Abstract
The cerebral cortex is composed of billions of morphologically and functionally distinct neurons. These neurons are produced and organized in a regimental fashion during development. The ability of neurons to encode and elicit complex cognitive and motor functions depends on their precise molecular processes, identity, and connectivity established during development. Elucidating the cellular and molecular mechanisms that regulate development of the neocortex has been a challenge for many years. The cerebral cortical neuronal subtypes are classified based on morphology, function, intrinsic synaptic properties, location, connectivity, and marker gene expression. Development of the neocortex requires an orchestration of a series of processes including the appropriate determination, migration and positioning of the neurons, acquisition of layer-specific transcriptional hallmarks, and formation of precise axonal projections and networks. Historically, fate mapping, genome-wide analysis, and transcriptome profiling have provided many opportunities for the characterization of neuronal subtypes. During the course of this review, we will address the regimental organization of the cerebral cortex, dissect the cellular subtypes that contribute to cortical complexity, and outline their molecular hallmarks to understand cellular diversity in the cerebral cortex with a focus on the excitatory neurons.
Keywords: Brain development, neural stem cells, neurogenesis, cerebral cortex
Neuroepithelium to Neural Stem Cell Transition and Beyond
Early during vertebrate embryonic development, neural fate is induced in the ectoderm.1 The consequent patterning of the neural plate results in the formation of the central nervous system. The process of neurulation induces formation of the neural tube, a pseudostratified epithelial sheet of neuroepithelial cells (NEPs). It is the NEPs that are the precursors of the central nervous system including cerebral cortex, which is formed over an extended period of development. Important biological questions remain about how the complex structure of the cerebral cortex, which is composed of diverse neuron subtypes, is generated from a simple epithelial sheet of cells to form the most complex tissue of the body. At embryonic day 9 (E9), the neuroepithelium gives rise to neural stem cells (NSCs) that line the luminal surface of the vesicles of the neural tube.2,3 In mice, NSCs are located in the ventricular zone (VZ) and the ends of their basal processes remain in contact with the outer (pial) surface of the neural tube. This apical-basal polarity, which spans the thickness of the neural tube, requires the integrity of adherens junctions to segregate the apical and basolateral cell membrane and adhere neighboring NSCs to each other. The importance of adherence in NSC polarity is exemplified by the knockdown of the adherens junction–associated protein Afadin (Af6). Af6 depletion leads to a loss of adherens junctions and disturbed cell polarity.4
At the onset of neurogenesis, the NEPs generate radial glial cells (RGCs) and short neural precursors.5,6 The somata of these cells remain within the VZ but migrate radially along the apical-basal process through the zone during cell division in a process referred to as interkinetic nuclear migration (INM).3,7–9 The location of the soma within the VZ is cell cycle dependent. During M-phase, the cell body is positioned apically at the luminal surface of the neural tube (Figure 1). As the cell progresses through G1-phase of the cell cycle, the cell body moves radially to the VZ boundary with the overlying subventricular zone (SVZ) and forming cerebral cortex. S-phase and DNA replication occur at the basal boundary of the VZ followed by migration of the cell body back to the luminal surface of the neural tube during G2 to initiate mitosis.7,10,11 Primary cilia in the apical membrane project into the vesicles and detect factors and signals in the fluid filling the neural tube and these support apical-basal polarity. The orientated cell polarity is important for determining the structure of the cerebral cortex. Disruption of the small GTPase, ADP ribosylation factor–like GTPase 13B (Arl13b), results in loss of cell polarity and the cortical wall is generated in an inverted fashion. M-phase of Arl13b-deficient RGCs is no longer restricted to the luminal surface but also occurs at the basal, pial surface and neurons migrate centripetally to the VZ.12
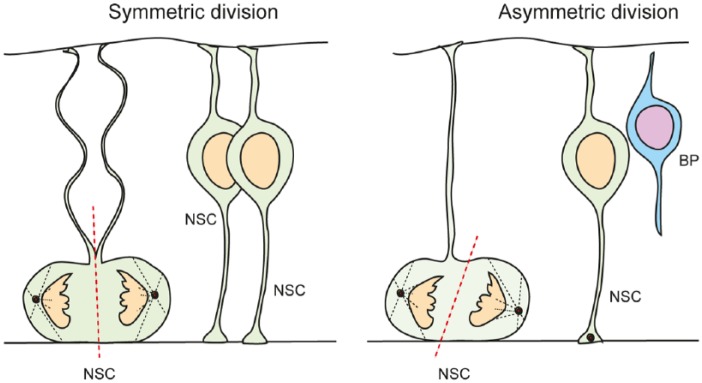
Figure 1.
Types of NSC divisions in the ventricular zone are determined by spindle orientation and the inheritance of cell fate determinants. Symmetric divisions generate 2 NSCs, whereas asymmetric division generates 1 NSC and 1 differentiating daughter cell. During neural expansion, most divisions are symmetric, whereas during neurogenesis, most divisions are asymmetric. BP indicates basal progenitor; NSC, neural stem cell.
During early phases of neurogenesis, embryonic days 10.5 to 11.5 (E10.5-11.5) in mice, NSCs undergo symmetric stem cell divisions, expanding the pool (Figure 1). This is referred to as the “neural expansion” phase of cortical development. Later, NSCs progressively undergo asymmetric cell divisions, allowing for both self-renewal and the generation of committed daughter cells (Figure 1). The transition from symmetric stem cell to asymmetric neurogenic divisions during neurogenesis is associated with a lengthening of primarily G1-phase of the cell cycle. However, the S-phase of the NSCs in the symmetric dividing, expansion phase is longer than of those in the asymmetric dividing neurogenic phases.7,13 Hence, although the precise function of INM and the changing in cell cycle phase length are not understood, it seems that they play an important role on the control of the sequential switching of NSCs from a symmetric self-renewing mode to the asymmetric division mode that drives the production of neurons.
During the neurogenic phase of cortical development, the self-renewal and generation of committed daughter cells have to be tightly controlled. Loss of self-renewing NSC daughter cells would purge the stem cell pool. Conversely, a failure to generate sufficient neuronal-determined precursors would severely affect neuronal composition and cortical layering. During early stages of cortical development, some asymmetric stem cell divisions generate one NSC daughter and a neuron directly. This is referred to as direct neurogenesis. However, as neurogenesis progresses, the daughter cell that is committed to differentiate and leaves the stem cell pool becomes a basal progenitor (BP) and migrates to the forming SVZ (Figure 1).7,13,14
Distinct Stem and Progenitor Populations Contribute to Cortical Development
Throughout neurogenesis, another VZ population of dividing cells called the short neural precursors contributes to the progenitor pool. Short neural progenitors have either a short or no basal process at all but retain the apical process and contact to the lumen of the neural tube. These cells are morphologically, ultrastructurally, and molecularly different from the NSCs and have been observed to undergo direct neurogenesis, generating neurons without passing through a BP state.5
In higher mammals including ferrets, primates, and humans, additional intermediate progenitor populations have evolved, and although they also reside in the SVZ, they have different morphologies and larger cell fate potentials compared with the classic BPs in mice.6,15,16 In fact, in primates, some of these intermediate progenitors even display NSC potential and are even referred to as outer RGCs (oRGCs).16 The oRGCs are morphologically distinct, unipolar, and retain only the basal process with no connection to the VZ and neural tube lumen (Figure 2).16–18 They also do not express the apical membrane constituents associated with VZ NSCs and RGC including prominin1 (CD133), Par3 family cell polarity regulator (Par3), or atypical protein kinase Cλ (aPKCλ).15 They have a long basal phospho-Vimentin (pVim)-positive process that extends toward the pia and retain the basal fiber throughout the duration of cell cycle.10 Similar to VZ RGCs, the soma of oRGCs also moves during cell divisions but this movement is distinct to the INM of VZ NSCs. The soma of oRGCs moves basally and once translocation is complete, they divide mostly by self-renewing, asymmetric divisions, and push the boundary of the outer SVZ (OSVZ) outward expanding the SVZ (Figure 2).16,19 Self-renewing oRGCs continue to proliferate, whereas the daughter cells differentiate into neurons.
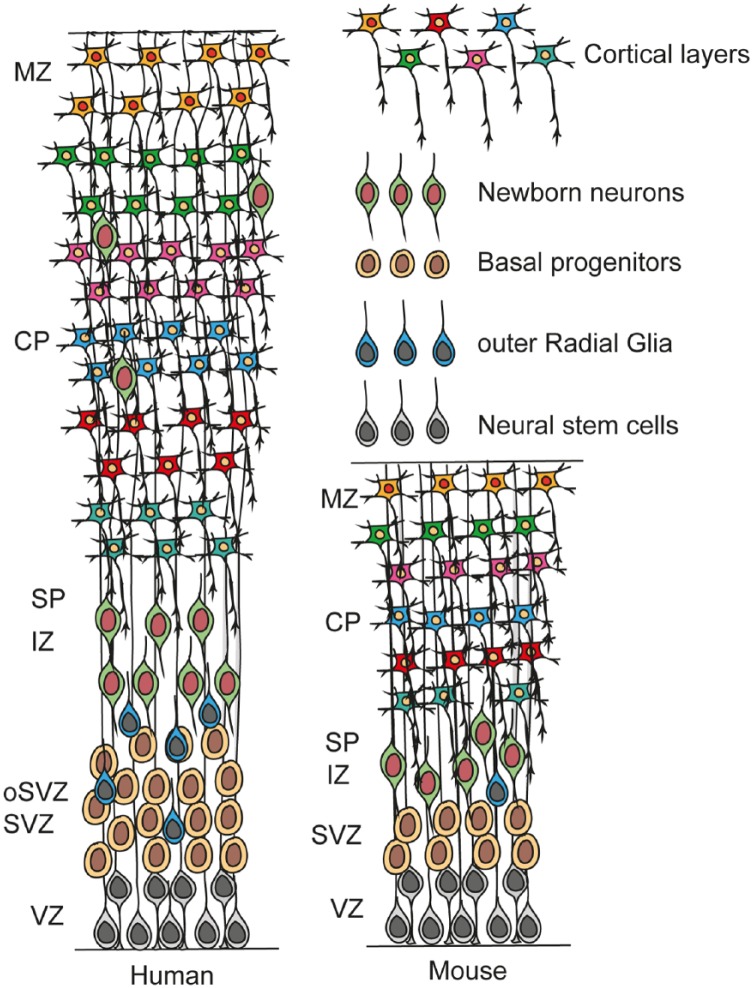
Figure 2.
Scheme illustrating the composition and laminar organization of the developing human cortex, in comparison with mouse cortex. The human cerebral cortex develops in a similar fashion to that of the mouse. One exception is the expansion of the subventricular zone (SVZ) to form the outer SVZ (oSVZ). The oSVZ in humans is the main zone of amplification. In addition to the neural stem cells (NSCs) and basal progenitors (BPs) of the developing mouse cerebral cortex, the human has addition progenitors, outer radial glial cells. CP indicates cortical plate; IZ, intermediate zone; MZ, marginal zone; SP, subplate; SVZ, subventricular zone; VZ, ventricular zone.
These SVZ progenitors in primates are the major source of expansion and neurogenesis in the cerebral cortex and are responsible for the massive evolutionary expansion of the cortical gray matter, neuron number, and cortical surface. Indirectly, these SVZ progenitors are responsible for the increase in functional capacity of the cerebral cortex in primates.20 The coexistence of oRGC cells and VZ RGCs demonstrates the distinct germinal zones in higher mammals, highlighting the mechanisms of increased neuron production, relevant for the formation of bigger brains (Figure 2). Here, we will focus on cortical development in the mouse and refer to excellent review focusing on primate and human cortical development.10
Basal progenitors are intermediate, transient amplifying cells that undergo 1 or 2 divisions before giving rise to neurons (Figure 2). The BPs are in one of the main zones of amplification and neurogenesis in the developing mouse cortex. As neurogenesis reaches completion, the NSCs start to generate other cell lineages, oligodendrocytes, astrocytes, and ependymal cells.21,22 This is referred to as the “gliogenesis phase” of cortical development. The transition from neurogenesis to gliogenesis is associated with a downregulation of the Golgi-derived apical trafficking and VZ NSCs lose tight junctions while keeping intact the adherens junctions.23,24 This is followed by the gradual expression of the astroglial hallmarks including glial fibrillary acidic protein (GFAP) in the mouse.22,24–26 Although the mechanisms of the neurogenic to gliogenic phase transition are not clearly understood, Notch signaling and its downstream targets, the bHLH (basic helix-loop-helix) transcription factors including the Hes proteins, and the growth factor Fgf10 are necessary for this transition.27–29
Because the generation of neurons from BPs results in the expansion of the neuronal progenitor pool enabling the production of many neurons from a restricted population of NSCs, their role is crucial in the expansion of the cortex.28,30,31 In the mouse, BPs can undergo symmetric divisions and generate 2 neuronal daughter cells.31 However, evidence suggests that some, if not all, may also undergo 1 or 2 rounds of self-renewing cell divisions.28,30,31 Basal progenitors are defined based on their position in the SVZ, their lack of polarized morphology and expression of the transcription factors, Eomesodermin (Eomes or Tbr2), Btg antiproliferation factor 2 (also called Tis21), Cut-like homeobox 1/2 (Cux1/Cux2), and special AT-rich sequence binding protein (Satb2) and the non-coding RNA Svet1.32–37 Because the different progenitor cell types are localized to different niches and thus likely exposed to different combinations of cues from their microenvironment, it is imperative to study the role of their niche in controlling their proliferation and fate commitment. This cellular heterogeneity requires a deeper understanding of the cell fate identities and commitments.6
Symmetric and Asymmetric Cell Divisions
Neural stem cells of the developing cerebral cortex display multiple modes of cell division. Initially, the major form of divisions is symmetric stem cell divisions, generating 2 daughter cells that retain stem cell potential and reenter cell cycle. As development progresses, the stem cell divisions are slowly superseded by asymmetric neurogenic divisions where 1 daughter remains a stem cell and reenters the cell cycle within the VZ, whereas the other is committed to differentiate and will leave the VZ (Figure 1). The third mode is the symmetric neurogenic division where both daughter cells will differentiate thereby depleting the stem cell pool. The balance between these different forms and outcomes of cell division are temporally and spatially regulated which is necessary to control correct cortical development.
The molecular basis of symmetric and asymmetric divisions and the transition from self-renewing to differentiating modes of cell division are not understood. It has become clear that the orientation of the mitotic spindle plays an important role in the type of division and the fate of the respective daughter cells generated (Figure 1). A cleavage plane bisecting the apical membrane of the NSCs, including inheritance of junctional complexes by both daughters, contributes heavily to maintenance of stem cell potential (Figure 1). During symmetric divisions of NSCs, the cleavage plane is oriented perpendicular to the ventricular surface (Figure 1).38 This spatial organization of the mitotic spindle requires a proper centrosome assembly, duplication and a precise interaction between planar cell polarity components, G protein signaling modulator 2 (Lgn), and Inscuteable (Insc).13,39–41 The partition of cell components involved in cell polarity, including the Par3 family cell polarity regulator (Par3/Par6), proteins between daughter cells is critical for differential cell fate determination. In symmetric stem cell divisions, the basal process is equally split between the daughter cells (Figure 1).42,43 The transcription factor empty spiracles homologue 2 (Emx2) is expressed by NSCs of the VZ and promotes perpendicular cleavage plane thereby promoting symmetric expansive cell divisions.44 Forced Emx2 expression in NSCs during cortical development increases clonal expansion and symmetric cell divisions.44
During asymmetric cell divisions, the cleavage plane is orientated parallel to the neural tube luminal surface (Figure 1). This results in an unequal partition of Par3 into the 2 daughter cells, and the sibling cell receiving less Par3 protein exits cell cycle and differentiates.42,43 In addition, asymmetric cell division is accompanied by an unequal distribution of fate determinants between the daughter cells. These components include mediators of Notch signaling, the Notch ligand delta-like 1 (Dll1), Mind bomb, and Numb.45,46 Segregation of Notch components including inhibitors of the pathway leads to differential Notch signaling between daughter cells. Notch signaling plays a critical role in NSC maintenance and differentiation by regulating cell proliferation and fate determination.45,47,48 Notch activates the expression of Hes genes which encode bHLH transcriptional regulators. Hes-related proteins repress expression of the proneurogenic transcription factors including neurogenins (Ngns) and Ascl1.49,50 Thus, activation of Notch signaling inhibits differentiation of NSCs by suppressing transcription factors required for neurogenesis.48 In addition, Notch signaling regulates cell cycle progression via regulation of Ascl1 expression. Ascl1 not only controls neurogenic differentiation but is also involved in entry of NSCs into cell cycle.51,52
In addition to Notch, some cytoplasmic proteins show differential distribution on asymmetric division. Staufen is a double-stranded RNA–binding protein which is pivotal in asymmetric cell fates in Drosophila neural development. Staufen is selectively segregated into the differentiating daughter cells on asymmetric self-renewing cell division.53 Staufen binds messenger RNAs that encode proteins crucial in cell cycle exit and differentiation. Furthermore, the transcription factor Pax6 promotes asymmetric neurogenic cell division.54 Pax6-mutant NSCs show a defective cell cycle exit and an increase in self-renewing capacity.54
In addition to molecular segregation, the orientation of the mitotic spindle plays an important role in fate determination. In NSCs, the mitotic spindle poles oscillate around their final positions before anaphase is initiated. This dynamic movement of the spindle seems to be important in determining the cleavage plane and then the segregation of intracellular components. Only subtle changes in spindle orientation can cause major shifts in the plane of cytokinesis and thereby the inheritance of membrane compartments and cell fate determinants.55 Mutations in the abnormal spindle-like microcephaly-associate (Aspm) gene severely affect cerebral cortical size and reduce the volume of the cerebral cortex in primates.56 Aspm is important for spindle orientation and control in the division modes of symmetric versus asymmetric cells.
Inheritance of the apical plasma membrane of NSCs has an influence on cell fate. During symmetric cell divisions, both daughters acquire apical membrane and junctional components. When only 1 daughter cell inherits the apical plasma membrane, for example, when the cleavage plane is parallel to the neural tube luminal surface, that daughter remains as an NSC, whereas the other sibling that does not receive apical membrane and adherens junctions from the mother cell will exit the VZ and commit to differentiation. The SNARE-mediated membrane fusion machinery controls NSC fate specification. A hypomorphic missense mutation in α-SNAP (α-soluble N-ethylmaleimide-sensitive fusion protein [NSF] attachment protein) causes NSCs to prematurely switch from symmetric proliferative to asymmetric neurogenic divisions.57 This is primarily due to an impaired apical protein localization affecting the Golgi-derived membrane traffic necessary for NSC proliferation.58 In addition, NSCs in these mice show distribution of apical β-catenin along the adherens junctions and phenocopying of conditional β-catenin null mutant mice. Hence, β-catenin plays a role not only in the control of cell cycle but also in the choice between symmetric and asymmetric divisions.59,60
Regulation and Cell Fate Commitment
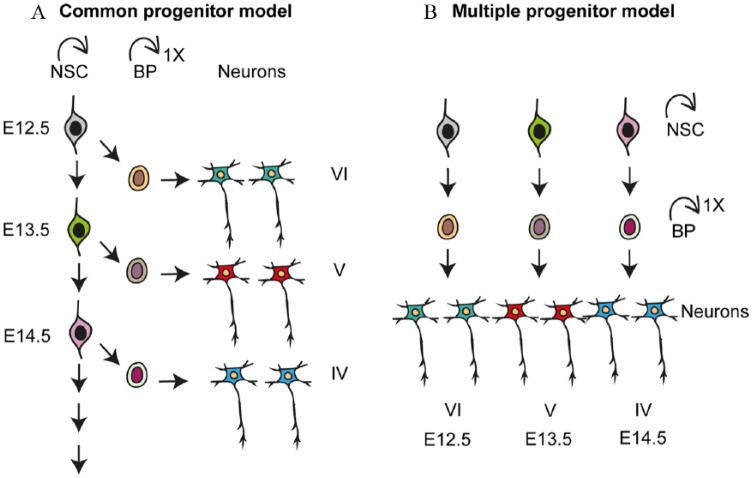
The stem and progenitor cells in the dorsal VZ of the anterior neural tube generate the multiple classes of projection neurons that make up the future cerebral cortex in sequential waves. In the dorsal cerebral cortex, neurogenesis commences around E10.5 in mice.25,26 The earliest born neurons segregate from the NSCs in the VZ and migrate radially to the pial surface forming the preplate. Later born neurons migrate into the preplate, splitting it into the marginal zone and the subplate (Figure 2). Throughout neurogenesis, newborn neurons migrate into the cortical plate (CP), through the preformed layers of earlier born neurons, and as such the early born neurons form the deep layers and the later born neurons form upper layers. The detailed molecular cascade that determines neuronal cell fate commitment, an excitatory neuron subtype specification, is largely unknown. Different models have been proposed to explain the temporal dynamics of neuronal specification in the dorsal cortex.61 The “common progenitor model” implies that a single type of NSC sequentially gives rise to the different neuron subtypes overtime during neurogenesis and that neuron fate is determined by time (Figure 3A). According to the “multiple progenitor model,” multiple stem cell types coexist and are predetermined to generate specific neuron subtypes.62 In the multiple progenitor model, the fate of the stem cell and the type of neuron generated are determined by the NSC type (Figure 3B). There is experimental evidence supporting both models.23,63
Figure 3.
Different models of neuronal subtype specification in developing neocortex. (A) In the common progenitor model, a single type of multipotent NSC sequentially gives rise to all neuronal subtypes during the course of development. Overtime, the fate potential of this NSC becomes increasingly restricted. Fate of the neuron is specified based on its birth date. (B) In the multiple progenitor model, multiple types of NSCs coexist and are, to some degree, predetermined to give rise to specific and restricted neuronal subtypes. Fate of the neuron is specified by the NSC type in this model. BP indicates basal progenitor; NSC, neural stem cell.
The Common Progenitor Model
McConnell demonstrated that NSC fate becomes restricted over time during development.64 By performing elegant heterochronic transplantation experiments initially in ferrets, they demonstrated that early developmental stage progenitors have a greater potential and can generate early and late neuronal subtypes when grafted into hosts. Conversely, late-stage progenitors have a more restricted potential and a reduced capacity to form early neuronal types in host embryos.64 This implies that NSCs lose their potential to generate deep layer neurons with time.64–66 In support of this model, clonal in vitro experiments showed the sequential generation of deep and upper layer neurons from NSCs supporting initial multipotency and subsequent fate restriction over time.64,67–69 Finally, retroviral labeling and lineage tracing of individual NSCs supported progressive fate restriction in vivo.70 More recently, genetic labeling in the developing mouse cerebral cortex following expression of the transcription factor Fezf2 (enriched in deep cortical layer V neurons) revealed that Fezf2-expressing NSCs generate deep, upper layer neurons and glial cells.63,71,72 Instructive roles of factors such as Fezf2 in NSCs can have major implications in cell fate commitment during neurogenesis. Ectopic expression of Fezf2 can direct NSCs to differentiate into deep layer neurons and reverse late fate commitment.63,73
The Multiple Progenitor Model
An alternative model for fate specification proposes independent, fate-restricted lineages of NSC that generate specific neuronal subtypes and have limited potentials (Figure 3B). Evidence suggests that many transcription factors expressed during cortical development instruct fate determination.72 The onset of expression of these transcription factors was proposed to coincide with the developmental time point at which specific neuronal subtypes are determined, indicative of the presence of predetermined NSC subtypes.62 Analysis of transgenic mice revealed that Cux1 and Cux2 are expressed in VZ and SVZ as early as E10.5, primarily in specific and fate-restricted NSCs. Genetic tracing of Cux1-positive progenitor cells mostly generated upper layer neurons.23,74 During early development, Cux1- and Cux2-positive NSCs undergo expansion and do not contribute to early layer neuronal differentiation.23,74 These seem to be restricted in fate while they undergo neurogenesis and produce only upper layer neurons. Conversely, follow-up experiments analyzing Cux2-positive cells by lineage tracing elucidated their role in generating both deep and upper layer neurons as well as the interneurons from the ventral telencephalon.63,66
Other experiments imply the coexistence of multipotent NSCs and their consequent fate restriction through the course of neurogenesis.23,75 It is possible that cells can be more restricted in their potential and change to alternate fates when subjected to different extrinsic cues. This may explain the switch between multipotent to restricted NSC states. Because the precise structure of the lineage trees for specific neuronal subtypes remains largely unknown in vivo, single-cell clonal analysis to identify markers of clusters of NSCs may contribute to this understanding of cell fate commitment. Both the “common and multiple progenitor models” do not negate the possibility of the other, and future high-resolution experiments are needed at the single-cell level to address NSC heterogeneity and dynamic potential.
Neuronal Diversity and Transcriptional Dynamics in Cortical Layering
The cerebral cortex is an isocortex composed of 6 clearly defined layers. The newborn excitatory neurons migrate out of the VZ along the radial processes of the NSCs (Figures 4 and 5). The immature neurons reach the CP by migrating through their earlier born siblings. On reaching the pial surface, the immature neurons leave the RGC process and differentiate and form neurons of their specific cortical layer. Hence, the isocortex of the cerebrum is formed in an inside-out fashion, with early born neurons forming the deep layers while the later born neurons generating the upper layers (Figure 5). The neuronal type and their location within the isocortex are critical for function. The interneurons of the cerebral cortex originate from the ventral telencephalon and migrate to their final destination in the cerebral cortex (we refer the reader to excellent recent reviews on cortical interneuron development and will not cover the topic here).76,77
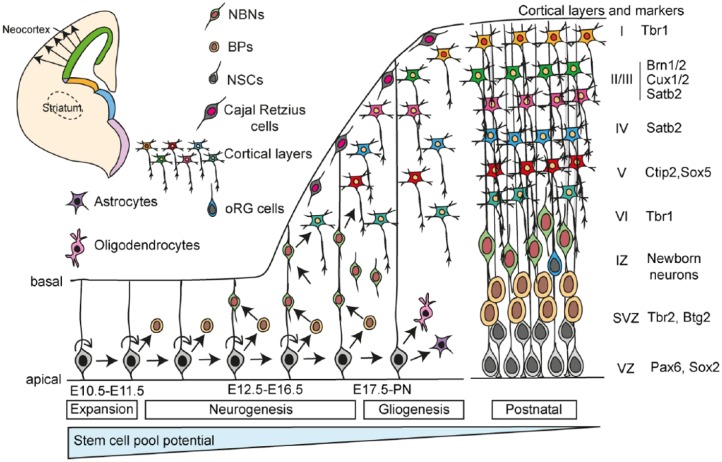
Figure 4.
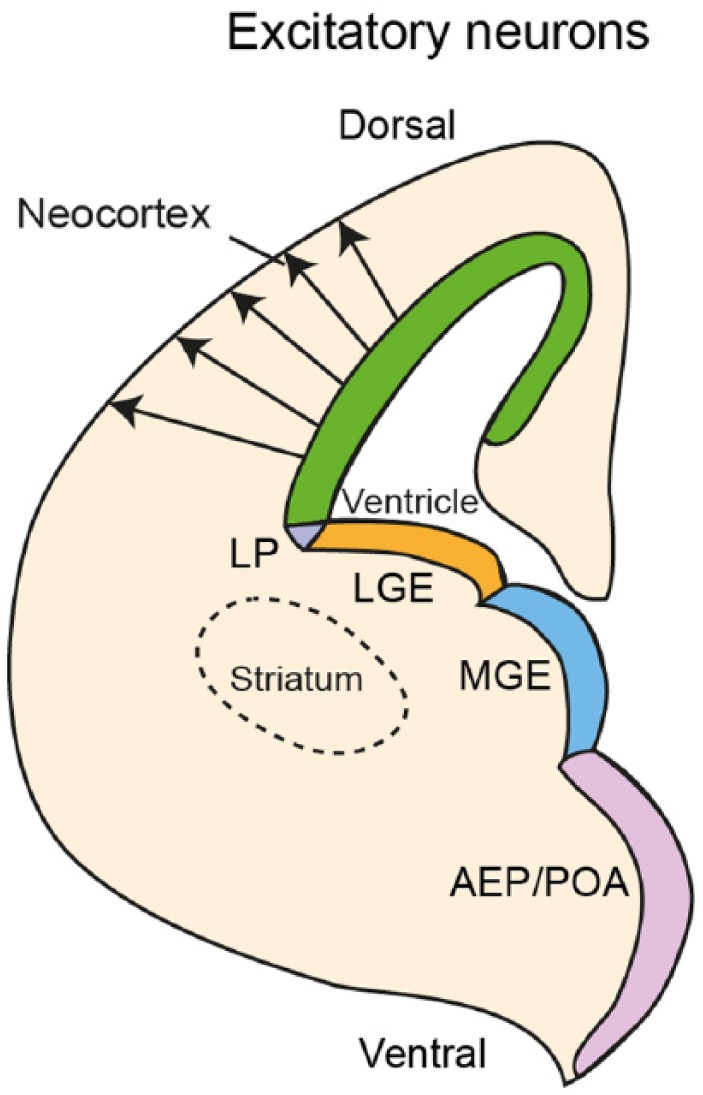
Origin of excitatory projection neurons of the cerebral cortex. Excitatory projection neurons originate from the ventricular zone of the dorsal telencephalon and migrate radially to the cortical plate. Inhibitory interneurons originate from the ventral telencephalon, especially from the MGE, AEP/POA.76,77 AEP/POA indicates anterior entopeduncular area of the subpallium/preoptic area; LGE, lateral ganglionic eminence; LP, lateral pallium; MGE, medial ganglionic eminence.
Figure 5.
Systematic formation of isocortex layers in the dorsal telencephalon. During early stages of cerebral cortical development (embryonic days E10.5-E11.5), NSCs predominantly undergo symmetric cells divisions to expand the NSC pool. This phase is referred to as the expansion phase. The first neurons to be formed are generated by direct neurogenesis of the NSCs. The Cajal-Retzius cells populate layer I of the isocortex and play important roles in establishing cortical architecture. During late embryogenesis (E12-E16.5), NSCs undergo increasingly more asymmetric divisions to generate 1 NSC (self-renewal) and 1 BP. The BPs generate the neurons. This is the neurogenic phase. Neurons are generated in a sequential, inside-out fashion and are specified by different transcription factors, some of which are shown. At later stages of development, NSCs generate the other cell types of the brain including astrocytes, oligodendrocytes, and ependymal cells (not shown). This is referred to as the gliogenic phase. The potential of the NSC pool reduces over time during development. This does not exclude that multiple restricted stem cells become activated and are lost at different times during cortical development. BPs indicate basal progenitors; IZ, intermediate zone; NBNs, newborn neurons; NSCs, neural stem cells; SVZ, subventricular zone; VZ, ventricular zone.
The major types of cortical projection neurons can be defined by their connectivity and projection patterns depending on whether they project through associative, commissural, and corticofugal projection fibers. Associative projection neurons project their axons within a single cerebral hemisphere connecting local areas or proximal gyri. Commissural, callosal projection neurons are localized primarily in layers II/III, V, and VI of the 6-layered isocortex. They extend their axons from 1 hemisphere to neurons in the contralateral hemisphere. The axons project through the corpus callosum, the major commissural connection between the hemispheres, or through the anterior or posterior commissures. The commissural neurons are further subdivided based on the projection destinations.72 Corticofugal neurons include the subcerebral neurons, which are the largest pyramidal neurons and extend projections to subcortical structures including the brainstem and spinal cord. The corticofugal projections include corticopontine, corticospinal, and corticotectal neurons.78 Another subtype of corticofugal neurons is the corticothalamic neurons, which populate the layer VI of the isocortex with a small population in layer V and extend their projections to different nuclei of the thalamus.72
Hence, the regulation of neuron subtype formation and the temporospatial control of neurogenesis are critical for brain function. Numerous neocortical determinants are expressed along the dorsolateral wall of the cortex, on the induction of neurogenesis. Key factors including forkhead box G1 (FoxG1), LIM homeobox 2 (Lhx2), Emx2, and Pax6 define and control the neocortical progenitor domains along the dorsal and ventral axis.24,72 Ablation of the dorsal progenitor domain determinants Pax6 and Emx2 results in expansion of the ventral domains.79 Pax6 and T-cell leukemia homeobox (Tlx) regulates the cell fate decisions in the VZ and the loss of function of these factors leads to a thicker superficial cortex. Hence, the NSCs and progenitors of the cerebral cortex are determined by their expression of axial-specifying factors but these fates are not fixed or restricted as loss of these determinants results in alternate fate acquisition.72
The transcription factors Tbr1, Ctip2, Sox5, Fezf2, Satb2, Cux1, Cux2, Brn1, Brn2, and others have been studied extensively and determined to be key determinants of neuronal specification.7,10,23,32,33,35,62,63,71,73 These transcription factors are often used as markers of specific cortical neuron populations and layers and are expressed in waves during cortical development. Some of these markers are expressed in specific neuronal subtypes within a layer or are expressed in more than one neuronal subtype and layer. For example, the Ets-related protein 81 (Er81/Etv1) is expressed in cortico-cortical and subcerebral projection neurons of layer V.80 Conversely, LIM domain only 4 (Lmo4) is selectively expressed in callosal neurons of layers II/III.
Some of these fate-determining and defining factors seem to be coexpressed initially and their expression becomes restricted and refined later in neuronal differentiation. For example, in mice, postmitotic deep layer neurons coexpress Ctip2 and Satb2 at E13.5.35,72,81 As development progresses, these deep layer neurons express either Ctip2 or Satb2 and become fate restricted to form subcerebral projection neurons or corticothalamic projection neurons, respectively.35,72,82,83 Ngn1 and Ngn2 are 2 proneural transcription factors and induce neurogenesis; however, Tbr1- and Er81-expressing deep layer neurons are still generated in their absence.84,85 It is likely that other proneural transcription factors are able to compensate for the loss of Ngns but the exact mechanism remains to be defined.
Epigenetic and Transcriptional Interplay During Neurogenesis
The gene expression in NSCs regulated not only by transcription factors but also by epigenetic mechanisms. DNA methylation and histone modifications are involved in spatial and temporal gene expression during neurogenesis and the switch from neuronal to glial fate.86–88 New methods for genome-wide methylation mapping facilitates investigation of epigenetic landscapes that control lineage commitments and fate decisions during neuronal specification. Epigenetic regulation of critical transcription factors in NSCs play important roles in the regulation of cell fate and neurogenesis. The expression of epigenetic regulators including the high-mobility group (HMG) proteins during early phases of cortical development regulate chromatin state and methyltransferase activity including enhancer of zeste 2 polycomb repressive complex 2 subunit (Ezh2).86–88 This suggests that the chromatin in early NSCs is in a more “open” state than that of later NSCs.87 The transcription factor Tbr2, which is critical for BP generation and differentiation, associates with the histone demethylase Jmjd3 (also called Kdm6b). Tbr2 directs Jmjd3-dependent chromatin remodeling to specific gene loci promoting the removal of H3K27me3 chromatin marks.89 This emphasizes the additional level of control that Tbr2 has on neuronal specification by regulating the epigenetic marks at specific promoter and enhancer sites.89
Hes5 is a pivotal mediator of Notch signaling and inducer of maintenance of NSCs by blocking proneural transcription factor expression. However, the expression of Hes5 also depends on glial cell missing homolog (Gcm) and active DNA demethylation during neurogenesis.90 Loss of Gcm prevents upregulation of Hes5 and the formation of definitive NSCs between E7.5 and E8.5.90 Pax6 interacts with BAF155 and BAF170, components of the ATP (adenosine triphosphate)–dependent multi-subunit mSWI/SNF nucleosome remodeling complexes in NSCs. At the onset of neurogenesis, BAF155 and BAF170 compete and modify euchromatin structure.91,92 This leads to the recruitment of the Pax6/RE1 silencing transcription factor (REST)-corepressor complex to the Pax6 targets, transducin-like enhancer of split 1 (Tle), Eomes, and Cux2, and repressing their expression. This prevents the formation of BPs and late NSCs.
During peak of neurogenesis, the chromatin remodeler sucrose nonfermenting–like protein 1 (Snf2l) represses expression of FoxG1, which leads to the derepression of the cell cycle regulator p21 and promotes neuronal differentiation by inducing cell cycle exit.91,92 During later stages of neurogenesis, the polycomb proteins repress Ngn1 expression to trigger the NSC fate switch from neurogenesis to astrogliogenesis.86 The NSC switch to gliogenesis is associated with the expression of the astrocytic protein GFAP. DNA methylation of the Gfap promoter prevents its premature activation. Notch signaling induces demethylation of the Gfap promoter through the transcription factor nuclear factor I (NFI), by dissociating associated DNA methyltransferases and thereby supports generation of astrocytes.28,93,94 Further analysis of the interplay between epigenetic and transcriptional dynamics during cortical development may contribute to a greater understanding of novel mechanisms and dysregulation during brain disorders.
Signaling Dynamics During Neurogenesis
Various signaling pathways impinge on downstream effectors and regulate NSC fate decisions during neurogenesis. Among these pathways are Notch, Wnt, Shh, fibroblast growth factors (FGFs), transforming growth factor β (TGF-β), retinoic acid (RA), and Hippo. How the cross talk between these signaling pathways and the integration of their signals on target genes governs complex cell fate choices is unclear.
Notch was first discovered in Drosophila, and the signaling pathway is evolutionarily highly conserved.95 It plays critical roles in NSC maintenance and differentiation. Conditional loss of Notch receptors results in the precocious differentiation, impaired survival, and aberrant migration of NSCs.45,48 Notch signaling regulates neurogenesis through lateral signaling between neighboring cells. In the presence of Notch ligands, delta-1 (Dll1) and Jagged1 (Jag1), the receptor is cleaved at S2 and S3 sites by ADAM metallopeptidase domain 10 (ADAM10) and γ-secretase, respectively.96,97 This results in the release of the Notch intracellular domain (NICD). The NICD has nuclear localization signals and mediates the transcription of Hes genes via DNA-binding factor recombination signal-binding protein for immunoglobulin kappa J region (RBPJ) complex.46,98 On interaction of NICD with RBPJ, the preformed repressor complex on target genes is disrupted and replaced by a coactivator complex including mastermind-like protein-1 (Maml) and histone acetyltransferase (HAT, p300). These changes facilitate the recruitment of RNA polymerase II and initiate transcription of target genes including Hes1 and Hes5.99 The Hes factors in turn downregulate expression of the genes encoding the proneural bHLH factors Ngn and Achaete-Scute family BHLH transcription factor 1b (Ascl).100,101 Hence, active Notch signaling maintains NSCs in a proliferative state and represses differentiation in the mammalian brain.49,101,102 The expression of Hes1 and Hes5 oscillates in the VZ due to an auto-inhibitory feedback loop.28,103 The oscillations in the expression of the Hes factors in turn induce oscillatory expression of proneural factors.50,97,104 This dynamic expression seems to be critical in modulating the functions of the proneural transcription factors in controlling proliferation and neurogenic differentiation.49,101,102 Although the source of Notch ligands is not clear, NSCs, BPs, and newborn neurons express Deltas and Jaggeds as well as the E3 ligase Mind bomb that potentially promote Notch signaling in the NSCs.105,106
Wnt signaling is involved in the patterning and development of many tissues including the nervous system.107,108 Wnt1 and Wnt3a are expressed by cells at the dorsal midline of the developing neural tube. In the absence of Wnt1, midbrain structures fail to form and Wnt3a-mutant mice do not form a hippocampus likely due to the reduced proliferation of hippocampal precursors.109 Wnt ligands bind the receptor complex Frizzled/LRP5/6 leading to stabilization of cytoplasmic β-catenin. β-catenin translocates to the nucleus and binds to target genes via LEF/TCF factors and recruits histone acetyltransferases.110 Transgenic overexpression of β-catenin induces enhanced proliferation of cortical neural progenitors leading to an increase in cortical neurons and surface area.111 During early neurogenesis, Wnts play an active role in symmetric divisions, whereas later, during neurogenesis, Wnts are implicated in neuronal differentiation through expression of N-myc, which in turn represses the Notch signaling.108 Wnt signaling can also induce the expression of Ngn1 and NeuroD1 thereby counteracting Notch signaling and promoting neuronal differentiation.86,108 Thus, Wnt and Notch compete to regulate proneural gene expression and the maintenance and differentiation of NSCs. The differential dynamics of signaling pathways impinging on same or different downstream effectors could be cell type or phase specific.
Fibroblast growth factor signaling has been long known to be involved in area specification in the brain.111 Many Fgfs are expressed in the developing cerebral cortex. Fgf3, 8, 15, 17, and 18 are expressed along the rostral midline of the neocortex in the commissural plate between E9.5 and E12.5, suggesting the presence of a rostral, Fgf-secreting signaling center.112 The Fgf signals play important roles in anterior-posterior patterning of NSCs and promote proliferation.113,114 In addition, Fgf signaling can regulate Hes1 transcription thereby synergizing with and promoting Notch signaling. Fgf18 is expressed in the CP between E13.5 and E16.5, although its role remains unclear.114 Three of the classical receptor tyrosine kinase Fgf receptors (Fgfr1-3) are expressed by NSCs.113,115 Fgfr1 is expressed higher by rostral NSCs than caudal NSCs, whereas Fgfr2 and Fgfr3 are expressed higher caudally than rostrally.116 In mice, loss of Fgfr1 function results in the loss of rostral identity, indicating that Fgf1 acts as a secreted rostral morphogen. Conversely, Fgf2 is expressed higher in the dorsal forebrain than in the ventral, thus contributing to the dorsoventral patterning of the developing brain.114 Loss of function of Fgf2 changes dorsal cortex specification.114 Pea3-ETS transcription factors are downstream of the Fgf signaling pathway and ectopic expression of Fgf18 induces their expression with phenotypic changes in neuronal migration.115 Pea3-ETS transcription factors are expressed in gradients high rostral to low caudally implying a role in axial patterning in the cerebral cortex.117
Transforming growth factor β/bone morphogenetic protein (BMP) signaling pathways play important roles in neurogenesis. Both TGF-β and BMP are expressed in the dorsal cerebral cortex during embryonic neurogenesis and regulate proliferation, survival, differentiation, and migration in the cerebral cortex.118 The BMP binds to the BMP receptor (BMPR1) on the cell surface and induces phosphorylation of Smad family transcription factors.119 The BMP signaling inhibits neuronal differentiation and promotes glial differentiation during corticogenesis.120 Both BMP and Notch may converge on some cellular processes, for example, they could impinge on some similar targets such as Hes3 and inhibitor of DNA-binding factor genes (Ids), as observed during adult neurogenesis.50
Retinoic acid, a derivative of vitamin A, is involved in neuronal differentiation.121,122 Retinoic acid binds nuclear receptors of the RA receptor family (RARs α, β, and γ) that regulate the expression of target genes that contain an RA response element.121 The interaction of RA with the RAR bound as a repressor complex to target genes releases corepressor proteins and recruits histone acetyltransferases.123 However, RA can also induce rapid and transient activation of a cascade of kinases including the MAPK and ERK pathways which contribute to coregulation of the RAR target genes by phosphorylation of cofactors and histones.123,124 Dietary depletion of vitamin A in pregnant mothers results in embryonic defects, including delayed development and reduced cortical hemispheres, with a reduction in neuron-specific class III β tubulin (β-tubulinIII) expression and lower levels of Harvey rat sarcoma viral oncogene (HRas) protein in the intermediate zone and CP regions. The reduction in HRas levels is rescued by supplementing the embryos with RA indicating a stabilization of HRas by RA.125,126 Retinoic acid deficiency affects neuronal migration to cortical layers V to III during development.127 This impaired migration also results in neuronal fate switching to layer II neuron subtypes.127 The RA pathway also cross-talks with Wnt signaling at the level of β-catenin.128 The Wnt-RA axis is most prominent at the rostral end of the developing cerebral cortex, implying a potential role of RA in arealization of the forebrain.13
Hippo signaling regulates size and homeostasis in many organs and tissues.129 The Hippo signaling pathway is a cascade of kinases that converge onto the control of the transcriptional coregulators Yap and Taz.130 The Hippo kinases and Yes-associated protein/transcriptional coactivator with PDZ-binding motif (Yap1/Taz) are regulated at different levels by different stimuli including G protein–coupled receptor signaling, cell adhesion, mechanical stress, and changes in cellular energy status.131 The Hippo kinase cascade can be activated by activation of the macrophage-stimulating 1/2 (Mst1/2) and large tumor suppressor 1/2 (Lats1/2) kinases. These serine/threonine kinases phosphorylate Yap1 and Taz. Phospho-Yap/Taz is targeted to degradation. If Lats1/2 is inactive, then Yap/Taz is dephosphorylated and translocated to the nucleus where they interact with multiple transcriptional regulators.130 Yap/Taz interacts with β-catenin and Smads and thus coregulates both the Wnt and TGF-β pathways to regulate gene expression.130 The TEA domain transcription factors (Tead) are key targets and mediators of the Hippo pathway and critical effectors of Hippo-regulated target gene expression.125,129 In NSCs, the Hippo pathway plays a niche role and regulates the communication between neighboring cells. The expression of fat tumor suppressor homologue (Fat4) and Dachsous (Dchs), the upstream receptor, and ligand of the pathway increases NSC proliferation and reduces differentiation.132 However, the targets and the exact mechanism of the Hippo pathway in NSCs and cortical development remain unclear. Hence, future analysis of the Hippo pathway and its control of NSC maintenance, commitment, and differentiation could uncover novel interactions and functions.
Although the origins of many of the factors described above are not clear, the cerebral spinal fluid is an obvious source. Growth factors and morphogens released into the cerebral spinal fluid can influence NSC proliferation and fate. Some of these factors are produced and released by the choroid plexus and their expression is dynamic during cortical development (reviewed by Lehtinen and Walsh133 and Johansson134). In addition, neuronal inputs from subcortical regions of the brain have also been shown to influence neural progenitor proliferation and maintenance.135,136 This suggests that not only the local environment of the developing cerebral cortex is affecting the production of neurons in the dorsal cortex but more distant brain regions may also play a critical role in the control of cortical NSC fate.
In summary, rather unsurprisingly, development of the brain and particularly the cerebral cortex incorporates many different signaling pathways. Here, we just cover a few but due to the complexity of the cerebral cortex and the need for precise NSC proliferation, fate commitment and differentiation, the balance, and interaction of these pathways will be critical. Hence, a deeper understanding of the signaling pathways and their underlying downstream mechanisms is required to develop a model of how NSCs integrate different signals to regulate development of the brain.
Conclusions
In this review, we have tried to highlight some of the main developmental processes and signaling mechanisms controlling cerebral cortical development. From decades of work, it is clear that transcription factors and signaling are key regulators of NSC generation of the cerebral cortex. However, there remains much to be learnt about how these pathways interact and converge to impose the precise regulation needed to form the complex structure of the cortical isocortex from a simple pseudostratified sheet of NEPs. State-of-the-art technologies employing high-throughput single-cell RNA sequencing platforms have allowed a considerable increase in resolution to the single-cell level. These techniques provide a comprehensive understanding of single cells isolated from the brain, facilitating the extrapolation of intrinsic molecular architecture to function. With the advent of high-throughput single-cell omics and lineage tracing in vivo, the future looks demanding but bright and exciting for elucidating the mechanism of development of the cerebral cortex.
Footnotes
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Swiss National Science Foundation, the SystemsX.ch through project NeuroStemX, and the University of Basel.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: TM and VT wrote the initial manuscipt. TM and VT edited the final version of the manuscript. TM and VT approved the final version to be published. VT acquired funding.
ORCID iD: Verdon Taylor  https://orcid.org/0000-0003-3497-5976
https://orcid.org/0000-0003-3497-5976
References
- 1. Tam PP, Loebel DA. Gene function in mouse embryogenesis: get set for gastrulation. Nat Rev Genet. 2007;8:368–381. [DOI] [PubMed] [Google Scholar]
- 2. Lee HK, Lee HS, Moody SA. Neural transcription factors: from embryos to neural stem cells. Mol Cells. 2014;37:705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Paridaen JT, Huttner WB. Neurogenesis during development of the vertebrate central nervous system. EMBO Rep. 2014;15:351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhadanov AB, Provance DW, Jr, Speer CA, et al. Absence of the tight junctional protein AF-6 disrupts epithelial cell-cell junctions and cell polarity during mouse development. Curr Biol. 1999;9:880–888. [DOI] [PubMed] [Google Scholar]
- 5. Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci. 2006;26:1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stancik EK, Navarro-Quiroga I, Sellke R, Haydar TF. Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. J Neurosci. 2010;30:7028–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tan X, Shi SH. Neocortical neurogenesis and neuronal migration. Wiley Interdiscip Rev Dev Biol. 2013;2:443–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kosodo Y, Suetsugu T, Suda M, et al. Regulation of interkinetic nuclear migration by cell cycle-coupled active and passive mechanisms in the developing brain. EMBO J. 2011;30:1690–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taverna E, Huttner WB. Neural progenitor nuclei IN motion. Neuron. 2010;67:906–914. [DOI] [PubMed] [Google Scholar]
- 10. Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takahashi T, Nowakowski RS, Caviness VS., Jr. Cell cycle parameters and patterns of nuclear movement in the neocortical proliferative zone of the fetal mouse. J Neurosci. 1993;13:820–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Higginbotham H, Guo J, Yokota Y, et al. Arl13b-regulated cilia activities are essential for polarized radial glial scaffold formation. Nat Neurosci. 2013;16:1000–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang X, Nardelli J. Cellular and molecular introduction to brain development. Neurobiol Dis. 2016;92:3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Taverna E, Gotz M, Huttner WB. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu Rev Cell Dev Biol. 2014;30:465–502. [DOI] [PubMed] [Google Scholar]
- 15. Fietz SA, Kelava I, Vogt J, et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci. 2010;13:690–699. [DOI] [PubMed] [Google Scholar]
- 16. Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. [DOI] [PubMed] [Google Scholar]
- 17. Kelava I, Reillo I, Murayama AY, et al. Abundant occurrence of basal radial glia in the subventricular zone of embryonic neocortex of a lissencephalic primate, the common marmoset Callithrix jacchus. Cereb Cortex. 2012;22:469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang X, Tsai JW, LaMonica B, Kriegstein AR. A new subtype of progenitor cell in the mouse embryonic neocortex. Nat Neurosci. 2011;14:555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hansen DV, Rubenstein JL, Kriegstein AR. Deriving excitatory neurons of the neocortex from pluripotent stem cells. Neuron. 2011;70:645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neuroscience. 2011;31:3683–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kriegstein AR, Gotz M. Radial glia diversity: a matter of cell fate. Glia. 2003;43:37–43. [DOI] [PubMed] [Google Scholar]
- 22. Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. [DOI] [PubMed] [Google Scholar]
- 23. Franco SJ, Muller U. Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron. 2013;77:19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. [DOI] [PubMed] [Google Scholar]
- 25. Hartfuss E, Galli R, Heins N, Gotz M. Characterization of CNS precursor subtypes and radial glia. Dev Biol. 2001;229:15–30. [DOI] [PubMed] [Google Scholar]
- 26. Malatesta P, Hack MA, Hartfuss E, et al. Neuronal or glial progeny: regional differences in radial glia fate. Neuron. 2003;37:751–764. [DOI] [PubMed] [Google Scholar]
- 27. Bansod S, Kageyama R, Ohtsuka T. Hes5 regulates the transition timing of neurogenesis and gliogenesis in mammalian neocortical development. Development. 2017;144:3156–3167. [DOI] [PubMed] [Google Scholar]
- 28. Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A. 2004;101:3196–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sahara S, O’Leary DD. Fgf10 regulates transition period of cortical stem cell differentiation to radial glia controlling generation of neurons and basal progenitors. Neuron. 2009;63:48–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. [DOI] [PubMed] [Google Scholar]
- 31. Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. [DOI] [PubMed] [Google Scholar]
- 32. Cubelos B, Sebastian-Serrano A, Kim S, et al. Cux-2 controls the proliferation of neuronal intermediate precursors of the cortical subventricular zone. Cereb Cortex. 2008;18:1758–1770. [DOI] [PubMed] [Google Scholar]
- 33. Englund C, Fink A, Lau C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nieto M, Monuki ES, Tang H, et al. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J Comp Neurol. 2004;479:168–180. [DOI] [PubMed] [Google Scholar]
- 35. Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur J Neurosci. 2005;21:658–668. [DOI] [PubMed] [Google Scholar]
- 36. Zimmer C, Tiveron MC, Bodmer R, Cremer H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb Cortex. 2004;14:1408–1420. [DOI] [PubMed] [Google Scholar]
- 37. Tarabykin V, Stoykova A, Usman N, Gruss P. Cortical upper layer neurons derive from the subventricular zone as indicated by Svet1 gene expression. Development. 2001;128:1983–1993. [DOI] [PubMed] [Google Scholar]
- 38. Lancaster MA, Knoblich JA. Spindle orientation in mammalian cerebral cortical development. Curr Opin Neurobiol. 2012;22:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Konno D, Shioi G, Shitamukai A, et al. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat Cell Biol. 2008;10:93–101. [DOI] [PubMed] [Google Scholar]
- 40. Peyre E, Jaouen F, Saadaoui M, et al. A lateral belt of cortical LGN and NuMA guides mitotic spindle movements and planar division in neuroepithelial cells. J Cell Biol. 2011;193:141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Postiglione MP, Juschke C, Xie Y, Haas GA, Charalambous C, Knoblich JA. Mouse inscuteable induces apical-basal spindle orientation to facilitate intermediate progenitor generation in the developing neocortex. Neuron. 2011;72:269–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bultje RS, Castaneda-Castellanos DR, Jan LY, Jan YN, Kriegstein AR, Shi SH. Mammalian par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Costa MR, Wen G, Lepier A, Schroeder T, Gotz M. Par-complex proteins promote proliferative progenitor divisions in the developing mouse cerebral cortex. Development. 2008;135:11–22. [DOI] [PubMed] [Google Scholar]
- 44. Heins N, Cremisi F, Malatesta P, et al. Emx2 promotes symmetric cell divisions and a multipotential fate in precursors from the cerebral cortex. Mol Cell Neurosci. 2001;18:485–502. [DOI] [PubMed] [Google Scholar]
- 45. Gaiano N, Fishell G. The role of notch in promoting glial and neural stem cell fates. Annu Rev Neurosci. 2002;25:471–490. [DOI] [PubMed] [Google Scholar]
- 46. Dong Z, Yang N, Yeo SY, Chitnis A, Guo S. Intralineage directional notch signaling regulates self-renewal and differentiation of asymmetrically dividing radial glia. Neuron. 2012;74:65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. [DOI] [PubMed] [Google Scholar]
- 48. Lutolf S, Radtke F, Aguet M, Suter U, Taylor V. Notch1 is required for neuronal and glial differentiation in the cerebellum. Development. 2002;129:373–385. [DOI] [PubMed] [Google Scholar]
- 49. Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. Essential roles of notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. 2010;30:3489–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Imayoshi I, Shimogori T, Ohtsuka T, Kageyama R. Hes genes and neurogenin regulate non-neural versus neural fate specification in the dorsal telencephalic midline. Development. 2008;135:2531–2541. [DOI] [PubMed] [Google Scholar]
- 51. Alvarez-Rodriguez R, Pons S. Expression of the proneural gene encoding Mash1 suppresses MYCN mitotic activity. J Cell Sci. 2009;122:595–599. [DOI] [PubMed] [Google Scholar]
- 52. Parras CM, Schuurmans C, Scardigli R, Kim J, Anderson DJ, Guillemot F. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 2002;16:324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vessey JP, Amadei G, Burns SE, Kiebler MA, Kaplan DR, Miller FD. An asymmetrically localized Staufen2-dependent RNA complex regulates maintenance of mammalian neural stem cells. Cell Stem Cell. 2012;11:517–528. [DOI] [PubMed] [Google Scholar]
- 54. Heins N, Malatesta P, Cecconi F, et al. Glial cells generate neurons: the role of the transcription factor Pax6. Nat Neurosci. 2002;5:308–315. [DOI] [PubMed] [Google Scholar]
- 55. Kosodo Y, Roper K, Haubensak W, Marzesco AM, Corbeil D, Huttner WB. Asymmetric distribution of the apical plasma membrane during neurogenic divisions of mammalian neuroepithelial cells. EMBO J. 2004;23:2314–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kouprina N, Pavlicek A, Mochida GH, et al. Accelerated evolution of the ASPM gene controlling brain size begins prior to human brain expansion. PLoS Biol. 2004;2:E126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chae TH, Kim S, Marz KE, Hanson PI, Walsh CA. The hyh mutation uncovers roles for alpha Snap in apical protein localization and control of neural cell fate. Nat Genet. 2004;36:264–270. [DOI] [PubMed] [Google Scholar]
- 58. Sheen VL, Ganesh VS, Topcu M, et al. Mutations in ARFGEF2 implicate vesicle trafficking in neural progenitor proliferation and migration in the human cerebral cortex. Nat Genet. 2004;36:69–76. [DOI] [PubMed] [Google Scholar]
- 59. Zechner D, Fujita Y, Hulsken J, et al. beta-Catenin signals regulate cell growth and the balance between progenitor cell expansion and differentiation in the nervous system. Dev Biol. 2003;258:406–418. [DOI] [PubMed] [Google Scholar]
- 60. Chenn A, Walsh CA. Increased neuronal production, enlarged forebrains and cytoarchitectural distortions in beta-catenin overexpressing transgenic mice. Cereb Cortex. 2003;13:599–606. [DOI] [PubMed] [Google Scholar]
- 61. Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 2008;18:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Custo Greig LF, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14:755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Guo C, Eckler MJ, McKenna WL, McKinsey GL, Rubenstein JL, Chen B. Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes. Neuron. 2013;80:1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McConnell SK. Fates of visual cortical neurons in the ferret after isochronic and heterochronic transplantation. J Neurosci. 1988;8:945–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Desai AR, McConnell SK. Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development. 2000;127:2863–2872. [DOI] [PubMed] [Google Scholar]
- 66. Han W, Sestan N. Cortical projection neurons: sprung from the same root. Neuron. 2013;80:1103–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Eiraku M, Sasai Y. Mouse embryonic stem cell culture for generation of three-dimensional retinal and cortical tissues. Nat Protoc. 2011;7:69–79. [DOI] [PubMed] [Google Scholar]
- 68. Gaspard N, Bouschet T, Hourez R, et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–357. [DOI] [PubMed] [Google Scholar]
- 69. Shen Q, Wang Y, Dimos JT, et al. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. [DOI] [PubMed] [Google Scholar]
- 70. Tan SS, Breen S. Radial mosaicism and tangential cell dispersion both contribute to mouse neocortical development. Nature. 1993;362:638–640. [DOI] [PubMed] [Google Scholar]
- 71. Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Nat Acad Sci U S A. 2005;102:17184–17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. [DOI] [PubMed] [Google Scholar]
- 73. Kwan KY, Sestan N, Anton ES. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development. 2012;139:1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Franco SJ, Gil-Sanz C, Martinez-Garay I, et al. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 2012;337:746–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gotz M, Williams BP, Bolz J, Price J. The specification of neuronal fate: a common precursor for neurotransmitter subtypes in the rat cerebral cortex in vitro. Eur J Neurosci. 1995;7:889–898. [DOI] [PubMed] [Google Scholar]
- 76. Hu JS, Vogt D, Sandberg M, Rubenstein JL. Cortical interneuron development: a tale of time and space. Development. 2017;144:3867–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wamsley B, Fishell G. Genetic and activity-dependent mechanisms underlying interneuron diversity. Nat Rev Neurosci. 2017;18:299–309. [DOI] [PubMed] [Google Scholar]
- 78. Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. [DOI] [PubMed] [Google Scholar]
- 79. Muzio L, DiBenedetto B, Stoykova A, Boncinelli E, Gruss P, Mallamaci A. Conversion of cerebral cortex into basal ganglia in emx2(−/−) pax6(Sey/Sey) double-mutant mice. Nat Neurosci. 2002;5:737–745. [DOI] [PubMed] [Google Scholar]
- 80. Hevner RF, Daza RA, Rubenstein JL, Stunnenberg H, Olavarria JF, Englund C. Beyond laminar fate: toward a molecular classification of cortical projection/pyramidal neurons. Dev Neurosci. 2003;25:139–151. [DOI] [PubMed] [Google Scholar]
- 81. Di Lullo E, Kriegstein AR. The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci. 2017;18:573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Alcamo EA, Chirivella L, Dautzenberg M, et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. [DOI] [PubMed] [Google Scholar]
- 83. Srinivasan K, Leone DP, Bateson RK, et al. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proc Natl Acad Sci U S A. 2012;109:19071–19078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Schuurmans C, Armant O, Nieto M, et al. Sequential phases of cortical specification involve neurogenin-dependent and -independent pathways. EMBO J. 2004;23:2892–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- 86. Hirabayashi Y, Suzki N, Tsuboi M, et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. [DOI] [PubMed] [Google Scholar]
- 87. Kishi Y, Fujii Y, Hirabayashi Y, Gotoh Y. HMGA regulates the global chromatin state and neurogenic potential in neocortical precursor cells. Nat Neurosci. 2012;15:1127–1133. [DOI] [PubMed] [Google Scholar]
- 88. Pereira JD, Sansom SN, Smith J, Dobenecker MW, Tarakhovsky A, Livesey FJ. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci U S A. 2010;107:15957–15962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sessa A, Ciabatti E, Drechsel D, et al. The Tbr2 molecular network controls cortical neuronal differentiation through complementary genetic and epigenetic pathways. Cereb Cortex. 2017;27:3378–3396. [DOI] [PubMed] [Google Scholar]
- 90. Hitoshi S, Ishino Y, Kumar A, et al. Mammalian Gcm genes induce Hes5 expression by active DNA demethylation and induce neural stem cells. Nat Neurosci. 2011;14:957–964. [DOI] [PubMed] [Google Scholar]
- 91. Tuoc TC, Boretius S, Sansom SN, et al. Chromatin regulation by BAF170 controls cerebral cortical size and thickness. Dev Cell. 2013;25:256–269. [DOI] [PubMed] [Google Scholar]
- 92. Yip DJ, Corcoran CP, Alvarez-Saavedra M, et al. Snf2l regulates Foxg1-dependent progenitor cell expansion in the developing brain. Dev Cell. 2012;22:871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Fan G, Martinowich K, Chin MH, et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132:3345–3356. [DOI] [PubMed] [Google Scholar]
- 94. Namihira M, Kohyama J, Semi K, et al. Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev Cell 2009;16:245–255. [DOI] [PubMed] [Google Scholar]
- 95. Blaschuk KL, Ffrench-Constant C. Developmental neurobiology: notch is tops in the developing brain. Curr Biol. 1998;8:R334–R337. [DOI] [PubMed] [Google Scholar]
- 96. Brou C, Logeat F, Gupta N, et al. A novel proteolytic cleavage involved in notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. [DOI] [PubMed] [Google Scholar]
- 97. Mumm JS, Schroeter EH, Saxena MT, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of notch1. Mol Cell. 2000;5:197–206. [DOI] [PubMed] [Google Scholar]
- 98. Basak O, Taylor V. Identification of self-replicating multipotent progenitors in the embryonic nervous system by high notch activity and Hes5 expression. Eur J Neurosci. 2007;25:1006–1022. [DOI] [PubMed] [Google Scholar]
- 99. Zhang R, Engler A, Taylor V. Notch: an interactive player in neurogenesis and disease. Cell Tissue Res. 2017;371:73–89. [DOI] [PubMed] [Google Scholar]
- 100. Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. [DOI] [PubMed] [Google Scholar]
- 101. Hatakeyama J, Bessho Y, Katoh K, et al. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–5550. [DOI] [PubMed] [Google Scholar]
- 102. Fiuza UM, Arias AM. Cell and molecular biology of notch. J Endocrinol. 2007;194:459–474. [DOI] [PubMed] [Google Scholar]
- 103. Ochiai W, Nakatani S, Takahara T, et al. Periventricular notch activation and asymmetric Ngn2 and Tbr2 expression in pair-generated neocortical daughter cells. Molec Cell Neurosci. 2009;40:225–233. [DOI] [PubMed] [Google Scholar]
- 104. Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. [DOI] [PubMed] [Google Scholar]
- 105. Kawaguchi D, Yoshimatsu T, Hozumi K, Gotoh Y. Selection of differentiating cells by different levels of delta-like 1 among neural precursor cells in the developing mouse telencephalon. Development. 2008;135:3849–3858. [DOI] [PubMed] [Google Scholar]
- 106. Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. [DOI] [PubMed] [Google Scholar]
- 107. Harrison-Uy SJ, Pleasure SJ. Wnt signaling and forebrain development. Cold Spring Harb Perspect Biol. 2012;4:a008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kuwahara A, Hirabayashi Y, Knoepfler PS, et al. Wnt signaling and its downstream target N-myc regulate basal progenitors in the developing neocortex. Development. 2010;137:1035–1044. [DOI] [PubMed] [Google Scholar]
- 109. Inestrosa NC, Varela-Nallar L. Wnt signalling in neuronal differentiation and development. Cell Tissue Res. 2015;359:215–223. [DOI] [PubMed] [Google Scholar]
- 110. Wrobel CN, Mutch CA, Swaminathan S, Taketo MM, Chenn A. Persistent expression of stabilized beta-catenin delays maturation of radial glial cells into intermediate progenitors. Dev Biol. 2007;309:285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Fukuchi-Shimogori T, Grove EA. Neocortex patterning by the secreted signaling molecule FGF8. Science. 2001;294:1071–1074. [DOI] [PubMed] [Google Scholar]
- 112. Bachler M, Neubuser A. Expression of members of the Fgf family and their receptors during midfacial development. Mech Develop. 2001;100:313–316. [DOI] [PubMed] [Google Scholar]
- 113. Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–569. [DOI] [PubMed] [Google Scholar]
- 114. Rash BG, Lim HD, Breunig JJ, Vaccarino FM. FGF signaling expands embryonic cortical surface area by regulating notch-dependent neurogenesis. J Neurosci. 2011;31:15604–15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Iwata T, Hevner RF. Fibroblast growth factor signaling in development of the cerebral cortex. Dev Growth Differ. 2009;51:299–323. [DOI] [PubMed] [Google Scholar]
- 116. Sansom SN, Livesey FJ. Gradients in the brain: the control of the development of form and function in the cerebral cortex. Csh Perspect Biol. 2009;1:a005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hasegawa H, Ashigaki S, Takamatsu M, et al. Laminar patterning in the developing neocortex by temporally coordinated fibroblast growth factor signaling. J Neurosci. 2004;24:8711–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Rodriguez-Martinez G, Velasco I. Activin and TGF-β effects on brain development and neural stem cells. CNS Neurol Disord Drug Targets. 2012;11:844–855. [DOI] [PubMed] [Google Scholar]
- 119. Ebendal T, Bengtsson H, Soderstrom S. Bone morphogenetic proteins and their receptors: potential functions in the brain. J Neurosci Res. 1998;51:139–146. [DOI] [PubMed] [Google Scholar]
- 120. Gomes FC, Sousa Vde O, Romao L. Emerging roles for TGF-beta1 in nervous system development. Int J Dev Neurosci. 2005;23:413–424. [DOI] [PubMed] [Google Scholar]
- 121. Gudas LJ, Wagner JA. Retinoids regulate stem cell differentiation. J Cell Physiol. 2011;226:322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–123. [DOI] [PubMed] [Google Scholar]
- 124. Rochette-Egly C. Retinoic acid signaling and mouse embryonic stem cell differentiation: cross talk between genomic and non-genomic effects of RA. Biochim Biophys Acta. 2015;1851:66–75. [DOI] [PubMed] [Google Scholar]
- 125. Park JC, Jeong WJ, Kim MY, Min D, Choi KY. Retinoic-acid-mediated hRas stabilization induces neuronal differentiation of neural stem cells during brain development. J Cell Sci. 2016;129:2997–3007. [DOI] [PubMed] [Google Scholar]
- 126. Chuang JH, Tung LC, Lin Y. Neural differentiation from embryonic stem cells in vitro: an overview of the signaling pathways. World J Stem Cells. 2015;7:437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Easwaran V, Pishvaian M, Salimuddin Byers S. Cross-regulation of beta-catenin-LEF/TCF and retinoid signaling pathways. Curr Biol. 1999;9:1415–1418. [DOI] [PubMed] [Google Scholar]
- 128. Haushalter C, Asselin L, Fraulob V, Dolle P, Rhinn M. Retinoic acid controls early neurogenesis in the developing mouse cerebral cortex. Dev Biol. 2017;430:129–141. [DOI] [PubMed] [Google Scholar]
- 129. Lin KC, Park HW, Guan KL. Regulation of the hippo pathway transcription factor TEAD. Trends Biochem Sci. 2017;42:862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Panciera T, Azzolin L, Cordenonsi M, Piccolo S. Mechanobiology of YAP and TAZ in physiology and disease. Nat Rev Mol Cell Biol. 2017;18:758–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–192. [DOI] [PubMed] [Google Scholar]
- 132. Cappello S, Gray MJ, Badouel C, et al. Mutations in genes encoding the cadherin receptor-ligand pair DCHS1 and FAT4 disrupt cerebral cortical development. Nat Genet. 2013;45:1300–1308. [DOI] [PubMed] [Google Scholar]
- 133. Lehtinen MK, Walsh CA. Neurogenesis at the brain-cerebrospinal fluid interface. Annu Rev Cell Dev Biol. 2011;27:653–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Johansson PA. The choroid plexuses and their impact on developmental neurogenesis. Front Neurosci. 2014;8:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Gerstmann K, Pensold D, Symmank J, et al. Thalamic afferents influence cortical progenitors via ephrin a5-EphA4 interactions. Development. 2015;142:140–150. [DOI] [PubMed] [Google Scholar]
- 136. Dehay C, Savatier P, Cortay V, Kennedy H. Cell-cycle kinetics of neocortical precursors are influenced by embryonic thalamic axons. J Neurosci. 2001;21:201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]