA, indicating that alternative Klotho mRNA is an NMD target transcript.
Figure 2. Blocking nonsense-mediated mRNA decay, using cycloheximide or by silencing UPF1, results in the accumulation of the alternative Klotho mRNA transcript in HK-2 cells.
(A) RT-PCR for the 2 Klotho transcripts in HK-2 cells incubated with 100 μg/ml cycloheximide for 0, 2, 4, or 6 hours, showing accumulation of the alternative Klotho transcript. (B) Densitometric quantification of A, expressed as the ratio of the alternative and membrane-bound Klotho mRNAs (values are provided in Supplemental Table 1). (C) qPCR analysis for both Klotho transcripts, using ΔCt = Ct(alternative Klotho transcript) – Ct(membrane-bound Klotho transcript), which confirms the RT-PCR results. (D) RT-PCR for the 2 Klotho transcripts in HK-2 cells after Upf1 or scrambled siRNA transfection for 48 or 120 hours, showing accumulation of the alternative Klotho transcript after 120 hours. (E) Densitometric quantification of D, expressed as the ratio of the alternative and membrane-bound Klotho mRNAs (values are provided in Supplemental Table 2). (F) qPCR analysis for both Klotho transcripts, using ΔCt = Ct(alternative Klotho transcript) – Ct(membrane-bound Klotho transcript), confirms the RT-PCR results. *P < 0.05, **P < 0.01, ***P < 0.001, as tested by one-way ANOVA with post-hoc Bonferroni correction. All individual data points represent means of 3 independent experiments, performed in triplicate (plotted with mean ± SD).
RNA IP for UPF1.
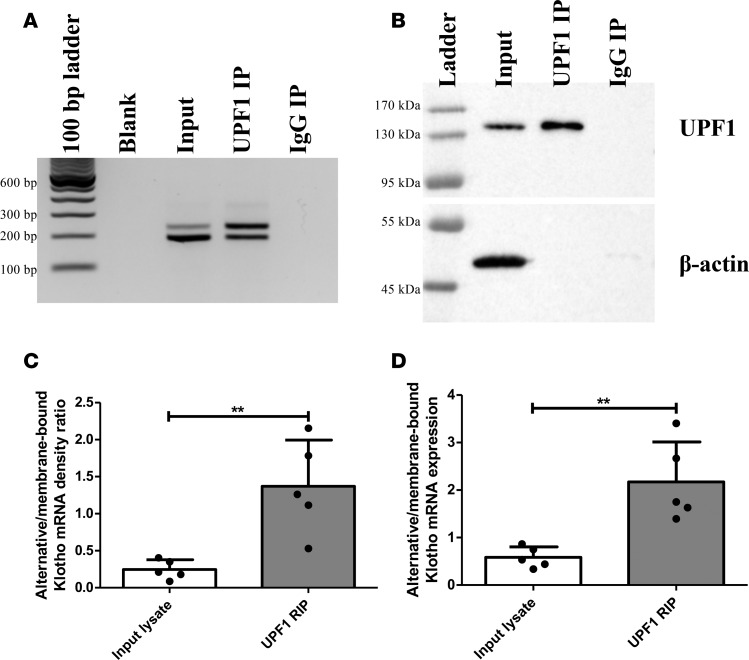
As UPF1 is not displaced by ribosomes from PTC-containing mRNAs, another method of identifying NMD targets is by RNA IP (RIP) for UPF1 and comparing the abundance of bound PTC transcripts to their non-PTC counterparts (34). In HK-2 lysate, alternative Klotho mRNA was bound to UPF1 more abundantly than membrane-bound Klotho mRNA, as compared with input lysate (Figure 3A) and detected by RT-PCR/densitometry (ratios 0.25 ± 0.13 versus 1.37 ± 0.63, P = 0.004) (Figure 3C) or by qPCR (ratios 0.59 ± 0.22 versus 2.17 ± 0.84, P = 0.004) (Figure 3D). An IgG control did not co-IP mRNA or protein, while β-actin was only detected in the input lysate, indicating that UPF1 IP was specific (Figure 3, A and B; see Supplemental Figure 2 for full Western blots of 3 independent UPF1 RIP experiments). The finding that enriching for UPF1 protein (Figure 3B) increases the abundance of alternative Klotho mRNA as compared with the membrane-bound Klotho mRNA also suggests that alternative Klotho mRNA is an NMD target.
Figure 3. RNA IP of HK-2 cell lysate reveals enrichment of the alternative Klotho mRNA associated with UPF1.
(A) RT-PCR analysis for both Klotho transcripts in HK-2 cell lysate (input), after IP for UPF1, and after IP with rabbit IgG, showing enrichment for the alternative Klotho mRNA in UPF1-associated RNA. (B) Western blot analysis for UPF1 and β-actin in HK-2 cell lysate and after IP with rabbit anti-UPF1 or rabbit IgG, showing enrichment for UPF1 and depletion of other proteins, as indicated by absence of β-actin in IP fractions. (C) Densitometric quantification of A, expressed as the ratio of the alternative and membrane-bound Klotho mRNAs. (D) qPCR analysis for both Klotho transcripts, using ΔCt = Ct(alternative Klotho transcript) – Ct(membrane-bound Klotho transcript), confirms the RT-PCR results. **P < 0.01, as tested by Student’s t test. All individual data points represent values from independent experiments (with mean ± SD).
Polysome fractionation.
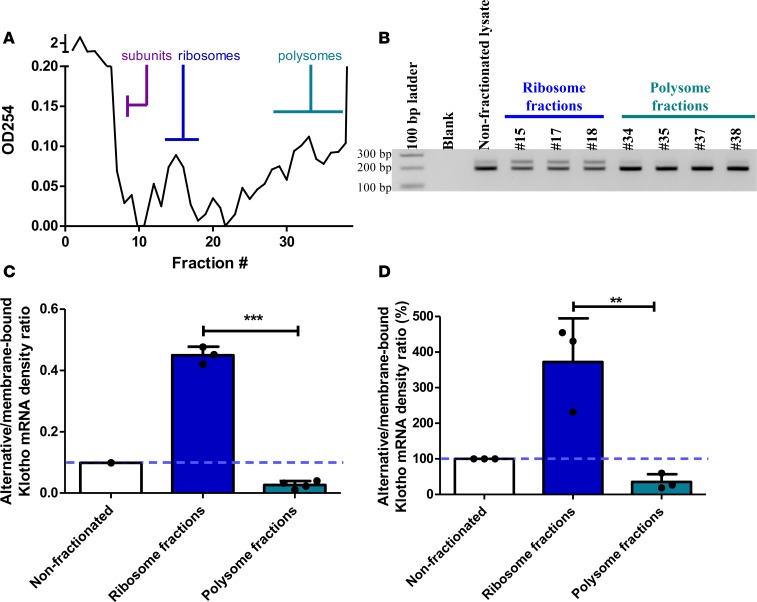
To determine whether translation of both transcripts occurs, we fractionated HK-2 lysates using a sucrose gradient. Based on the absorbance profile at OD254, we identified fractions enriched for single ribosomes or polysomes (Figure 4A). Densitometric quantification of Figure 4B is shown in Figure 4C, while Figure 4D depicts means of replication experiments, standardized to input values. Membrane-bound Klotho mRNA was associated with both ribosome and polysome fractions, indicative of active translation. Alternative Klotho mRNA, however, was most abundantly associated with single ribosomes and was depleted in polysome-enriched fractions (372% ± 122% of input ratio in single ribosome fractions and 35% ± 22% of input ratio in polysome fractions, P = 0.009) (Figure 4, B–D), indicating that the pioneer round of translation occurs but active protein translation does not.
Figure 4. Polysome fractionation of HK-2 cell lysate on sucrose gradients reveals enrichment of the alternative Klotho mRNA associated with single ribosomes and depletion in polysome-associated fractions.
(A) Representative absorbance spectrum of sucrose gradient fractions at 254 nm allows for identification of fractions enriched for free material, single ribosome subunits, single ribosomes, and polysomes. (B) RT-PCR analysis for both Klotho transcripts in nonfractionated HK-2 cell lysate, in single ribosome–associated fractions, and in polysome-associated fractions, showing enrichment for the alternative Klotho mRNA on single ribosomes and depletion in polysomes. (C) Densitometric quantification of the experiment in B, expressed as the ratio of the alternative and membrane-bound Klotho mRNAs. Individual data points refer to individual fractions (with mean ± SD). (D) Overall densitometric quantification of 3 replicate experiments, expressed as the ratio of the alternative and membrane-bound Klotho mRNAs, standardized to the nonfractionated lysate. **P < 0.01, ***P < 0.001, as tested by Student’s t test. Individual data points represent means of independent experiments with averages of 2–4 fractions in the same range (with means ± SD).
Detection of alternative Klotho mRNA using RNA in situ hybridization.
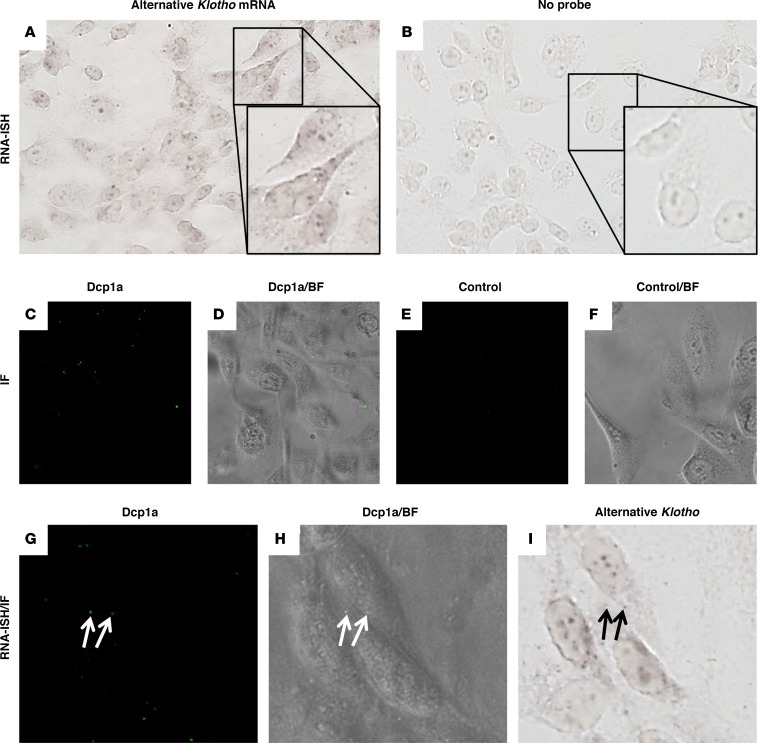
Using an RNA in situ hybridization (ISH) probe specific for alternative Klotho mRNA, we demonstrated specific expression of alternative Klotho mRNA in HK-2 cells as compared with controls without probe (Figure 5, A and B). We then assessed whether this transcript is detected in P bodies, the organelles where NMD transcripts are degraded by exoribonculeases and decapping enzymes. Using decapping enzyme 1a (Dcp1a) as P body marker (Figure 5, C–F), we found that some P bodies colocalized with alternative Klotho mRNA ISH signal (Figure 5, G–I). The lack of colocalization in other P bodies may be the consequence of mRNA degradation. It should also be noted that P bodies are, to an extent, a heterogeneous set of organelles, and perhaps only a specific subset would be involved in the degradation of this transcript.
Figure 5. RNA in situ hybdridization for the alternative Klotho mRNA transcript in HK-2 cells reveals colocalization with some P bodies.
(A) RNA in situ hybridization for the alternative Klotho mRNA transcript indicates expression in HK-2 cells. (B) Control cells hybridized without probe. Insets magnify individual cells. (C) Immunofluorescence for Dcp1a identifies P bodies in HK-2 cells, (D) merged with bright-field image. (E) Immunofluorescence without primary antibody, (F) merged with bright-field image. (G) Immunofluorescence for Dcp1a in HK-2 cells, (H) merged with bright-field image. (I) RNA ISH reveals that some P bodies in the HK-2 cells in G and H colocalize with the signal for the alternative Klotho mRNA transcript (arrows). Images depicted in panels A, B, and I were digitally contrast-enhanced to 150% in Microsoft Powerpoint. Original magnifications are 400×; insets are 2-fold the magnification of A and B.
Detection of soluble Klotho in vitro.
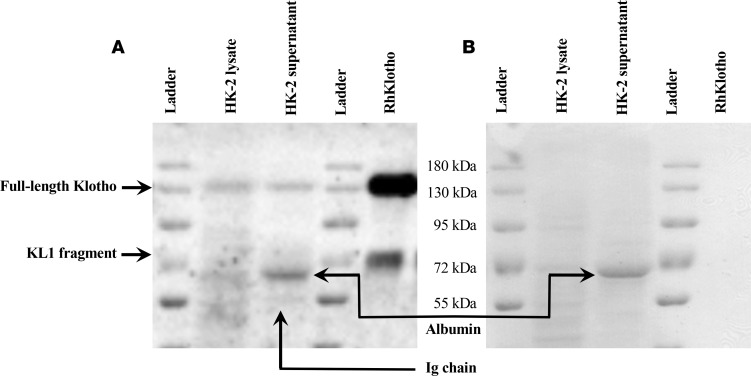
As established, the alternative Klotho mRNA is an NMD target and does not undergo protein translation, which suggests that the putative secreted Klotho protein would not be produced. We therefore assessed which soluble Klotho proteins we would find in a human in vitro cell culture model in which Klotho is naturally produced. To that end, we let HK-2 cells condition serum-free culture medium for 48 and 72 hours, after which we concentrated the culture medium and performed a Western blot for Klotho using KM2076, which detects the KL1 domain (in either the membrane-bound protein or in soluble Klotho proteins containing the KL1 domain). As shown in Figure 6, full-length soluble Klotho (130 kDa) is detected as the recombinant protein and in both HK-2 cell lysate and supernatant. The KL1 domain is detected at the expected height of about 70 kDa in the recombinant human Klotho sample. HK-2 cell supernatant did not exhibit immunoreactivity with any proteins of that size. As shown in independent experiments depicted in Figure 6 and Supplemental Figure 3, two smaller bands were consistently detected at 65 kDa and at 55 kDa, which were determined to be, respectively, (residual) albumin and a likely immunoglobulin chain due to immunoreactivity with the secondary antibody. In short, we did not detect a truncated, secreted Klotho protein in HK-2 cell supernatant.
Figure 6. A putative secreted Klotho protein is not detected in HK-2 cell supernatant.
(A) Western blot for Klotho on HK-2 cell lysate, 48 hour–conditioned supernatant, and recombinant human (Rh) Klotho. Note the 130 kDa bands corresponding to full-length Klotho in HK-2 cells and in supernatant, similar to 130 kDa positive control recombinant Klotho. While a 70 kDa KL1 band was detected in the positive control, no such band was detected in HK-2 cell supernatant. Note that the relative densities of the recombinant protein and KL1 fragment are unrelated to the expected secondary cleavage of 130 kDa soluble Klotho generated by HK-2 cells. Smaller bands were determined to correspond to albumin and a likely immunoglobulin chain, as detailed in Supplemental Figure 3. (B) Ponceau S protein staining showing a prominent band at 65 kDa in HK-2 supernatant, corresponding to albumin, which is likely residual BSA from FCS. This band is further characterized in Supplemental Figure 3.
Dysregulation of relative Klotho mRNA abundance in renal disease.
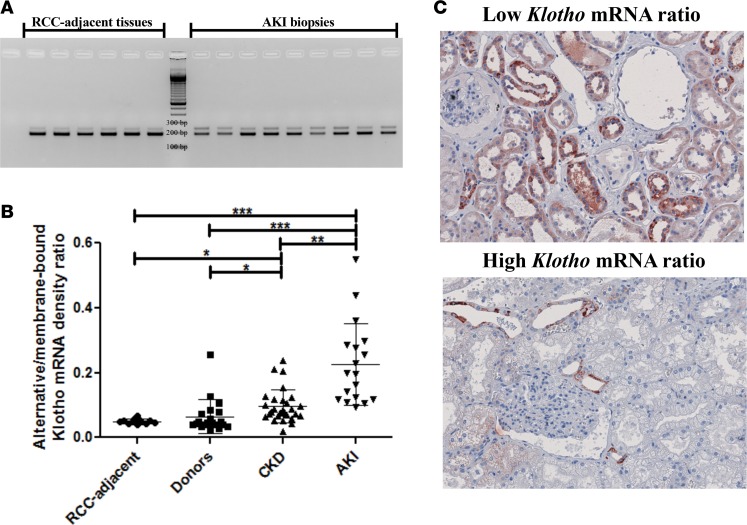
To assess whether the previous findings have any clinical relevance, we investigated the relative abundance of both mRNA transcripts in human kidneys and noticed a lot of variation, with ratios ranging from 0.03 ± 0.01 to 0.20 ± 0.02, in addition to a possible correlation between less alternative Klotho mRNA (lower ratio) and less renal damage (Supplemental Figure 4). We therefore investigated healthy tissue adjacent to resected renal cell carcinoma (RCC) (n = 12), biopsies from brain-dead and living kidney donors (n = 21), CKD (n = 28), and acute kidney injury (AKI) (n = 18). Patient characteristics are reported in Table 1. The RCC-adjacent samples displayed a very low alternative/membrane-bound Klotho mRNA ratio (0.05 ± 0.01), which was similar to kidney donors (0.05 [0.04–0.07]). A modest, but significant increase was observed in CKD samples (0.07 ± 0.03, P = 0.003) and a marked, significant increase was found in AKI samples (0.22 ± 0.11, P < 0.001) (random samples depicted in Figure 7A; quantification of all samples in Figure 7B). Furthermore, comparing Klotho protein expression between 3 kidneys with low and 3 kidneys with high Klotho mRNA ratios, we found lower Klotho protein expression in kidneys with a higher relative abundance of alternative Klotho mRNA transcript (Figure 7C). No difference was observed between living and brain-dead donor kidneys (Supplemental Figure 5).
Table 1. Patient characteristics.
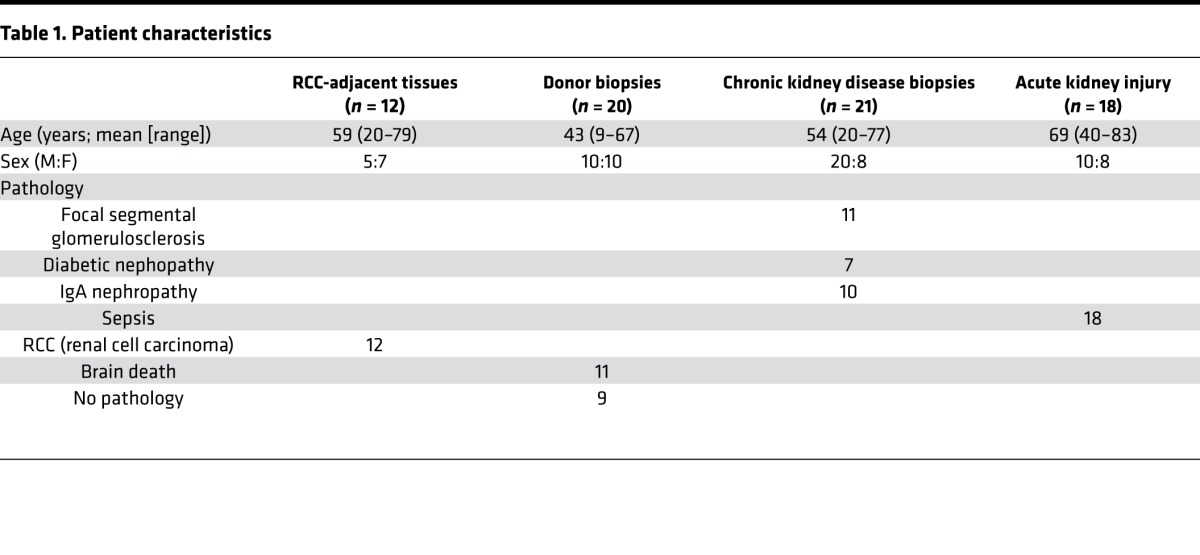
Figure 7. The relative abundance of the Klotho gene transcripts is dysregulated in vivo in renal disease.
(A) RT-PCR analysis of healthy renal cortices and acute kidney injury samples for both Klotho transcripts. (B) Densitometric quantification of renal cell carcinoma–adjacent (RCC-adjacent) healthy renal cortices (n = 12), donor kidney biopsies (n = 20), biopsies from kidneys afflicted with chronic kidney disease (CKD) (n = 28), and biopsies from kidneys suffering from acute kidney injury (AKI) (n = 18). (C) IHC for Klotho on kidneys with high alternative/membrane-bound Klotho mRNA ratios (n = 3) and kidneys with low alternative/membrane-bound Klotho mRNA ratios (n = 3) reveals lower Klotho protein expression in the kidneys with a relatively higher alternative Klotho mRNA abundance. Original magnifications: 200×. *P < 0.05, **P < 0.01, ***P < 0.001, as tested by Kruskal-Wallis test with Dunn’s post-hoc correction. Individual data points are plotted with mean ± SD.
Dysregulation of relative Klotho mRNA abundance in vitro.
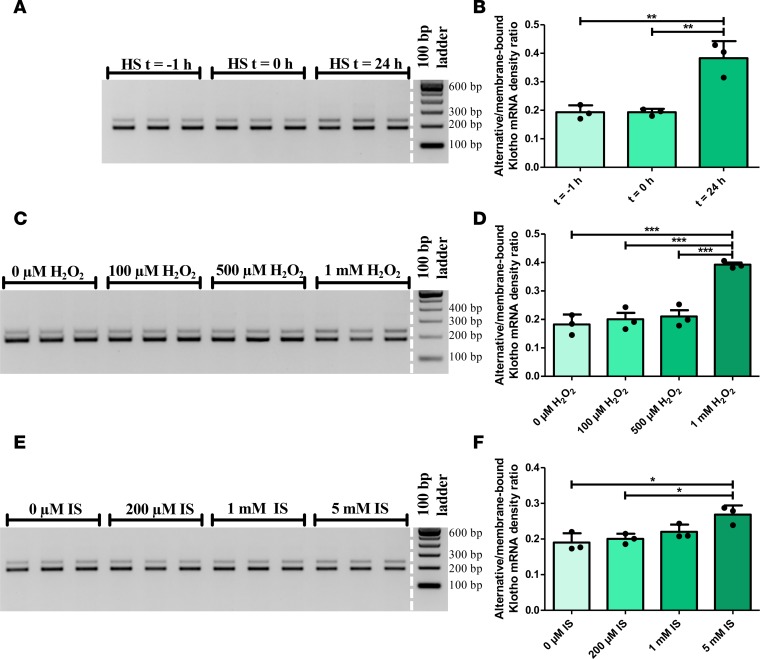
To assess whether the difference observed in patients constitutes a general damage response, we investigated different in vitro damage models to see whether an increase in the alternative/membrane-bound Klotho mRNA ratio would be observed. Using heat shock (45 minutes at 43°C) as a physical stressor, we noted an increase in ratio, 24 hours after heat shock compared with 1 hour before heat shock (0.19 ± 0.02 versus 0.38 ± 0.06, P = 0.003) (Figure 8, A and B). Using stimulation with H2O2 as an in vitro model for oxidative stress, we found an increase in ratio using 1 mM H2O2 (0.18 ± 0.04 versus 0.39 ± 0.01, P < 0.001) (Figure 8, C and D). Incubating with indoxyl sulfate (IS) for 48 hours as an in vitro model for uremia, the ratio was found to increase using 5 mM IS (0.19 ± 0.03 versus 0.27 ± 0.03, P = 0.015) (Figure 8, E and F). As a proof of principle, we also observed a shift in relative Klotho mRNA transcript abundance strongly favoring the alternative transcript upon stimulation with IS for 96 hours (Supplemental Figure 6).
Figure 8. The relative abundance of the Klotho gene transcripts is dysregulated in vitro after different stimuli.
(A) RT-PCR analysis for both Klotho transcripts after application of a heat shock as a physical stressor (45 min at 43°C) to HK-2 cells, that were lysed before the heat shock (t = –1 hour), after the heat shock (t = 0 hour), or 24 hours later. (B) Densitometric quantification of A, showing an increase in the alternative/membrane-bound Klotho mRNA ratio after 24 hours. (C) RT-PCR analysis for both Klotho transcripts after stimulation with different concentrations of H2O2 for 24 hours, to induce oxidative stress in HK-2 cells. (D) Densitometric quantification of C, showing an increase in the alternative/membrane-bound Klotho mRNA ratio after stimulation. (E) RT-PCR analysis for both Klotho transcripts after stimulation of HK-2 cells with different concentrations of indoxyl sulfate for 48 hours, as a model for uremia. (F) Densitometric quantification of E, showing a dose-dependent increase in the alternative/membrane-bound Klotho mRNA ratio after stimulation. Depicted are individual data points representing mean of 3 independent experiments, performed in triplicate (plotted with mean ± SD). *P < 0.05, **P < 0.01, ***P < 0.001, as tested by one-way ANOVA with Bonferroni’s post-hoc test.
Discussion
The major findings of this study are the identification of the alternative Klotho mRNA transcript as an NMD target and dysregulation of its splicing as a result of renal damage in vitro and in vivo.
We have demonstrated that the PTC-containing alternative Klotho mRNA transcript accumulates upon inhibition of NMD, is enriched after IP for NMD core factor UPF1, and does not actively undergo protein translation. These findings indicate that alternative Klotho mRNA is an NMD target that does not translate to protein in the kidney. This has several implications. First of all, if alternative Klotho mRNA does not produce a protein, the concepts of secreted Klotho protein and the physiology of its secretion are in question. To our best knowledge, the human secreted Klotho protein has not yet been identified (13), nor did we detect it, so we posit that it may be best to consider kidney-derived secreted Klotho protein nonexistent and the alternative mRNA transcript as an NMD target in human physiology.
Secondly, in the absence of secreted Klotho, the proteins commonly designated as circulating or soluble Klotho are likely derived from cleaved membrane-bound Klotho only, and proteolytic cleavage would gain relevance as the process generating circulating Klotho (13, 14, 35–37). An experiment performed by Hu et al. is in line with cleaved Klotho as the sole contributor to circulating Klotho; Hu et al. show that inhibition of both α- and β-secretases for 48 hours in mice virtually depletes circulating Klotho (38).
The question of why Klotho gene splicing results in 2 transcripts — of which one is continuously degraded — is not easily answered, but it can be placed in context. NMD is widely used to compensate for the inaccuracy of splicing, since about a third of the total number of transcripts is estimated to contain a PTC (39). Furthermore, selective pressure for introns to produce PTCs in the event of intron retention may exist (40). Additionally, a process termed regulated unproductive splicing and translation (RUST) may fine-tune protein expression levels more precisely than occurs at the transcriptional level by producing nonsense transcripts (41).
We have selected different approaches to substantiate the involvement of NMD in the fate of alternative Klotho mRNA beyond sequence-based prediction. It is challenging to study NMD in a physiological setting, and most studies are performed by overexpressing PTC-containing mRNAs. The resultant high expression levels — and, in turn, our constitutively low physiological alternative Klotho mRNA expression levels — are likely the reason behind comparatively modest effect sizes in our CHX and UPF1 silencing experiments. While not impossible to perform our experiments with transfected Klotho plasmids with and without PTCs, it is challenging having to use the large, native 50 kb gene, since splicing is eliminated in cDNA translation. As expected, “secreted Klotho” cDNA leads to protein production for this reason (10, 42, 43), which is likely nonphysiological, in spite of the biological effects of overexpressed secreted Klotho, essentially amounting to KL1 domain effects (3, 44–50). Notably, the antibody we used to detect Klotho (KM2076) would detect the KL1 domain in any KL1-containing form of Klotho (13, 51). Additionally, our choice to study naturally occurring PTC–containing mRNA makes our study more relevant for physiology, and the non-PTC counterpart functions as an internal control.
On a related note, using a physiological cell system of Klotho production, we did not detect a truncated, KL1 domain–containing, secreted Klotho protein in HK-2 cell supernatant. While smaller proteins were detected, we were able to identify one band as detected by the secondary antibody and the other band as residual BSA, which could not be washed away. KM2076 has been shown before to have some affinity for albumin if abundantly present (52). While not detecting secreted Klotho is not conclusive evidence of nonexistence and proving a negative is problematic on multiple levels, we think that the lines of evidence demonstrating that the alternative Klotho mRNA is not actively translated, undergoes NMD instead, and any hypothetical protein product could not be detected, strongly support our conclusion. One study by Massó et al., however, merits some further discussion, as they aimed to study the putative mouse secreted klotho protein by generating an antibody, K113, against the murine unique amino acid sequence (23). They detected a protein on Western blot that was expressed at a level in the brain 9-fold higher than in kidney. Adding to that, qPCR data by Massó et al. indicate that, in the brain, the membrane-bound klotho transcript is expressed at a level that was 78-fold higher compared with the alternative transcript. Since klotho is well-documented to be expressed at a much lower level in the brain compared with the kidney, these findings are difficult to interpret. We should also consider that the expression pattern for this antibody has not yet been studied, it has not been used for detecting the putative secreted klotho in blood or other extracellular compartments, nor has it been validated using Klotho-/- samples. A possible relationship between the alternative klotho transcript in mouse and the protein detected by K113 is therefore yet to be established, especially since previous studies using validated antibodies have not resulted in the detection of circulating secreted Klotho (13).
As a technical note, given the relative variability in expression level of both Klotho mRNA transcripts, actual (membrane-bound) Klotho levels are variably overestimated, especially Klotho downregulation in AKI. To circumvent indistinguishable amplification of both transcripts, primers flanking the exon 3–exon 4 boundary and a probe spanning that boundary are the best tools for accurate human Klotho expression qPCR analysis. While we slightly underestimated the relative abundance of alternative Klotho mRNA by employing a competitive RT-PCR, this is not an uncommon approach (40) and results were highly consistent and reproducible, while qPCR invariably yielded highly similar results, albeit with slightly more variation due to separate amplification reactions.
Furthermore, an interesting finding in our study is that, in both CKD and AKI, the relative abundance of alternative Klotho mRNA is increased. Because depletion of circulating Klotho levels is found after bilateral nephrectomy (38) and after kidney-specific deletion of Klotho (32), the kidney has been identified as principal contributor to systemic Klotho levels, adding to the relevance of our findings in renal cells and tissues. Caution, however, should be exercised in extrapolating our secreted Klotho results to other tissues. It is difficult to discriminate between dysfunction of NMD and dysregulation of splicing, processes that are particularly intertwined. However, the downregulation of membrane-bound Klotho mRNA relative to the increase in alternative Klotho mRNA suggests that at least a dysregulation in splicing is involved. This could essentially amount to downregulation of membrane-bound Klotho protein. Further supporting this notion, we indeed found lower Klotho protein levels in kidneys with a relatively higher alternative Klotho mRNA abundance, although various other mechanisms, such as promoter hypermethylation, are likely to contribute as well. It is currently unclear why Klotho is rapidly downregulated in renal disease while upregulation would be protective (31), but we may have identified the existence of alternative Klotho mRNA and dysregulation of its splicing as possible mechanisms contributing to Klotho downregulation, in addition to transcriptional regulation (16, 53, 54), epigenetic regulation (55–57), and miRNAs (58, 59).
Moreover, identification of alternative Klotho mRNA as an NMD target may cast a different light on published studies. For example, Lu et al. found that ovarian carcinoma patients positive for alternative Klotho mRNA have a higher tumor grade (17), which is interesting in light of the generally accepted view of Klotho as a tumor suppressor (44, 46, 60, 61), including in ovarian carcinoma (62, 63). However, higher alternative Klotho expression levels may not implicate effects of Klotho protein in whichever form and may even indicate dysregulated splicing or NMD dysfunction, which would lead to ER stress and may therefore reflect general cellular health.
To conclude, we have shown that the human alternative Klotho mRNA is a target for NMD and does not undergo protein translation, indicating that the supposed kidney-derived secreted Klotho does not exist. Furthermore, in vitro and in vivo renal damage result in increased alternative Klotho mRNA and in decreased membrane-bound Klotho mRNA levels, likely due to dysregulated splicing, essentially constituting an additional mechanism that may contribute to downregulation of Klotho in renal disease. Additional research is needed to elucidate the regulation of this process and for strategies aimed at improving Klotho gene splicing efficiency and increasing expression of the protective membrane-bound Klotho protein.
Methods
Cell culture.
HK-2 cells (provided by Theo Borghuis, Division of Medical Biology, UMCG) were cultured in growth medium consisting of low-glucose (1 g/l) DMEM supplied with 10% FCS, 1% 100 IE penicillin/100 μg streptomycin solution, and 1% 200 mM L-glutamine solution (all from Lonza). Human renal cortical tubular epithelial cells (RTECs) were isolated and cultured as previously described (64). Cells were incubated at 37°C and 5% CO2. For CHX experiments, cells were serum-starved for 24 hours before medium was changed to growth medium supplied with 100 μg/ml CHX (MilliporeSigma) for 0, 2, 4, or 6 hours. Heat shock was performed at 43°C for 45 minutes using a water bath and continuous temperature monitoring by submerging sealed culture plates. HK-2 cells were harvested before heat shock, 1 hour after initiation of heat shock and 24 hours later. Oxidative stress was induced in vitro by adding 0 μM, 100 μM, 500 μM, and 1 mM H2O2 (Merck) in growth medium to HK-2 cells for 24 hours. IS (I3875, MilliporeSigma) was used to stimulate cells for 48 or 96 hours in concentrations of 0 μM, 200 μM, 1 mM, and 5 mM in serum-containing growth medium. All stimulation experiments were preceded by 24 hours of serum starvation. To study the presence of Klotho in conditioned medium, T-75 flasks with HK-2 cells at subconfluence were washed with HBSS (Lonza) and incubated with serum-free culture medium for 48 or 72 hours, after which medium was collected and cells were lysed. Supernatants (10 ml) were concentrated using Vivaspin 20 and Vivaspin 2 filters (Vivaproducts) to ~50 μl.
RNA interference.
Silencer Select siRNAs against Upf1 (s11926; Ambion, Thermo Fisher Scientific) or scrambled negative control siRNAs and lipofectamine RNAiMAX (Thermo Fisher Scientific) were mixed in Opti-MEM reduced serum medium (Gibco, Thermo Fisher Scientific), before transfection of HK-2 cells suspended in DMEM/10% FCS. The final siRNA concentration was 10 nM. Cells were retransfected after 72 hours.
RT-PCR.
Cells and tissues were lysed in TRIzol, followed by RNA extraction using chloroform, precipitation with 2-propanol, washing in 70% ethanol (all from Merck), and solubilizing in nuclease-free water (Braun). Synthesis of cDNA was performed using random hexamer primers and Superscript II (Invirogen). To investigate the relative abundance of the alternative and membrane-bound Klotho transcripts, PCR was performed using the following primers: a forward primer specific for exon 3 of the human Klotho gene (5′-CTAAGCCAGGACAAGATG-3′) and a reverse primer specific for exon 4 of the human Klotho gene (5′-TCAGGTCGGTAAACTGAG-3′) and a PCR program of 40 cycles of 30 sseconds at 94°C, 30 seconds at 59°C, and 30 seconds at 72°C. PCR products were separated on a 2% agarose gel. Densitometry was performed using the ImageLab 4.0.1 software (Bio-Rad).
qPCR.
Primer/probe sets were used specifically for the membrane-bound Klotho mRNA (TaqMan assay Hs00935388_m1, Thermo Fisher Scientific) and for the alternative Klotho mRNA (forward: 5′-AACTACATTCAAGTAAGTCAGC-3′, reverse: 5′-CAGAGTGGTATCTACTAGTG-3′, probe: 5′-56-FAM/TCAGCAGTC/ZEN/TCACCAAGCCCT/31ABkFQ-3′, IDT). The following program was used on an ABI7900HT (Applied Biosystems, USA): 2 minutes at 50°C, 10 minutes at 95°C, followed by 40 cycles of 15 seconds at 95°C and 60 seconds at 60°C. Relative expression levels were calculated using the 2(-ΔCt) method. TATA box-binding protein (TBP) was used as a normalization gene.
DNA sequencing.
Bands were excised from 2% agarose gels under a UV light, and DNA was extracted using the Zymoclean Gel DNA Recovery Kit (Zymo Research). Briefly, agarose gel pieces were liquefied in agarose dissolving buffer at 54°C, after which DNA was collected on a spin-column. DNA was washed on a column and eluted from the column, after which it was sequenced by LGC Genomics GmbH. DNA sequences were analyzed using the CLC Main Workbench (Qiagen).
RIP.
RIP was performed according to ref. 25 with minor modifications. HK-2 cells from 2 confluent 75 cm2 flasks were lysed for 20 minutes on ice, using 1.2 ml of 10 mMol Tris/10 mM NaCl/2 mM EDTA/0.5% Triton X-100 (pH 7.5) supplied with Halt protease inhibitor cocktail (1861279, Thermo Fisher Scientific) and RiboLock RI RNase inhibitor (EO0381, Thermo Fisher Scientific), followed by centrifugation at 16,000 g for 15 minutes at 4°C. Input samples (50 μl for protein, mixed with 50 μl 2× Laemmli sample buffer, and 100 μl for RNA isolation, treated with RiboLock RI RNase inhibitor and Turbo DNase [AM2238, Ambion] for 5 minutes at 37°C) were set apart. The remainder of the lysate was rendered isotonic by correcting the sodium concentration, and 500 μl was incubated with 7 μg rabbit anti-UPF1 antibody (03-191, Merck Millipore) or 7 μg rabbit IgG for 90 minutes. Then, 64 μl of protein G Dynabeads (10004D, Thermo Fisher Scientific) were washed in wash buffer (150 mM NaCl/50 mM Tris/0.05% NP-40, pH 7.5) and lysis buffer, and then incubated for 1 hour in lysis buffer with 1% BSA and 0.1% yeast transfer RNA (tRNA) (15401-011, Thermo Fisher Scientific). After washing twice with lysis buffer, 32 μl of beads was added to the lysates and were incubated for 90 minutes at 4°C. Beads were then isolated magnetically and were washed 6 times for 5 minutes in 1 ml Net-2 buffer (150 mM NaCl/50 mM Tris/0.1% Triton X-100, pH 7.5) at 4°C. One third of the beads was then mixed with 2× Laemmli sample buffer and two thirds were used for DNAse treatment and RNA isolation. The work-up of IgG fraction-associated RNA for cDNA synthesis and subsequent PCR was corrected to the volume of the UPF1 RIP fractions, rather than the RNA concentration.
Western blotting.
Samples in 1× Laemmli buffer were heated for 5 minutes at 95°C, followed by SDS-PAGE of 10 μl of input lysate or RIP samples on 8% polyacrylamide gels for 90 minutes at 100 V. Proteins were then transferred to a methanol-activated PVDF membrane at 100 V for 90 minutes. Ponceau staining was performed to assess protein transfer and sample quality. Membranes were blocked in 5% nonfat dried milk in TBST for 30 minutes at room temperature. Incubation with goat anti-UPF1 (1:200; A300-038A, Bethyl Laboratories) was performed overnight in 5% nonfat dried milk in tris-buffered saline/0.05% tween-20 (TBST) at 4°C. After washing in TBST, membranes were incubated with rabbit anti–goat-HRP (1:1500; P0449, Dako) for 1 hour at room temperature and washed with TBST. β-Actin (mouse anti–β-actin-HRP, sc-47778, Santa Cruz Biotechnology Inc.) was used as a loading control and as a control for RIP fraction purity. Western blotting for Klotho was performed using monoclonal antibody KM2076 (TransGenic Inc.) 1:500 and rabbit anti–rat-HRP (P0450, Dako), on nitrocellulose, using 10 μl of concentrated supernatant sample, 40 μg of HK-2 cell lysate, and 5 ng of recombinant human Klotho (R&D Systems). Blots were reprobed with anti-albumin (AHP102, Serotec) and rabbit anti–sheep-HRP (P0163, Dako), which were provided by Fransien van Dijk (Department of Pharmacokinetics, Toxicology and Targeting, Groningen Research Institute of Pharmacy, UMCG). Imaging was performed using chemiluminescence (SuperSignal West Femto Chemiluminescent Substrate or SuperSignal West Pico Chemiluminescent Substrate [Thermo Fisher Scientific] and a ChemiDoc MP Imaging System [Bio-Rad]). Densitometry was performed using the ImageLab 4.0.1 software.
IHC.
Formalin-fixed, paraffin-embedded renal tissue sections (4 μm) were deparaffinized and rehydrated. Endogenous peroxidase activity was blocked, and antigens were retrieved by boiling sections for 15 minutes in 1 mM EDTA (pH 8). Slides were rinsed with PBS prior to incubation with rat anti-Klotho (clone KM2076, TransGenic Inc.), dilution 1:100, in 5% BSA solution for 1 hour at room temperature, followed by incubation with secondary antibody rabbit anti–rat-HRP (1:300; AI-4001, Vector Labs) for 45 minutes. Sections were then incubated with an anti-rabbit and multiple HRP-conjugated polymers (Dako) for 30 minutes. Peroxidase activity was detected with 3-amino-9-ethylcarbazole (AEC, Dako). Slides were counterstained with Mayer’s hematoxylin (Merck). All slides were scanned using a Hamamatsu Nanozoomer 2.0HT (Hamamatsu Photonics), and representative images were selected.
Polysome fractionation.
Polysome fractionation was performed according to ref. 65 with minor modifications. Sucrose gradients were prepared on the day before fractionation by layering 7%, 17%, 27%, 37%, and 47% sucrose solutions in lysis buffer (20 mM HEPES, 15 mM MgCl2, 200 mM KCl, and 100 μg/mL CHX) in 38.5 ml Ultra-Clear centrifuge tubes (344058, Beckman-Coulter). Gradients were stored overnight at 4°C to become continuous. HK-2 cells in 6 confluent 75 cm2 culture flasks were washed with PBS and incubated with CHX (100 μg/ml) at 37°C for 15 minutes to freeze the ribosomes on their mRNA strands. Cells were then washed with PBS, placed on ice, and lysed using 1 ml lysis buffer consisting of 20 mM HEPES, 15 mM MgCl2, 200 mM KCl, 1% Triton X-100, and 100 μg/ml CHX. Cells were collected using a scraper, were mechanically disrupted by pipetting, and were centrifuged at 14,000 g for 5 minutes at 4°C. Supernatants were then pipetted onto the sucrose gradients, which were placed in a swinging bucket SureSpin 630 rotor and were centrifuged for 4 hour at 23,000 g in a Sorvall Discovery 90SE ultracentrifuge (Thermo Fisher Scientific). Fractions of 1 ml were then collected, and the absorbance at 254 nm was measured on the NanoDrop (Thermo Fisher Scientific). RNA was isolated from fractions using TRIzol, and cDNA was synthesized from fractions with sufficient RNA content, followed by PCR on single ribosome- and polysome-enriched fractions.
RNA ISH.
HK-2 cells were grown in Lab-Tek chamber slides (Thermo Fisher Scientific) and fixed using 4% paraformaldehyde/5% acetic acid in PBS for 15 minutes. Cells were washed with PBS and incubated with rabbit anti–Dcp1a-A488 (1:100; Ab208275, Abcam) for 1 hour, after which imaging was performed on a Zeiss AxioObserver Z1 microscope equipped with TissueFAXS Image Analysis Software (TissueGnostics). Cells were then rinsed in PBS and treated with 0.1% pepsin/10 mM HCl for 1 minute at 37°C. Cells were then dehydrated and incubated with a 40 nM probe specific for the alternative Klotho mRNA (/5DigN/AGTGGTATCTACTAGTGATAGG/3Dig_N, Exiqon, Denmark) in 50% formamide/2× SSC/10% dextran sulfate/PBS. The probes were denatured at 80°C for 2 minutes and then hybridized for 30 minutes at 54°C on a ThermoBrite (Abbott), after which slides were rinsed in 0.1% Tween-20/2× SSC and rinsed 3 times with 0.1× SSC at 65°C. Cells were then incubated twice for 5 minutes at room temperature in 0.15 M NaCl/0.1 M maleic acid (pH 7.5), followed by 15 minutes at 37°C in 1% blocking reagent (11096176, Roche Diagnostics) and 2% normal rabbit serum in 0.15 M NaCl/0.1 M maleic acid. Then, cells were incubated with the same buffer supplied with mouse anti-DIG antibody (1:2,000; 11333062910, Roche Diagnostics) for 1 hour at 37°C. Cells were then incubated with rabbit anti-mouse (1:20; Z0259, Dako) for 30 minutes and with alkaline phosphatase–conjugated anti-alkaline phosphatase (1:50; D0651, Dako) for 30 minutes, followed by development of the chromogenic reaction using NBT, BCIP, and levamisole (Roche Diagnostics) overnight. Imaging was performed on a NanoZoomer 2.0HT (Hamamatsu).
Statistics.
Data were tested for normality using the Shapiro-Wilk test. Normally distributed data were reported as mean ± SD, and nonnormally distributed data were reported as median (interquartile range), unless stated otherwise. Depending on the distribution, differences between groups were assessed by a two-tailed Student’s t test or one-way ANOVA, or by Mann-Whitney U or Kruskal-Wallis test. These tests were followed by Bonferroni’s or Dunn’s post-hoc test, respectively, to correct for multiple comparisons. P < 0.05 was considered significant. For statistical analysis, SPSS version 18.0.3 (IBM) or GraphPad Prism 5.0 (GraphPad Software) was used.
Study approval.
Post-mortem kidney biopsies were performed in 18 patients with sepsis and renal failure (66). Adult patients who died of sepsis in the ICU were included in the study. Control patients underwent total nephrectomy as a result of RCC. Exclusion criteria were preexisting CKD, active autoimmune disorder with renal involvement, and treatment with immune-suppressive medication. For all patients, the next of kin was given an oral and written explanation about the study, and written informed consent was taken if permission was given. The Medical Ethics Review Committee (METC) of the UMCG reviewed and approved this study protocol (METc 2011/372, Protocol ID 2011.AKI). Kidney biopsies were taken within 35 minutes postmortem. The patient was placed in prone position, and the kidney was located by ultrasound. The biopsy device (Tru Core2 Biopsy Instrument, Angiotech; 14 G × 20 cm, 763114200x) was introduced through a small skin incision, and multiple kidney cortex biopsies were taken under ultrasound guidance.
For donor kidney biopsies, after informed consent was obtained from the patient or on his/her behalf from relatives, biopsies were taken before the start of cold ischemia. For RCC-adjacent renal tissue harvesting, the kidney was carefully inspected by the surgeon after nephrectomy, and kidney cortex biopsies were harvested distant from the renal cancer lesion. All kidney biopsies were fixed in 10% formalin and afterwards embedded in paraffin. Additionally, material was snap frozen in liquid nitrogen and stored at –80°C. CKD biopsies (leftover after completion of diagnostic procedures) were similarly used in accordance with the Dutch law on medical research on humans.
Author contributions
RM conceived and performed the experiments, analyzed data, and wrote the manuscript. GH performed analyses, analyzed data, and revised the manuscript. JM, MVM, AD, and HGL collected human tissue and revised the manuscript. JLH conceived the experiments and revised the manuscript.
Supplementary Material
Acknowledgments
We thank Anke van den Berg for advice and Marian Bulthuis, Bea Rutgers, Monique Lodewijk, Wierd Kooistra, and Siobhan Conroy for technical assistance. This work was supported by a consortium grant from the Dutch Kidney Foundation (NIGRAM, CP10.11; see Supplemental Acknowledgments for consortium details), the UMCG GSMS MD/PhD program, and a Dutch Kidney Foundation Kolff grant (Kolff 13OKJ35). Fluorescence imaging was performed at the UMCG Imaging Center, supported by the Netherlands Organization for Health Research and Development (ZonMW grant 40-00506-98-9021). Part of this work was presented as a poster presentation at the ASN Kidney Week 2015.
Version 1. 10/19/2017
Electronic publication
Footnotes
License: This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2017;2(20):e94375. https://doi.org/10.1172/jci.insight.94375.
Contributor Information
Rik Mencke, Email: r.mencke@umcg.nl.
Geert Harms, Email: g.harms@umcg.nl.
Jill Moser, Email: j.moser@path.umcg.nl.
Matijs van Meurs, Email: m.van.meurs@umcg.nl.
Arjan Diepstra, Email: a.diepstra@umcg.nl.
References
- 1.Kuro-o M, et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390(6655):45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- 2.Kurosu H, et al. Suppression of aging in mice by the hormone Klotho. Science. 2005;309(5742):1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou L, Li Y, Zhou D, Tan RJ, Liu Y. Loss of Klotho contributes to kidney injury by derepression of Wnt/β-catenin signaling. J Am Soc Nephrol. 2013;24(5):771–785. doi: 10.1681/ASN.2012080865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu MC, et al. Klotho and phosphate are modulators of pathologic uremic cardiac remodeling. J Am Soc Nephrol. 2015;26(6):1290–1302. doi: 10.1681/ASN.2014050465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubal DB, et al. Life extension factor klotho prevents mortality and enhances cognition in hAPP transgenic mice. J Neurosci. 2015;35(6):2358–2371. doi: 10.1523/JNEUROSCI.5791-12.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ravikumar P, et al. αKlotho deficiency in acute kidney injury contributes to lung damage. J Appl Physiol. 2016;120(7):723–732. doi: 10.1152/japplphysiol.00792.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi M, et al. αKlotho Mitigates Progression of AKI to CKD through Activation of Autophagy. J Am Soc Nephrol. 2016;27(8):2331–2345. doi: 10.1681/ASN.2015060613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsumura Y, Aizawa H, Shiraki-Iida T, Nagai R, Kuro-o M, Nabeshima Y. Identification of the human klotho gene and its two transcripts encoding membrane and secreted klotho protein. Biochem Biophys Res Commun. 1998;242(3):626–630. doi: 10.1006/bbrc.1997.8019. [DOI] [PubMed] [Google Scholar]
- 9.Kurosu H, et al. Regulation of fibroblast growth factor-23 signaling by klotho. J Biol Chem. 2006;281(10):6120–6123. doi: 10.1074/jbc.C500457200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrow EG, Davis SI, Summers LJ, White KE. Initial FGF23-mediated signaling occurs in the distal convoluted tubule. J Am Soc Nephrol. 2009;20(5):955–960. doi: 10.1681/ASN.2008070783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urakawa I, et al. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444(7120):770–774. doi: 10.1038/nature05315. [DOI] [PubMed] [Google Scholar]
- 12.Kuro-OM, Moe OW. FGF23-αKlotho as a paradigm for a kidney-bone network. Bone. 2017;100:4–18. doi: 10.1016/j.bone.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 13.Imura A, et al. Secreted Klotho protein in sera and CSF: implication for post-translational cleavage in release of Klotho protein from cell membrane. FEBS Lett. 2004;565(1-3):143–147. doi: 10.1016/j.febslet.2004.03.090. [DOI] [PubMed] [Google Scholar]
- 14.Chen CD, Podvin S, Gillespie E, Leeman SE, Abraham CR. Insulin stimulates the cleavage and release of the extracellular domain of Klotho by ADAM10 and ADAM17. Proc Natl Acad Sci USA. 2007;104(50):19796–19801. doi: 10.1073/pnas.0709805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azuma M, et al. Promoter methylation confers kidney-specific expression of the Klotho gene. FASEB J. 2012;26(10):4264–4274. doi: 10.1096/fj.12-211631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forster RE, et al. Vitamin D receptor controls expression of the anti-aging klotho gene in mouse and human renal cells. Biochem Biophys Res Commun. 2011;414(3):557–562. doi: 10.1016/j.bbrc.2011.09.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu L, Katsaros D, Wiley A, de la Longrais IA, Puopolo M, Yu H. Klotho expression in epithelial ovarian cancer and its association with insulin-like growth factors and disease progression. Cancer Invest. 2008;26(2):185–192. doi: 10.1080/07357900701638343. [DOI] [PubMed] [Google Scholar]
- 18.Suvannasankha A, et al. FGF23 is elevated in multiple myeloma and increases heparanase expression by tumor cells. Oncotarget. 2015;6(23):19647–19660. doi: 10.18632/oncotarget.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiraki-Iida T, et al. Structure of the mouse klotho gene and its two transcripts encoding membrane and secreted protein. FEBS Lett. 1998;424(1-2):6–10. doi: 10.1016/S0014-5793(98)00127-6. [DOI] [PubMed] [Google Scholar]
- 20.Mizuno I, Takahashi Y, Okimura Y, Kaji H, Chihara K. Upregulation of the klotho gene expression by thyroid hormone and during adipose differentiation in 3T3-L1 adipocytes. Life Sci. 2001;68(26):2917–2923. doi: 10.1016/S0024-3205(01)01092-X. [DOI] [PubMed] [Google Scholar]
- 21.Ravikumar P, et al. α-Klotho protects against oxidative damage in pulmonary epithelia. Am J Physiol Lung Cell Mol Physiol. 2014;307(7):L566–L575. doi: 10.1152/ajplung.00306.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bektas A, Schurman SH, Sharov AA, Carter MG, Dietz HC, Francomano CA. Klotho gene variation and expression in 20 inbred mouse strains. Mamm Genome. 2004;15(10):759–767. doi: 10.1007/s00335-004-2375-3. [DOI] [PubMed] [Google Scholar]
- 23.Massó A, et al. Secreted and Transmembrane αKlotho Isoforms Have Different Spatio-Temporal Profiles in the Brain during Aging and Alzheimer’s Disease Progression. PLoS ONE. 2015;10(11):e0143623. doi: 10.1371/journal.pone.0143623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohyama Y, et al. Molecular cloning of rat klotho cDNA: markedly decreased expression of klotho by acute inflammatory stress. Biochem Biophys Res Commun. 1998;251(3):920–925. doi: 10.1006/bbrc.1998.9576. [DOI] [PubMed] [Google Scholar]
- 25.Zünd D, Gruber AR, Zavolan M, Mühlemann O. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3’ UTRs. Nat Struct Mol Biol. 2013;20(8):936–943. doi: 10.1038/nsmb.2635. [DOI] [PubMed] [Google Scholar]
- 26.Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133(2):314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tucker Zhou TB, King GD, Chen C, Abraham CR. Biochemical and functional characterization of the klotho-VS polymorphism implicated in aging and disease risk. J Biol Chem. 2013;288(51):36302–36311. doi: 10.1074/jbc.M113.490052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esapa CT, et al. N-ethyl-N-Nitrosourea (ENU) induced mutations within the klotho gene lead to ectopic calcification and reduced lifespan in mouse models. PLoS ONE. 2015;10(4):e0122650. doi: 10.1371/journal.pone.0122650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koh N, et al. Severely reduced production of klotho in human chronic renal failure kidney. Biochem Biophys Res Commun. 2001;280(4):1015–1020. doi: 10.1006/bbrc.2000.4226. [DOI] [PubMed] [Google Scholar]
- 30.Hu MC, et al. Klotho deficiency causes vascular calcification in chronic kidney disease. J Am Soc Nephrol. 2011;22(1):124–136. doi: 10.1681/ASN.2009121311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu MC, Shi M, Zhang J, Quiñones H, Kuro-o M, Moe OW. Klotho deficiency is an early biomarker of renal ischemia-reperfusion injury and its replacement is protective. Kidney Int. 2010;78(12):1240–1251. doi: 10.1038/ki.2010.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindberg K, et al. The kidney is the principal organ mediating klotho effects. J Am Soc Nephrol. 2014;25(10):2169–2175. doi: 10.1681/ASN.2013111209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carter MS, et al. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270(48):28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 34.Kurosaki T, et al. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev. 2014;28(17):1900–1916. doi: 10.1101/gad.245506.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bloch L, et al. Klotho is a substrate for alpha-, beta- and gamma-secretase. FEBS Lett. 2009;583(19):3221–3224. doi: 10.1016/j.febslet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Loon EP, et al. Shedding of klotho by ADAMs in the kidney. Am J Physiol Renal Physiol. 2015;309(4):F359–F368. doi: 10.1152/ajprenal.00240.2014. [DOI] [PubMed] [Google Scholar]
- 37.Chen CD, et al. Identification of cleavage sites leading to the shed form of the anti-aging protein klotho. Biochemistry. 2014;53(34):5579–5587. doi: 10.1021/bi500409n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu MC, et al. Renal Production, Uptake, and Handling of Circulating αKlotho. J Am Soc Nephrol. 2016;27(1):79–90. doi: 10.1681/ASN.2014101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA. 2003;100(1):189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaillon O, et al. Translational control of intron splicing in eukaryotes. Nature. 2008;451(7176):359–362. doi: 10.1038/nature06495. [DOI] [PubMed] [Google Scholar]
- 41.Green RE, et al. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics. 2003;19 Suppl 1:i118–i121. doi: 10.1093/bioinformatics/btg1015. [DOI] [PubMed] [Google Scholar]
- 42.Smith RC, et al. Circulating αKlotho influences phosphate handling by controlling FGF23 production. J Clin Invest. 2012;122(12):4710–4715. doi: 10.1172/JCI64986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kato Y, et al. Establishment of the anti-Klotho monoclonal antibodies and detection of Klotho protein in kidneys. Biochem Biophys Res Commun. 2000;267(2):597–602. doi: 10.1006/bbrc.1999.2009. [DOI] [PubMed] [Google Scholar]
- 44.Abramovitz L, et al. KL1 internal repeat mediates klotho tumor suppressor activities and inhibits bFGF and IGF-I signaling in pancreatic cancer. Clin Cancer Res. 2011;17(13):4254–4266. doi: 10.1158/1078-0432.CCR-10-2749. [DOI] [PubMed] [Google Scholar]
- 45.Wolf MT, An SW, Nie M, Bal MS, Huang CL. Klotho up-regulates renal calcium channel transient receptor potential vanilloid 5 (TRPV5) by intra- and extracellular N-glycosylation-dependent mechanisms. J Biol Chem. 2014;289(52):35849–35857. doi: 10.1074/jbc.M114.616649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ligumsky H, et al. Tumor Suppressor Activity of Klotho in Breast Cancer Is Revealed by Structure-Function Analysis. Mol Cancer Res. 2015;13(10):1398–1407. doi: 10.1158/1541-7786.MCR-15-0141. [DOI] [PubMed] [Google Scholar]
- 47.Liu H, et al. Augmented Wnt signaling in a mammalian model of accelerated aging. Science. 2007;317(5839):803–806. doi: 10.1126/science.1143578. [DOI] [PubMed] [Google Scholar]
- 48.Chen B, Wang X, Zhao W, Wu J. Klotho inhibits growth and promotes apoptosis in human lung cancer cell line A549. J Exp Clin Cancer Res. 2010;29:99. doi: 10.1186/1756-9966-29-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang B, et al. Klotho inhibits the capacity of cell migration and invasion in cervical cancer. Oncol Rep. 2012;28(3):1022–1028. doi: 10.3892/or.2012.1865. [DOI] [PubMed] [Google Scholar]
- 50.Liu X, et al. Differential Regulatory Role of Soluble Klothos on Cardiac Fibrogenesis in Hypertension. Am J Hypertens. 2016;29(10):1140–1147. doi: 10.1093/ajh/hpw062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cha SK, Ortega B, Kurosu H, Rosenblatt KP, Kuro-OM, Huang CL. Removal of sialic acid involving Klotho causes cell-surface retention of TRPV5 channel via binding to galectin-1. Proc Natl Acad Sci USA. 2008;105(28):9805–9810. doi: 10.1073/pnas.0803223105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim JH, et al. Klotho May Ameliorate Proteinuria by Targeting TRPC6 Channels in Podocytes. J Am Soc Nephrol. 2017;28(1):140–151. doi: 10.1681/ASN.2015080888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsu SC, et al. Resveratrol increases anti-aging Klotho gene expression via the activating transcription factor 3/c-Jun complex-mediated signaling pathway. Int J Biochem Cell Biol. 2014;53:361–371. doi: 10.1016/j.biocel.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 54.Hsu SC, et al. Testosterone increases renal anti-aging klotho gene expression via the androgen receptor-mediated pathway. Biochem J. 2014;464(2):221–229. doi: 10.1042/BJ20140739. [DOI] [PubMed] [Google Scholar]
- 55.Sun CY, Chang SC, Wu MS. Suppression of Klotho expression by protein-bound uremic toxins is associated with increased DNA methyltransferase expression and DNA hypermethylation. Kidney Int. 2012;81(7):640–650. doi: 10.1038/ki.2011.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Irifuku T, et al. Inhibition of H3K9 histone methyltransferase G9a attenuates renal fibrosis and retains klotho expression. Kidney Int. 2016;89(1):147–157. doi: 10.1038/ki.2015.291. [DOI] [PubMed] [Google Scholar]
- 57.King GD, Rosene DL, Abraham CR. Promoter methylation and age-related downregulation of Klotho in rhesus monkey. Age (Dordr) 2012;34(6):1405–1419. doi: 10.1007/s11357-011-9315-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He XJ, et al. Up-regulated miR-199a-5p in gastric cancer functions as an oncogene and targets klotho. BMC Cancer. 2014;14:218. doi: 10.1186/1471-2407-14-218. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Mehi SJ, Maltare A, Abraham CR, King GD. MicroRNA-339 and microRNA-556 regulate Klotho expression in vitro. Age (Dordr) 2014;36(1):141–149. doi: 10.1007/s11357-013-9555-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolf I, et al. Klotho: a tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast cancer. Oncogene. 2008;27(56):7094–7105. doi: 10.1038/onc.2008.292. [DOI] [PubMed] [Google Scholar]
- 61.Zhou X, et al. Klotho, an anti-aging gene, acts as a tumor suppressor and inhibitor of IGF-1R signaling in diffuse large B cell lymphoma. J Hematol Oncol. 2017;10(1):37. doi: 10.1186/s13045-017-0391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lojkin I, et al. Reduced expression and growth inhibitory activity of the aging suppressor klotho in epithelial ovarian cancer. Cancer Lett. 2015;362(2):149–157. doi: 10.1016/j.canlet.2015.03.035. [DOI] [PubMed] [Google Scholar]
- 63.Yan Y, Wang Y, Xiong Y, Lin X, Zhou P, Chen Z. Reduced Klotho expression contributes to poor survival rates in human patients with ovarian cancer, and overexpression of Klotho inhibits the progression of ovarian cancer partly via the inhibition of systemic inflammation in nude mice. Mol Med Rep. 2017;15(4):1777–1785. doi: 10.3892/mmr.2017.6172. [DOI] [PubMed] [Google Scholar]
- 64.van Ark J, et al. Circulating alpha-klotho levels are not disturbed in patients with type 2 diabetes with and without macrovascular disease in the absence of nephropathy. Cardiovasc Diabetol. 2013;12:116. doi: 10.1186/1475-2840-12-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Esposito AM, et al. Eukaryotic polyribosome profile analysis. J Vis Exp. 2010;(40):1948. doi: 10.3791/1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aslan A, et al. The renal angiopoietin/Tie2 system in lethal human sepsis. Crit Care. 2014;18(2):423. doi: 10.1186/cc13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.