Summary
Epithelial cells are polarized along their apical-basal axis by the action of the small GTPase Cdc42, which is known to activate the aPKC kinase at the apical domain. However, loss of aPKC kinase activity was reported to have only mild effects on epithelial cell polarity. Here, we show that Cdc42 also activates a second kinase, Pak1, to specify apical domain identity in Drosophila and mammalian epithelia. aPKC and Pak1 phosphorylate an overlapping set of polarity substrates in kinase assays. Inactivating both aPKC kinase activity and the Pak1 kinase leads to a complete loss of epithelial polarity and morphology, with cells losing markers of apical polarization such as Crumbs, Par3/Bazooka, or ZO-1. This function of Pak1 downstream of Cdc42 is distinct from its role in regulating integrins or E-cadherin. Our results define a conserved dual-kinase mechanism for the control of apical membrane identity in epithelia.
Keywords: epithelial polarity, Cdc42, Drosophila, aPKC, Pak1, apical domain, organoids, ZO-1
Graphical Abstract

Highlights
-
•
aPKC is not the only kinase responsible for epithelial cell polarization in Drosophila
-
•
Loss of another Cdc42 effector, Pak1 kinase, causes a moderate polarity phenotype
-
•
aPKC and Pak1 kinases act redundantly to maintain apical domain identity
-
•
The dual-kinase mechanism of aPKC and Pak1 is conserved in mammalian epithelia
Downstream of Cdc42, the atypical protein kinase C (aPKC) is thought to be the main effector at the apical domain. Aguilar-Aragon et al. identify a second kinase, Pak1, as being an additional essential effector downstream of Cdc42 at the apical domain in both Drosophila and mammalian epithelial cells.
Introduction
Epithelial tissues make up much of the human body, forming layers and tubes that separate the internal cavities of the body from the external environment. Epithelial tissues also are the site of origin for most human cancers, and progression of tumors to malignancy involves disruption of epithelial morphology. Epithelial cells exhibit polarization along their apical-basal axis, which is essential for both the form and function of the epithelium. How epithelial cell polarity is orchestrated at the molecular level, and how it may become disrupted in human cancer is still poorly understood (Harris et al., 2009, Macara and McCaffrey, 2013, Martin-Belmonte and Perez-Moreno, 2011, St Johnston and Ahringer, 2010, Wirtz-Peitz and Zallen, 2009).
Pioneering genetic studies in model organisms such as the budding yeast Saccharomyces cerevisiae, the worm Caenorhabditis elegans, and the fruit fly Drosophila melanogaster have identified a set of cell polarity determinants that are essential for the polarization of all other molecules, organelles, and cytoskeletal elements in the cell (Thompson, 2013). In particular, the small GTP-binding protein (GTPase) Cdc42 is a key regulator of cell polarity in many species. In epithelial cells, Cdc42 forms a complex with Par6 and the kinase aPKC (Garrard et al., 2003, Genova et al., 2000, Hutterer et al., 2004, Joberty et al., 2000, Ohno, 2001, Peterson et al., 2004, Petronczki and Knoblich, 2001, Wodarz et al., 2000, Yamanaka et al., 2001) that is recruited to the plasma membrane by either Bazooka (Baz/Par3) or the Crumbs (Crb) complex (Crb-Sdt/PALS1-PALS1-associated tight junction [PATJ]) to define the apical membrane domain (Benton and St Johnston, 2003, Fletcher et al., 2012, Hurd et al., 2003, Joberty et al., 2000, Penkert et al., 2004, Tanentzapf and Tepass, 2003). Null mutants in either cdc42, par6, or aPKC result in a complete loss of the apical domain and consequent rounding up and extrusion of cells in Drosophila epithelia (Fletcher et al., 2012, Hutterer et al., 2004, Petronczki and Knoblich, 2001, Rolls et al., 2003, Wodarz et al., 2000); however, recent work demonstrated that kinase-impaired mutants in aPKC did not completely disrupt apical-basal polarity in Drosophila epithelia (Kim et al., 2009) (Figure S1). This surprising finding suggests that aPKC has an essential scaffold function, whereas its kinase activity is non-essential.
Here, we show that apical membrane identity also requires Pak1 kinase activity, in addition to aPKC kinase activity, downstream of Cdc42. This is a distinct function for Pak1 from its previously reported roles in regulating integrins or E-cadherin (Conder et al., 2007, del Pozo et al., 2000, Dummler et al., 2009, Harden et al., 1996, Lucanic and Cheng, 2008, Pirraglia et al., 2010, Santiago-Medina et al., 2013, Tomar and Schlaepfer, 2010). Pak1 appears to function similarly to aPKC, phosphorylating an overlapping set of targets and acting in a genetically semiredundant fashion. These findings clarify how apical domain identity is defined in epithelial cells.
Results and Discussion
We sought to identify additional effectors of Cdc42 that may mediate its function in specifying apical domain identity in epithelial cells. We systematically examined the epithelial loss-of-function phenotype of several alternative Cdc42 effectors. These effectors included the actin nucleating Wasp-Arp2/3 complex; the myotonic dystrophy-related Cdc42-binding kinase (MRCK) or Genghis Khan (Gek) in Drosophila; and the kinases Pak1, Pak3, and Pak4. Silencing expression of these effectors by RNAi in the Drosophila follicular epithelium had no effect on epithelial polarity, except in the case of the kinase Pak1, whose knockdown caused a mild polarity phenotype (Figure 1A). We examined the phenotype of pak1 null mutant clones in follicle cells, which precisely phenocopied the RNAi knockdown phenotype, producing a mild disruption of epithelial polarity reminiscent of a mild cdc42 loss of function (Figure 1B). We validated the RNAi screen using mutant clones for each gene or, in the case of Pak3, an additional previously validated RNAi line (Felix et al., 2015) (Figures S2A and S2B). This result suggests that Cdc42 may activate Pak1 kinase activity to maintain apical identity in epithelial cells. In support of this view, expression of a constitutively active form of Cdc42 (V12) is sufficient to drive recruitment of Pak1-GFP to the plasma membrane, along with the aPKC kinase (Figure 1C).
Figure 1.
An RNAi Screen for Cdc42 Effectors Contributing to Epithelial Polarization Identifies Pak1
(A) RNAi knockdown of Wasp-Arp2/3 complex, Pak3, Pak4, or MRCK/Gek does not have polarity phenotype, whereas Pak1 knockdown causes a partial epithelial polarity disruption.
(B) The cdc423 mutant phenotype is similar to but stronger than that of Pak1 loss of function. Note that RNAi knockdown of Pak1 or induction of pak114 null mutant clones throughout the epithelium causes a mild disruption of aPKC.
(C) Pak1-GFP is recruited to the plasma membrane by active Cdc42. Coexpression of Cdc42V12 with Pak1-GFP results in translocation of Pak1-GFP from cytoplasmic punctae to the plasma membrane.
(D) Epithelial polarity is not affected in follicle cells expressing RNAi against β-integrins (mys) or in the triple Rac mutant (rac1, rac2, mtl).
We note that Pak1 is well known to act as an effector of Rac and Cdc42 in basal integrin-Src signaling for cell migration (del Pozo et al., 2000, Dummler et al., 2009, Lucanic and Cheng, 2008, Santiago-Medina et al., 2013, Tomar and Schlaepfer, 2010); it also affects basal F-actin bundles in Drosophila epithelial cells (Conder et al., 2007, Harden et al., 1996). However, the role of Pak1 at the apical domain of epithelial cells is not a consequence of disruption of basal integrins or integrin-Rac signaling, because the loss of β-integrin (myospheroid [mys]) or the absence of all three Rac genes (rac1, rac2, mtl) (Hakeda-Suzuki et al., 2002, Ng et al., 2002) does not affect epithelial polarity in Drosophila (Figures 1D and S2C). Pak1 also has been implicated in regulating E-cadherin endocytosis to control lumen size (Pirraglia et al., 2010), but this role cannot account for the function that we observe in maintaining apical identity.
We reasoned that Cdc42 may function by activating both the aPKC kinase and the Pak1 kinase at the apical domain to specify apical identity. If the two kinases acted in a redundant fashion, then it would explain why inactivating either aPKC kinase activity or Pak1 activity fails to completely disrupt epithelial polarity. To examine this possibility, we treated follicle cells with highly specific kinase inhibitors for both aPKC and Pak1 (Deacon et al., 2008, Kjær et al., 2013, Rodriguez et al., 2017), which, when used together, are able to disrupt the apical plasma membrane domain (Figures 2A–C). Note that Pak1 or aPKC inhibitor treatment does not cause increased phenotypic severity in Pak1-RNAi expressing cells or aPKC kinase-impaired clones, respectively (Figures S2D and S2E). To confirm the synergy between loss of aPKC and Pak1 kinase activity, we compared the phenotypes of aPKC kinase-impaired mutant cells alone with aPKC kinase-impaired mutant cells in which pak1 expression was silenced by RNAi. We find that the loss of Pak1 dramatically enhanced the aPKC kinase-impaired mutant phenotype, resulting in a complete loss of the apical plasma membrane domain and flattening of cell morphology (Figure 2D). When GFP-marked clones of cells lacking both aPKC kinase activity and Pak1 kinase were induced, the apical domain was disrupted cell autonomously within the clone, whereas the basolateral domain spread around the entire plasma membrane within the clone (Figure 2E). We confirmed these findings by overexpressing myr-Lgl-GFP, a membrane-associated form of Lgl that was previously shown to be an aPKC kinase inhibitor (Bell et al., 2015), with and without Pak1-RNAi (Figure 2F). The myr-Lgl-GFP phenotype resembles the aPKC kinase-impaired phenotype, alone or when expressed together with Pak1-RNAi (Figure 2F). These results demonstrate that aPKC kinase activity and Pak1 kinase activity act semiredundantly downstream of Cdc42 to maintain epithelial cell polarity in the ovaries of Drosophila.
Figure 2.
aPKC and Pak1 Act Redundantly to Maintain Apical Domain Identity
(A–C) aPKC (A), Crb-GFP (B), and Baz/Par3 (C) localize apically in control follicle cells treated with DMSO, but are partially lost upon treatment with either aPKC kinase inhibitors or Pak1 kinase inhibitors, and are completely lost upon treatment with both inhibitors.
(D) aPKC localizes apically in control follicle cells, is partially lost in aPKCpsu265 mutant follicle cells or upon RNAi knockdown of Pak1, and is completely lost upon RNAi knockdown of Pak1 in aPKCpsu265 mutant follicle cells.
(E) RNAi knockdown of Pak1 in aPKCpsu265 mutant follicle cells (GFP+ cells) causes the loss of aPKC from the apical domain and the ectopic spreading of Dlg.
(F) RNAi knockdown of Pak1 in myr-Lgl-GFP expressing follicle cells (an inhibitor of aPKC kinase) causes the loss of aPKC from the apical domain.
We next considered how aPKC and Pak1 may act in a redundant fashion. We note that aPKC and Pak1 have similar consensus sites (Rennefahrt et al., 2007), and we found that Pak1 can act upon many aPKC targets in kinase assays, including Crb, Baz, Par6, and Lgl (Figure 3A). To confirm these findings in vivo, we examined phosphorylation of Baz with an anti-p-Baz antibody, and found that staining is reduced only upon inhibition of both aPKC and Pak1 kinases (Figure 3B). We also examined a GFP-tagged form of the Lgl substrate, which is known to be excluded from the apical domain of epithelial cells by phosphorylation on its lipid-binding motif (Figure 3C). Treatment with the aPKC inhibitor drug fails to cause apical localization of Lgl-GFP, as does treatment with the Pak1 inhibitor alone, but the combination of aPKC and Pak1 inhibitors was able to relocalize Lgl to the apical domain (Figure 3C). These results support the notion that both aPKC and Pak1 can act in parallel to phosphorylate an overlapping set of substrates.
Figure 3.
aPKC and Pak1 Both Contribute to Phosphorylation of Known aPKC Targets Such as Baz and Lgl
(A) Consensus motifs for aPKC and Pak1 are similar, and both kinase domains (KDs; top) can phosphorylate known aPKC targets (Crb, Par6, Baz/Par3, Lgl) in vitro (bottom).
(B) Phospho-Baz (p-Baz) staining of UAS.Baz-GFP expressing follicular epithelia in the presence of aPKC inhibitor, Pak1 inhibitor, or both. Note that loss of p-Baz requires inhibition of both kinases.
(C) Lgl localization in UAS.Lgl-GFP expressing follicular epithelia in the presence of aPKC inhibitor, Pak1 inhibitor, or both. Note that apical localization of Lgl requires inhibition of both kinases.
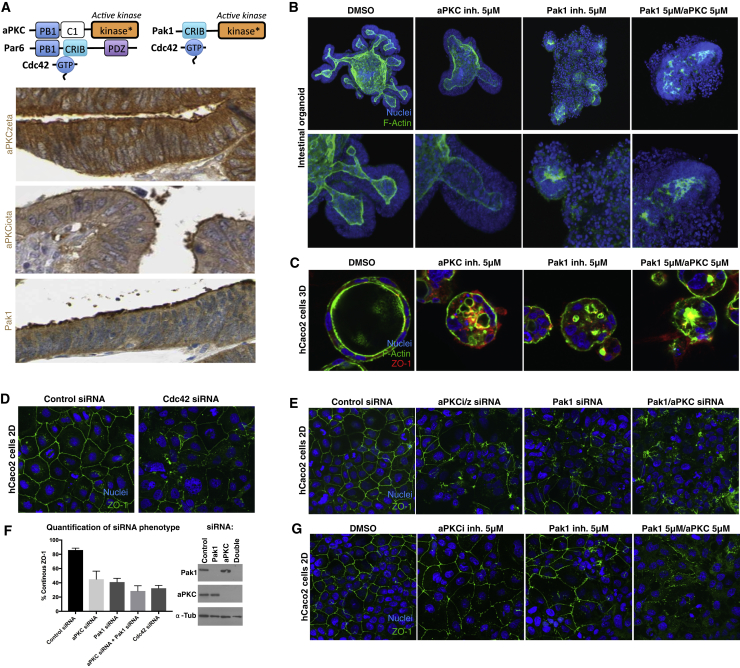
Finally, we examined whether our findings in Drosophila were conserved in mammalian epithelial cells. In support of this idea, both aPKC isoforms (iota and zeta) and Pak1 localized similarly to the apical domain of human intestinal epithelial cells (Figure 4A). We found that treatment with aPKC and Pak1 inhibitors completely disrupted the ability of epithelial cells to form proper polarized structures in mouse intestinal organoids, human Caco2 intestinal cells, and canine kidney epithelial cells (MDCK), both cultured in 3D (Figures 4B, 4C, and S4). We further cultured Caco2 cells in 2D, where we found that small interfering RNA (siRNA) silencing Cdc42 expression or both aPKC and Pak1 expression disrupts epithelial integrity as marked by the continuous localization of ZO-1 in an apical ring (Figures 4D–4F). The aPKC and Pak1 siRNA phenotype can be rescued by overexpression of cDNA against Pak1 or aPKC, respectively (Figures S3A and S3B). Treatment with aPKC and Pak1 inhibitors disrupts the localization of ZO-1 in the same fashion (Figures 4G, S3C, and S3D). These results indicate that the system we have uncovered in Drosophila has a conserved and fundamental role in maintaining epithelial cell polarity in mammalian epithelia.
Figure 4.
Requirement for aPKC and Pak1 Kinases in Mammalian Colonic Epithelial Cell Polarity
(A) Schematic diagram of aPKC and Pak1 activation by Cdc42. Note that two aPKC zeta/iota homologs and Pak1 localize apically in human colonic epithelia.
(B) Mouse intestinal organoids grown in Matrigel and treated with both aPKC and Pak1 inhibitors exhibit loss of epithelial polarity more strongly in the double treatments than in the single treatments.
(C) aPKC and Pak 1 inhibitor treatment in 3D Caco-2 cultures results in disorganized Caco-2 epithelial cysts.
(D) Cdc42 siRNA in 2D plated Caco-2 cells results in a strong disruption of the apical marker ZO-1.
(E) Double siRNA of aPKC and Pak1 results in a much stronger effect on ZO-1 than single knockdowns in Caco-2 cells, thus mimicking Cdc42 depletion.
(F) Quantification of the effect of various siRNA treatments on the organization of ZO-1 and immunoblots showing the efficiency of the Pak1 and aPKC knockdowns in Caco-2 cells.
(G) aPKC and Pak 1 inhibitor phenocopies siRNA treatment in 2D Caco-2 cultures.
Our findings reveal that Cdc42 acts to polarize the apical domain of epithelial cells via two kinases: aPKC and Pak1. We note a striking analogy with Cdc42 function in the yeast S. cerevisiae, where it also acts via two Pak-family kinases: Ste20 and Cla4. Thus, Pak kinase activation appears to be the ancestral function for Cdc42 GTPases in eukaryotes, whereas aPKC activation may have evolved later in metazoans to diversify Cdc42 function. Our data show that both kinases contribute to the polarization of epithelial cells, and we find that chemically inhibiting both aPKC and Pak1 disrupts epithelial polarity in human epithelial cells in a manner that resembles the progression of adenocarcinomas. Thus, our results indicate that loss of apical domain identity could be a critical contributor to the progression of epithelial cancers.
Experimental Procedures
Mitotic clones in follicle cells were generated using the FLP (Flipase)/FRT (Flipase recombination target) system and were marked either positively (presence of GFP; MARCM) or negatively (absence of GFP). Third instar larvae were heat shocked once for 1 hr at 37°C and dissected 3 days after eclosion. Expression of upstream activation sequence (UAS) transgenic lines was achieved with the tj.Gal4 line, the actin “flip-out” system, and the MARCM system. For “flip-out” clones, third instar larvae were heat shocked at 37°C for 20 min and dissected 3 days after eclosion.
Immunostaining of Ovaries
Ovaries were dissected in PBS, fixed for 20 min in 4% paraformaldehyde in PBS, washed for 30 min in PBS/0.1% Triton X-100 (PBT), and blocked for 15 min in 5% normal goat serum (NGS)/PBT. Primary antibodies were diluted in NGS/PBT and samples were incubated overnight at 4°C. Secondary antibodies were used for 2 hr at room temperature and then mounted on slides in Vectashield (Vector Laboratories). Images were taken with a Leica SP5 confocal microscope and processed with Adobe Photoshop.
The primary antibodies used were rabbit anti-PKCζ (C-20) (1:250, Santa Cruz Laboratories), mouse anti-Dlg (1:250, Developmental Studies Hybridoma Bank [DSHB]), rat anti-E-cadherin (DCAD2) (1:100, DSHB), rabbit anti-Baz (1:250, a gift from A. Wodarz), rabbit anti-pBazS980 (1:100, a gift from D. St Johnston), and mouse anti-βPS (1:100, DSHB).
The secondary antibodies used were goat Alexa Fluor 488, 546, or 647 (1:500, Invitrogen); Phalloidin (2.5:250, Life Technologies) to stain F-actin; and 4′,6-diamidino-2-phenylindole (DAPI;1 μg/mL, Life Technologies) to visualize nuclei.
Inhibitor Treatments in Ovaries
Wild-type egg chambers were cultured in imaging media containing Schneider’s Media (Invitrogen), insulin (Sigma), heat-inactivated fetal calf serum (FCS; GE Healthcare), trehalose (Sigma), adenosine deaminase (Roche), methoprene (Sigma), and Ecdysone (Sigma) with either aPKC inhibitor (0.01 mM, Cancer Research Technology CRT0066854 and CRT0103390), Pak1 inhibitor (0.01 mM, Sigma IPA3), or DMSO (Sigma) as a control for either 1 or 2 hr at room temperature. After treatment, samples were fixed and processed normally for imaging.
In Vitro Kinase Assay
For in vitro kinase assays, high-pressure liquid chromatography (HPLC)-purified peptide substrates were incubated with either 150 pg recombinant human Pak1 kinase domain (AbNOVA) or 0.1 μM recombinant human aPKCiota kinase domain (a gift from N. McDonald) for 30 min at 30°C in kinase reaction buffer (250 mM HEPES [(4-(2- hydroxyethyl)-1-piperazineethanesulfonic acid] pH 7.5, 50 mM MgCl2, 5 mM EGTA, 0.05% Brij35) containing 10 μM cold ATP and 3 μCi Y-P32 ATP (PerkinElmer). Samples were blotted on P81 phosphocellulose paper (Millipore) and washed 3 × 10 min in 1% phosphoric acid, then once in acetone. Incorporation of Y-P32 was quantified in counts per minute by scintillation (Beckman LS 6500).
The peptides used in this study were as follows:
Crb VMARNKRATRGTYSPSA (1865.97 Da)
Baz EHFSRDALGRRSISEKHHAALD (2532.27 Da)
Par6 FDAEFRRWSFKRNEAEQ (2216.06 Da)
Lgl LSRRKSFKKSLRESFRKLR (2422.91 Da).
Peptides were diluted with deionized water to working dilutions (1 mg/mL) and stored at −20°C. Histone H1 (Millipore), a known substrate for Pak1, was used as a control.
Mammalian Cell Culture and Transfection
Human Caco-2 adenocarcinoma colon cells were grown in DMEM (GIBCO 41966) containing l-glutamine and sodium pyruvate supplemented with 10% heat-inactivated FCS, 100 μg/mL streptomycin, and 100 μg/mL penicillin. Cells were maintained in a 37°C incubator at 5% atmospheric CO2. siRNA transfections were performed using Lipofectamine RNAiMax transfection reagent (Invitrogen) in antibiotic-free media. Briefly, cells were seeded in 6-well plates and treated with the siRNA/transfection mix 2 hr post-seeding. A final concentration of 50–100 nM siRNA was used for transfection. Media was changed the following morning and another round of siRNA transfection was performed before the cells were trypsinised 4 hr later and reseeded upon 10-mm coverslips in a 48-well plate. Cells were left for a total of 72 hr before being processed for either immunofluorescence or immunoblotting. The siRNA oligonucleotides targeted were as follows:
Pak1: 5′-CAUCAAAUAUCACUAAGUC-3′, 5′-CAACAAAGAACAAUCACUA-3′, 5′-AGAAAUACCAGCACUAUGA-3′, and 5′-GUGAAAUGCUCUCGGCUAU-3′
aPKC: 5′-GGGUACAGACAGAGAAGCAUU-3′ and 5′- GUGUUUGAGCAGGCAUCCAUU-3′
Cdc42: 5′-CGGAAUAUGUACCGACUGU-3′ and 5′-GAUGACCCCUCUACUAUUG-3′ or non-targeting control siRNAs (Thermo Fisher Scientific or Sigma).
For the siRNA rescue experiment, Caco-2 cells were transfected with a single siRNA oligonucleotide and left for 48 hr before being transfected with Rat cDNA for 24 hr. Cells were fixed after 72 hr post-siRNA transfection. The Rat cDNA was refractory to the human-specific siRNA oligos.
Antibodies, Image Acquisition, and Quantification for Mammalian Cell Culture
Cells were washed with 1× PBS and fixed, either in 4% paraformaldehyde in PBS for 15 min before permeabilising with 0.3% Triton X-100 in blocking buffer (PBS containing 0.5% BSA, 10 mM glycine, and 0.1% sodium azide) or −20°C methanol, for 5 min before quenching with PBS. Samples were then washed and incubated in blocking buffer for 1 hr before staining. Primary and secondary antibodies were incubated in blocking buffer. Coverslips were mounted using ProLong Antifade Reagent (Invitrogen).
The primary antibody used was mouse ZO-1 (Life Technologies); the secondary antibodies were from Invitrogen, and used at 1:500 for 2 hr at room temperature along with DAPI. Samples were imaged with a Leica SP5 confocal microscope and processed using Adobe Photoshop.
Quantification of ZO-1 staining in mammalian cell culture was scored as either continuous, in which the junctional ZO-1 staining formed a complete ring around the cell, or discontinuous, in which the ZO-1 staining was repeatedly broken or fragmented. Cells were assessed over three independent experiments counting 400–700 cells per condition. For the rescue experiments, the quantification of ZO-1 staining was assessed either in untransfected cells using the method outlined above or in transfected cells, scoring all of the transfected cells and assessing the percentage. This was assessed over three independent experiments counting 200–400 cells per condition. For 3D cultures, the quantification was scored as either single lumen-containing cysts or multi-lumen-containing cysts, assessed over three independent experiments counting 100–200 cysts per experiment.
For 3D morphogenesis of Caco2 and MDCK cells, 8-well chambers (ibidi) were coated with a layer of growth factor-reduced Matrigel (BD Biosciences) and left to polymerize for 1 hr at 37°C. Caco-2 cells were plated in low glucose media with 2% Matrigel. After 48 hr, the medium was replaced with fresh medium containing 2% Matrigel. The media was replaced with low glucose media containing 2% Matrigel and 0.1 μg/mL cholera toxin 24 hr before fixation. The next day, Pak1 and aPKC inhibitors were added for 8 hr at a concentration of 5 μM before being fixed with PFA and permeabilized for 5 min using a BSA/SDS/Triton X-100 blocking solution (1% BSA, 0.1% NaN3, 20 mM glycine, 0.1% SDS, and 1% Triton X-100 in PBS). Samples were then washed and incubated as described above. Wells were covered with 80% glycerol before being imaged.
Statistical Analysis
Experiments were performed with at least three biological replicates. Prism software was used to plot the mean of the experimental data, and error bars represent the standard deviation. t Test for all of the conditions tested in this article was found to be p < 0.01.
Inhibitor Treatment of Mammalian Tissue Culture
2D mammalian inhibitor treatments were maintained for 4 hr at a concentration of 5 μM per inhibitor; 3D mammalian inhibitor treatments were carried out at the same concentration for a period of 8 hr. siRNA and inhibitor treatments were carried out with the same siRNA method, and on the day of fixation the inhibitor was added for 4 hr, as described.
Animal Procedures
All animal-regulated procedures were carried out according to Project License constraints (PPL 70/8560) and Home Office guidelines and regulations.
Acknowledgments
We thank Daniel St. Johnston, Franck Pichaud, Edward Manser, Peter Parker, Cancer Research Technology, Peptide Chemistry Facility, VDRC, and Bloomington for reagents. This work was supported by The Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001180), the UK Medical Research Council (FC001180), and the Wellcome Trust (FC001180), as well as a Wellcome Trust Investigator award (102853/B/13/Z).
Author Contributions
M.A.A. performed the experiments in Drosophila, A.E. performed the experiments in mammalian cell culture, V.F. performed the experiments in organoids, G.C.F. helped with some of the experiments, and B.J.T. and V.S.W.L. supervised the experiments. M.A.A. and B.J.T. conceived the experiments, and B.J.T. wrote the manuscript with input from M.A.A.
Declaration of Interests
The authors declare no competing interests.
Published: February 13, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.01.060.
Supplemental Information
References
- Bell G.P., Fletcher G.C., Brain R., Thompson B.J. Aurora kinases phosphorylate Lgl to induce mitotic spindle orientation in Drosophila epithelia. Curr. Biol. 2015;25:61–68. doi: 10.1016/j.cub.2014.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R., St Johnston D. A conserved oligomerization domain in Drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr. Biol. 2003;13:1330–1334. doi: 10.1016/s0960-9822(03)00508-6. [DOI] [PubMed] [Google Scholar]
- Conder R., Yu H., Zahedi B., Harden N. The serine/threonine kinase dPak is required for polarized assembly of F-actin bundles and apical-basal polarity in the Drosophila follicular epithelium. Dev. Biol. 2007;305:470–482. doi: 10.1016/j.ydbio.2007.02.034. [DOI] [PubMed] [Google Scholar]
- Deacon S.W., Beeser A., Fukui J.A., Rennefahrt U.E., Myers C., Chernoff J., Peterson J.R. An isoform-selective, small-molecule inhibitor targets the autoregulatory mechanism of p21-activated kinase. Chem. Biol. 2008;15:322–331. doi: 10.1016/j.chembiol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo M.A., Price L.S., Alderson N.B., Ren X.D., Schwartz M.A. Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. EMBO J. 2000;19:2008–2014. doi: 10.1093/emboj/19.9.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dummler B., Ohshiro K., Kumar R., Field J. Pak protein kinases and their role in cancer. Cancer Metastasis Rev. 2009;28:51–63. doi: 10.1007/s10555-008-9168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix M., Chayengia M., Ghosh R., Sharma A., Prasad M. Pak3 regulates apical-basal polarity in migrating border cells during Drosophila oogenesis. Development. 2015;142:3692–3703. doi: 10.1242/dev.125682. [DOI] [PubMed] [Google Scholar]
- Fletcher G.C., Lucas E.P., Brain R., Tournier A., Thompson B.J. Positive feedback and mutual antagonism combine to polarize Crumbs in the Drosophila follicle cell epithelium. Curr. Biol. 2012;22:1116–1122. doi: 10.1016/j.cub.2012.04.020. [DOI] [PubMed] [Google Scholar]
- Garrard S.M., Capaldo C.T., Gao L., Rosen M.K., Macara I.G., Tomchick D.R. Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6. EMBO J. 2003;22:1125–1133. doi: 10.1093/emboj/cdg110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genova J.L., Jong S., Camp J.T., Fehon R.G. Functional analysis of Cdc42 in actin filament assembly, epithelial morphogenesis, and cell signaling during Drosophila development. Dev. Biol. 2000;221:181–194. doi: 10.1006/dbio.2000.9671. [DOI] [PubMed] [Google Scholar]
- Hakeda-Suzuki S., Ng J., Tzu J., Dietzl G., Sun Y., Harms M., Nardine T., Luo L., Dickson B.J. Rac function and regulation during Drosophila development. Nature. 2002;416:438–442. doi: 10.1038/416438a. [DOI] [PubMed] [Google Scholar]
- Harden N., Lee J., Loh H.Y., Ong Y.M., Tan I., Leung T., Manser E., Lim L. A Drosophila homolog of the Rac- and Cdc42-activated serine/threonine kinase PAK is a potential focal adhesion and focal complex protein that colocalizes with dynamic actin structures. Mol. Cell. Biol. 1996;16:1896–1908. doi: 10.1128/mcb.16.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T.J., Sawyer J.K., Peifer M. How the cytoskeleton helps build the embryonic body plan: models of morphogenesis from Drosophila. Curr. Top. Dev. Biol. 2009;89:55–85. doi: 10.1016/S0070-2153(09)89003-0. [DOI] [PubMed] [Google Scholar]
- Hurd T.W., Gao L., Roh M.H., Macara I.G., Margolis B. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat. Cell Biol. 2003;5:137–142. doi: 10.1038/ncb923. [DOI] [PubMed] [Google Scholar]
- Hutterer A., Betschinger J., Petronczki M., Knoblich J.A. Sequential roles of Cdc42, Par-6, aPKC, and Lgl in the establishment of epithelial polarity during Drosophila embryogenesis. Dev. Cell. 2004;6:845–854. doi: 10.1016/j.devcel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Joberty G., Petersen C., Gao L., Macara I.G. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- Kim S., Gailite I., Moussian B., Luschnig S., Goette M., Fricke K., Honemann-Capito M., Grubmüller H., Wodarz A. Kinase-activity-independent functions of atypical protein kinase C in Drosophila. J. Cell Sci. 2009;122:3759–3771. doi: 10.1242/jcs.052514. [DOI] [PubMed] [Google Scholar]
- Kjær S., Linch M., Purkiss A., Kostelecky B., Knowles P.P., Rosse C., Riou P., Soudy C., Kaye S., Patel B. Adenosine-binding motif mimicry and cellular effects of a thieno[2,3-d]pyrimidine-based chemical inhibitor of atypical protein kinase C isoenzymes. Biochem. J. 2013;451:329–342. doi: 10.1042/BJ20121871. [DOI] [PubMed] [Google Scholar]
- Lucanic M., Cheng H.J. A RAC/CDC-42-independent GIT/PIX/PAK signaling pathway mediates cell migration in C. elegans. PLoS Genet. 2008;4:e1000269. doi: 10.1371/journal.pgen.1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara I.G., McCaffrey L. Cell polarity in morphogenesis and metastasis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20130012. doi: 10.1098/rstb.2013.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Belmonte F., Perez-Moreno M. Epithelial cell polarity, stem cells and cancer. Nat. Rev. Cancer. 2011;12:23–38. doi: 10.1038/nrc3169. [DOI] [PubMed] [Google Scholar]
- Ng J., Nardine T., Harms M., Tzu J., Goldstein A., Sun Y., Dietzl G., Dickson B.J., Luo L. Rac GTPases control axon growth, guidance and branching. Nature. 2002;416:442–447. doi: 10.1038/416442a. [DOI] [PubMed] [Google Scholar]
- Ohno S. Intercellular junctions and cellular polarity: the PAR-aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr. Opin. Cell Biol. 2001;13:641–648. doi: 10.1016/s0955-0674(00)00264-7. [DOI] [PubMed] [Google Scholar]
- Penkert R.R., DiVittorio H.M., Prehoda K.E. Internal recognition through PDZ domain plasticity in the Par-6-Pals1 complex. Nat. Struct. Mol. Biol. 2004;11:1122–1127. doi: 10.1038/nsmb839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson F.C., Penkert R.R., Volkman B.F., Prehoda K.E. Cdc42 regulates the Par-6 PDZ domain through an allosteric CRIB-PDZ transition. Mol. Cell. 2004;13:665–676. doi: 10.1016/s1097-2765(04)00086-3. [DOI] [PubMed] [Google Scholar]
- Petronczki M., Knoblich J.A. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat. Cell Biol. 2001;3:43–49. doi: 10.1038/35050550. [DOI] [PubMed] [Google Scholar]
- Pirraglia C., Walters J., Myat M.M. Pak1 control of E-cadherin endocytosis regulates salivary gland lumen size and shape. Development. 2010;137:4177–4189. doi: 10.1242/dev.048827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennefahrt U.E., Deacon S.W., Parker S.A., Devarajan K., Beeser A., Chernoff J., Knapp S., Turk B.E., Peterson J.R. Specificity profiling of Pak kinases allows identification of novel phosphorylation sites. J. Biol. Chem. 2007;282:15667–15678. doi: 10.1074/jbc.M700253200. [DOI] [PubMed] [Google Scholar]
- Rodriguez J., Peglion F., Martin J., Hubatsch L., Reich J., Hirani N., Gubieda A.G., Roffey J., Fernandes A.R., St Johnston D. aPKC cycles between functionally distinct PAR protein assemblies to drive cell polarity. Dev. Cell. 2017;42:400–415.e9. doi: 10.1016/j.devcel.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls M.M., Albertson R., Shih H.P., Lee C.Y., Doe C.Q. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J. Cell Biol. 2003;163:1089–1098. doi: 10.1083/jcb.200306079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Medina M., Gregus K.A., Gomez T.M. PAK-PIX interactions regulate adhesion dynamics and membrane protrusion to control neurite outgrowth. J. Cell Sci. 2013;126:1122–1133. doi: 10.1242/jcs.112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D., Ahringer J. Cell polarity in eggs and epithelia: parallels and diversity. Cell. 2010;141:757–774. doi: 10.1016/j.cell.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Tanentzapf G., Tepass U. Interactions between the crumbs, lethal giant larvae and bazooka pathways in epithelial polarization. Nat. Cell Biol. 2003;5:46–52. doi: 10.1038/ncb896. [DOI] [PubMed] [Google Scholar]
- Thompson B.J. Cell polarity: models and mechanisms from yeast, worms and flies. Development. 2013;140:13–21. doi: 10.1242/dev.083634. [DOI] [PubMed] [Google Scholar]
- Tomar A., Schlaepfer D.D. A PAK-activated linker for EGFR and FAK. Dev. Cell. 2010;18:170–172. doi: 10.1016/j.devcel.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz-Peitz F., Zallen J.A. Junctional trafficking and epithelial morphogenesis. Curr. Opin. Genet. Dev. 2009;19:350–356. doi: 10.1016/j.gde.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Grimm A., Knust E. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J. Cell Biol. 2000;150:1361–1374. doi: 10.1083/jcb.150.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T., Horikoshi Y., Suzuki A., Sugiyama Y., Kitamura K., Maniwa R., Nagai Y., Yamashita A., Hirose T., Ishikawa H. PAR-6 regulates aPKC activity in a novel way and mediates cell-cell contact-induced formation of the epithelial junctional complex. Genes Cells. 2001;6:721–731. doi: 10.1046/j.1365-2443.2001.00453.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.