Summary
Organoid technology provides the possibility of culturing patient-derived colon tissue and colorectal cancers (CRCs) while maintaining all functional and phenotypic characteristics. Labeling stem cells, especially in normal and benign tumor organoids of human colon, is challenging and therefore limits maximal exploitation of organoid libraries for human stem cell research. Here, we developed STAR (stem cell Ascl2 reporter), a minimal enhancer/promoter element that reports transcriptional activity of ASCL2, a master regulator of LGR5+ intestinal stem cells. Using lentiviral infection, STAR drives specific expression in stem cells of normal organoids and in multiple engineered and patient-derived CRC organoids of different genetic makeup. STAR reveals that differentiation hierarchies and the potential for cell fate plasticity are present at all stages of human CRC development. Organoid technology, in combination with the user-friendly nature of STAR, will facilitate basic research into human adult stem cell biology.
Keywords: organoids, intestine, colorectal cancer, stem cells, LGR5, ASCL2
Graphical Abstract

Highlights
-
•
ASCL2-responsive minigene (STAR) is specific for intestinal stem cells
-
•
STAR is compatible with user-friendly techniques like lentiviral infections
-
•
STAR enables stem cell labeling in normal and in cancer organoids of human colon
-
•
Cellular plasticity is present at all stages of colorectal cancer development
Oost et al. present an ASCL2-responsive minigene (STAR) that enables stem cell labeling in patient-derived colorectal cancer organoids, as well as in normal and benign colorectal tumor samples. The user-friendly nature of STAR applications in combination with organoid technology will facilitate basic research into human adult stem cell biology.
Introduction
The cancer stem cell model was postulated after initial transplantation experiments demonstrated that only small subpopulations of tumor cells, as few as 100, turned out to be tumorigenic while tens of thousands of cancer cells with alternative phenotypes did not (Al-Hajj et al., 2003, Bonnet and Dick, 1997, Lapidot et al., 1994). For colorectal cancer (CRC), the concept gained impact by the detection of stem cell-like cancer cells that shared functional (Kreso et al., 2013, O’Brien et al., 2007, Ricci-Vitiani et al., 2007) as well as molecular properties with normal stem cells, like co-expression of stem cell marker genes Lgr5 and Ascl2 (Dalerba et al., 2011). Using Lgr5 as a marker gene, the presence of cancer stem cells in colon cancer is now well established, revealing self-renewal and differentiation capacity for LGR5+ tumor cells (Cortina et al., 2017, de Sousa e Melo et al., 2017, Schepers et al., 2012, Shimokawa et al., 2017). Moreover, re-emergence of LGR5+ cancer stem cells after their selective targeting revealed extensive plasticity in CRCs (de Sousa e Melo et al., 2017, Shimokawa et al., 2017), underscoring the importance to understand the biological cues that mediate plasticity and stem cell function in tumors in order to design effective anti-cancer therapies. Although CRISPR-mediated labelings of cancer stem cells have been achieved in CRC organoids (Cortina et al., 2017, Shimokawa et al., 2017), it is currently still technically challenging to mark stem cells in human colorectal organoids, especially from normal tissue and benign tumors. As a result, it is currently unresolved to what extent stem cell characteristics evolve while colorectal tumors progress along the adenoma-carcinoma sequence.
The establishment of CRC organoid biobanks, derived from multiple patients, can capture the genetic diversity of the disease (Fujii et al., 2016, van de Wetering et al., 2015) and allow the faithful evaluation of personalized drug responses to therapies in a preclinical setting (Verissimo et al., 2016). As such, patient-derived CRC organoids have the potential to enable comparative studies how diverse mutational landscapes of cancer genomes from different patients affect the functional properties of cancer stem cells.
To include multiple organoid lines for basic research into human (cancer) stem cell biology, e.g., from many different patients, disease stages, and settings, including normal and benign colorectal tumors, we developed an ASCL2-responsive minigene called STAR (stem cell ASCL2 reporter). STAR is compatible with user-friendly strategies (e.g., lentiviral infections) to report stem cell activity in human colon organoids. Using STAR, we demonstrate that stem cell-driven differentiation hierarchies and cell fate plasticity are present at all stages of the adenoma-carcinoma-sequence of human CRC progression.
Results
Intestinal Stem Cell-Specific Reporters
β-Catenin/Tcf4 transcriptional activity is key for intestinal stem cell (ISC) fate but is also active in non-stem cells (Van der Flier et al., 2007, van Es et al., 2005). In search of an ISC-specific reporter, the transcription factor Ascl2 is of particular interest considering its role as the master regulator of ISC fate (van der Flier et al., 2009). Ascl2 synergistically enhances the expression level of a subset of Wnt/β-catenin target genes, thereby imposing a stem cell-specific expression pattern on combined Wnt/β-catenin and Ascl2 target genes such as Lgr5, Rnf43, EphB2, and notably Ascl2 itself. As a consequence of the positive transcriptional feedback loop of Ascl2 on its own gene transcription, Ascl2 converts the Wnt morphogen gradient that is present along the crypt-villus axes into an almost binary signal for ISC fate (Schuijers et al., 2015).
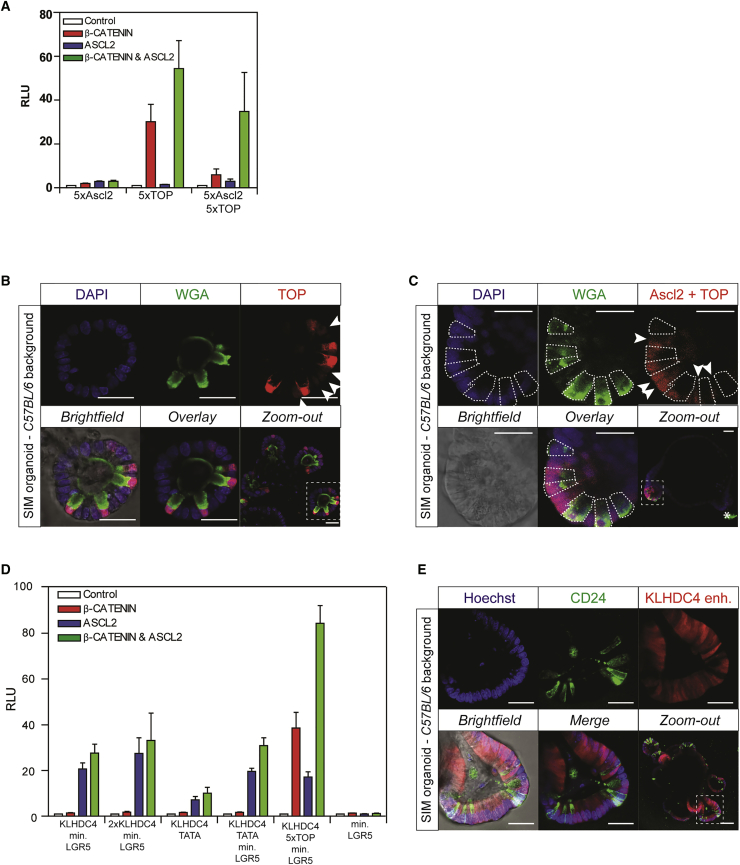
To test transcriptional activity of different ASCL2-dependent reporters, we performed luciferase assays in human embryonic kidney cells (HEK293T) that contain a fully functional yet inactive Wnt signaling pathway. As expected based on previous reports (Schuijers et al., 2015), we hardly observed any transcriptional activity of five optimal ASCL2 binding sites in the presence of active Wnt/β-catenin signaling, presence of ASCL2, or both combined (Figure 1A). Indeed, in line with the proposed model that β-catenin/TCF and ASCL2 can function as a tripartite transcriptional complex (Schuijers et al., 2015), the addition of five Wnt-responsive TCF optimal binding motifs (TOP) to ASCL2 sites provided a synergistic effect on transcriptional activity (Figure 1A). The minimal 150-bp promoter element of LGR5 (100-bp promoter region plus first 50-bp 5′-UTR) that includes an endogenous TATA box sequence demonstrates no transcriptional activity by itself (Figure 1A).
Figure 1.
ISC-Specific Reporters
(A) Transcriptional activity of indicated reporters measured by luciferase assay in HEK293T cells co-transfected with a dominant-positive mutant of β-catenin (S33Y), ASCL2, the combination, or none. A synthetic promoter with five optimal ASCL2 binding sites is non-responsive, whereas a TOP reporter (5× TCF sites) is predominantly sensitive to active WNT/β-catenin signaling. Combining ASCL2 and TCF sites (both 5×) demonstrates a synergistic activation upon presence of β-catenin and ASCL2. Luciferase activity was normalized to the “empty” pGL4.10 backbone. All transcriptional elements include TATA-box. RLU, relative luciferase units. Triplicates of a representative experiment are shown. Data are represented as mean ± SEM.
(B) Stably integrated TOP reporter (red) reveals Wnt activity in mouse small intestinal organoids. High levels of Wnt activity are observed in WGA+ Paneth cells (green), while only low levels are observed in CBC stem cells (arrows). DAPI (blue) marks nuclei.
(C) Stably integrated ASCL2-TOP reporter (red) reveals similar expression levels between WGA+ (green) Paneth cells (dashed lines) and intercalating CBC stem cells (arrows) at the bottom of crypt compartments in mouse small intestinal organoids. WGA (green) stains Paneth cells (dashed lines). DAPI (blue) marks nuclei. ∗Autofluorescence.
(D) Transcriptional activity of indicated reporters measured by luciferase assay in HEK293T cells in co-presence of a dominant-positive mutant of β-catenin (S33Y), ASCL2, both, or none. KLHDC4 enhancer shows ASCL2-dependent transcriptional activity and is insensitive to active Wnt/β-catenin signaling. Like TATA box, minimal LGR5 promoter on itself is non-responsive but facilitates transcriptional activity mediated by enhancer. Addition of TOP sites to enhancer yields β-catenin-responsive effects, as well as synergism to co-presence of ASCL2 and β-catenin. Triplicates of a representative experiment are shown. RLU, relative luciferase units. Data are represented as mean ± SEM.
(E) Stably integrated KLHDC4 reporter (red) reveals bright signal at crypt bottoms with the exclusion of CD24+ Paneth cells (green). Hoechst (blue) marks nuclei. Scale bars: 50 μm.
To validate expression patterns of functional reporters in intestinal epithelia, we generated small intestinal mouse (SIM) organoids with stable integration of the reporter plasmids. Although Wnt/β-catenin signaling is well known for its effects in stem (arrows) and progenitor cells, the TOP reporter predominantly shows strong Wnt activity in Paneth cells in agreement with its reported role in specializing terminally differentiated Paneth cells (Figure 1B) (Farin et al., 2012, van Es et al., 2005). In addition, supplementing TOP with ASCL2 sites confirmed enhanced reporter expression in Lgr5+ crypt base columnar (CBC) stem cells of SIM organoids (arrows), in line with the measurements in cell lines (Figure 1C). However, while the signal became much enhanced in CBC stem cells, i.e., almost equalized in relation to Paneth cells, the lack of exclusivity for CBC cells disqualifies this reporter for specific visualization and manipulation of Lgr5+ CBC stem cells.
To obtain stem cell specificity, we searched for enhancer elements with exclusive responsiveness to stem cell specific ASCL2 in publicly available chromatin immunoprecipitation (ChIP) sequencing data of ASCL2 and TCF4 (Schuijers et al., 2015). Intriguingly, while most binding elements revealed co-occupancy of both transcription factors, a single 700-bp enhancer element in closest proximity, and thus annotated, to the KLHDC4 gene stood out with significant levels of ASCL2 binding without any detectable TCF (Schuijers et al., 2015). Intriguing, the KLHDC4 gene itself is not enriched in ISCs, nor is it known to be important for stem cell biology. Moreover, the enhancer element, identified in the human genome, does not seem to be conserved among species (data not shown). Nevertheless, the KLHDC4 enhancer in combination with the minimal LGR5 promoter indeed revealed ASCL2-specific transcriptional activity in a luciferase assay without any responsiveness to Wnt/β-catenin (Figure 1D). To assess influence of the minimal LGR5 promoter on transcriptional activity, we replaced this sequence for synthetic TATA box sequences and obtained similar, albeit lower, ASCL2-dependent effects (Figure 1D). Importantly, introduction of the KLHDC4 enhancer that is driving expression of a fluorescent marker in SIM organoids, revealed high activity at the crypt base, flanked by non-positive CD24+ Paneth cells (Figure 1E).
Identification of Stem Cell Activity Using an ASCL2-Responsive Minigene
We observed that the core of the KLHDC4-related enhancer element contained 11 near-perfect repeats of 41 bp that are interspersed with three random base pairs. Each 41-bp repeat contained an optimal 10-bp ASCL2-binding motif (Figure 2A). To test whether these repeats are responsible for ASCL2-dependent transcriptional activation, we repeated the luciferase assay in 293T cells. Moreover, to validate ASCL2 specificity, we generated a mutant repeat with directed point mutations within the core of the ASCL2 binding motif (Figure 2A). As expected, the 41-bp repeats became transcriptionally active only in the presence of ASCL2. Moreover, its activity strongly increased upon multiplying the number of repeats, while the mutant versions remained inactive (Figure 2B). From now, on we refer to the 41-bp repeat as STAR.
Figure 2.
Identification of Stem Cell Activity Using an ASCL2-Responsive Minigene
(A) Representation of the KLHDC4 enhancer that contains 11 near-identical 41-bp repeats. Based on these repeats, reporters were generated consisting of normal repeats (STAR, red box) that includes an optimal binding motif for ASCL2 (red font) driving transcription of all possible genes of interest (GOIs). In contrast, repeats with mutations in ASCL2 binding motif (blue font) function as control elements named mSTAR (blue box).
(B) Transcriptional activity of indicated reporters measured by luciferase assay in HEK293T cells co-transfected with a dominant-positive mutant of β-catenin (S33Y), ASCL2, the combination, or none. STAR repeats are responsive to ASCL2 and insensitive to active WNT/β-catenin signaling. Moreover, mSTAR is inactive under all conditions, underscoring ASCL2-specific transcriptional activity of STAR repeats. As a positive control, combining TCF binding sites (TOP) with minimal ASCL2 binding sites renders responsiveness to WNT/β-catenin and provides synergistic activity with co-occupancy of β-catenin/TCF and ASCL2. All transcriptional elements are supplemented with min.LGR5 promoter. RLU, relative luciferase units. Triplicates of a representative experiment are shown. Data are represented as mean ± SEM.
(C) Pulldown of DNA oligos containing 4× STAR or 4× mSTAR repeats in CRC cell line LS174T followed by mass spectrometry to identify STAR-specific interacting proteins. Scatterplot indicates significant interactors (red circles) that show specific binding to the STAR repeat. As expected, bHLH transcription factor ASCL2, as well as its heterodimeric binding partners HEB (TCF12) and E2A (TCF3) show specific binding.
(D) Mass spectrometry-based identification of non-DNA-mediated protein-protein interaction partners of ASCL2 in LS174T CRC cells. Volcano plot indicates specific interactors with pulled-down flag-ASCL2.
(E) Left: Immunofluorescence cross-section image of stably integrated STAR minigene reveals STAR activity (red) in mouse small intestinal organoids. Middle: Enlargement of stem cell niche reveals specific expression in CBC stem cells (arrowheads) that directly flank post-mitotic Paneth cells stained with lysozyme (blue). Right: 3D rendering of the same organoid, clearly indicating the restrictive expression pattern of the STAR minigene in the direct vicinity of Paneth cells. DAPI (white) marks nuclei.
(F) Left: Immunofluorescence image of stably integrated STAR minigene in mouse small intestinal organoids derived from Lgr5-DTR-eGFP knock-in mice. Co-expression of STAR minigene (red) with Lgr5 (green) in CBC stem cells. Right panel: Enlargement of crypt compartment. Lysozyme (blue) stains Paneth cells. DAPI (white) marks nuclei. Scale bars: 50 μm (left) and 25 μm (right). See also Figure S1.
To confirm transcriptional activity of STAR repeats in combination with endogenous ASCL2 expression levels, we performed the luciferase assay in LS174T CRC cell line. These human CRC cells show robust expression levels of multiple Wnt-dependent ISC markers such as LGR5, RNF43, and ASCL2 (van de Wetering et al., 2002, Van der Flier et al., 2007). As expected, multiplying the number of STAR repeats enhanced activity, while mutant STAR repeats remained inactive. Strikingly, four or more (eight) STAR repeats showed activity that surpassed the original KLHDC4 enhancer element (Figure S1A).
For molecular validation of STAR repeat-driven transcription, we performed a DNA pull-down with STAR repeats in LS174T CRC cells for subsequent identification of all interacting proteins using mass spectrometry (Figure S1B). We indeed identify ASCL2-specific binding to the STAR repeats mediated by its consensus binding motif (Figures 2C and S1C). Moreover, the experiment shows that STAR DNA sequences interact with two general heterotypic binding partners of the E protein family of bHLH transcription factors, namely HEB (TCF12) and E2A (TCF3). Subsequently, we performed an ASCL2 pull-down in LS174 CRC cells to identify direct protein-protein interaction partners of ASCL2 by mass spectrometry. In agreement with previous reports (Johnson et al., 1992, Scott et al., 2000, van der Flier et al., 2009), HEB and E2A were observed to be the most common binding partners of ASCL2 (Figure 2D). Moreover, due to the presence of ethidium bromide during the experiment, we disrupt protein DNA-dependent interactions, suggesting that ASCL2 binds in a protein complex with HEB and/or E2A to the STAR repeat.
To test the STAR-mediated expression pattern in intestinal organoid cultures, we generated a STAR minigene that combines 41-bp STAR repeats with the minimal LGR5 promoter to drive expression of a fluorescent marker. As expected, introduction of a STAR minigene into SIM organoids resulted in restricted activity in CBC stem cells at the base of crypt structures with the notable exception of Lysozyme+ Paneth cells (Figure 2E). To confirm stem cell-specific STAR activity, we determined its expression pattern in relation to stem cell marker gene Lgr5. We isolated SIM organoids from the Lgr5-DTR-GFP mouse in which Lgr5+ CBC stem cells are marked with DTR-GFP (Tian et al., 2011). We observed co-expression of STAR in DTR-GFP+ Lgr5 stem cells as assessed by their GFP+ membranes, as well as exclusion of STAR expression in Lysozyme+ Paneth cells (Figure 2F). In contrast to the observed heterogeneity in organoids, STAR activity in clonal human CRC cell lines showed a more homogeneous expression pattern, where averaged levels of STAR activity correlated with previous reported ASCL2 expression levels for the different cell lines (Figure S1D) (Van der Flier et al., 2007). Notably, upon induced differentiation of these cancer cells using transient transfection of dnTCF4, we noticed that the activity of STAR correlates with downregulation of LGR5 expression on the single-cell level (Figure S1E). In summary, activity of the STAR minigene faithfully represents ISC activity that is mediated by the stem cell-specific transcription factor ASCL2.
STAR Minigene Reveals Stem Cell Activity in Human Organoids of Normal Colon
Considering the unmet demand for selective labeling of stem cells in colon organoids of human origin, we tested the expression pattern of lentiviral STAR minigenes in human colon organoid lines. We first validated its expression pattern in normal human colon organoids and noticed restricted expression in crypt-like structures reminiscent of the expression pattern in mouse small intestinal organoids and in agreement with the in vivo localization of Lgr5+ stem cell activity at the crypt base (Figure 3A). Moreover, STAR+ cells were intercalated between STAR-negative cells (arrowheads), most likely reflecting “Paneth cell-equivalent” deep crypt secretory (DCS) cells that support stem cell function in the colon (Rothenberg et al., 2012, Sasaki et al., 2016).
Figure 3.
STAR Minigene Reveals Stem Cell Activity in Human Organoids of Normal Colon
(A) Cross-section of crypt structure of normal human colon organoids with stable integration of the STAR minigene. Stem cells visualized by STAR activity (second panel in white) are confined to cells at the crypt base. In line with cellular positioning in in vivo crypt, STAR+ cells are intermingled with STARneg cells (arrowheads). Nuclei are marked with H2B-mNeonGreen expression (third panel in white). Right panel: Overlay of STAR expression (red) and nuclei (green). ∗Autofluorescence. Scale bar: 25 μm.
(B) Colony-forming efficiency of single STAR+ and STARneg cells, FACS isolated from mature organoid structures with crypt and villus-like structures. At day 5 and 10 after plating, STAR+ colon cells are most potent in organoid formation. Increasing activity of the WNT signaling pathway with WNT and R-spondin ligands enhances organoid formation efficiency of STAR+ colon cells, but not of STARneg cells. N = 3 independent experiments. Data are represented as mean ± SEM.
(C) Gene expression profiling of STAR+ versus STARneg cells from normal human colon organoids. Shown are the changes in expression levels of genes between STAR+ and STARneg cells.
(D) GSEA on gene expression dynamics of STAR+ versus STARneg colon cells with the previously published gene set that is specific for Lgr5+ ISCs. The strong ISC signature in STAR+ cells confirms that the STAR minigene indeed marks stem cells in normal human colon organoids. See also Figure S2.
Next, we determined outgrowth potential of fluorescence-activated cell sorting (FACS)-sorted STAR+ and STARneg cells from normal human colon organoids that contain crypt and villus-like structures. As expected, colony formation was most robust for colon cells with STAR-labeled stem cell activity, and almost negligible for non-STAR cells. Moreover, we noticed that the amount of WNT and R-spondin in the culture medium corresponded with the overall efficiency of colony formation for the STAR+ colon cells, but not for the non-STAR cells (Figure 3B). Independently, when normal human colon organoids are grown in previous reported expansion medium (Sato et al., 2011), we noticed significantly improved outgrowth for the non-STAR cells, indicative of fate plasticity within this non-stem cell population to dedifferentiate toward stem cell state upon stem cell-favorable culture conditions (Figure S2A).
Subsequently, we performed RNA sequencing on sorted colon cells with either presence or absence of STAR-labeled stem cell activity (Figure S2B). As expected, among the most upregulated genes in STAR+ colon cells is ASCL2, the master regulator of stem cell fate and the inducer of STAR activity (Figure 3C). In addition, known stem cell genes such as ZNRF3, EPHB3, and LRIG1 were upregulated. In contrast, colon cells without stem cell activity express differentiation markers such as BMP3, ALPI, TFFs, and Mucins. To assess the gene expression profile of STAR+ cancer stem cells in an unbiased manner, we performed a gene set enrichment analysis (GSEA) comparing our data with available gene signatures of ISCs (Muñoz et al., 2012). The gene expression signature of STAR+ colon cells resembles Lgr5+ ISCs to a large extent (Figure 3D), implying that the STAR-labeled cells in normal human colon organoids are indeed LGR5+ colon stem cells.
STAR Minigene Reveals Stem Cell Activity in Engineered Tumor Organoids
Tumor progression organoids (TPOs) are normal colon organoids in which various combinations of cancer mutations were introduced using CRISPR/Cas9, thereby recapitulating various stages of CRC development along the adenoma-carcinoma sequence (Drost et al., 2015). STAR activity across the panel of TPOs with identical genetic background enables us to study the functional properties of stem cells while colon tumors evolve from benign lesions to invasive cancers. All TPO lines of the adenoma-carcinoma sequence revealed presence of STAR activity in a heterogeneous expression pattern (Figure 4A), in line with previous reports that signaling pathways, including Wnt, still display heterogeneous activity patterns in malignant tissue (Prasetyanti et al., 2013). Especially the Quadruple mutant TPO line, i.e., APC, KRAS, TP53, and SMAD4, showed pronounced STAR activity confined to crypt-like budding structures, suggesting intense stem cell activity in TPO subtypes that is associated with invasive behavior (Drost et al., 2015, Fumagalli et al., 2017).
Figure 4.
STAR Minigene Reveals Stem Cell Activity in Engineered Tumor Organoids
(A) Tumor progression organoids (TPOs) that are representative of different stages along the adenoma-carcinoma sequence. Representative organoids of TPO subtypes are presented at day 10 after single-cell plating. Top row shows bright field. Middle row depicts STAR expression (white). Bottom row shows merge between STAR expression (red) and H2B-mNeon (green). Cellular heterogeneity within clonally grown organoids is clearly reflected by the restricted STAR expression pattern (dashed lines). Normal human colon and APC organoids were supplemented with Rock-inhibitor Y-27632 to prevent anoikis at single-cell stage. ∗Autofluorescence. Scale bars: 50 μm.
(B) Orthotopic transplantation of human TripleAPC/ KRAS/ TP53 and Quadruple mutant STAR organoids into the cecal wall of immune-deficient mice. Tumors were analyzed approximately 8–12 weeks after transplantation. TripleAPC/ KRAS/ TP53 remains tumor in situ and contains clusters of STAR+ cells, whereas the Quadruple mutant tumor shows an invasive morphology with STAR+ cells localized at the outer regions. STAR activity is depicted in red, and counterstaining with β-catenin in white. ∗Autofluorescence of necrotic debris. Scale bars: 100 μm.
(C) H2B-mNeon+ tumor cells from transplanted TripleAPC/ KRAS/ TP53 and Quadruple mutant tumor organoids were FACS sorted and gated on STAR activity (tagBFP2). Gating settings were defined using intestinal cells of the recipient mice.
(D) Scatterplot indicating the correlation between the expression profiles of STAR+ cancer cells from the Quadruple mutant tumor organoids that have been cultured in vitro versus grown in vivo as an orthotopic transplanted tumor. Red dots represent mRNA transcripts from a previously described ISC-specific gene signature (Muñoz et al., 2012). See also Figure S3.
Since the relative level of STAR activity in TPO subtypes might be influenced by culture media with customized growth factor supplements, we xenotransplanted the TPO lines into the cecum of mice in order to test whether the proportion of cancer cells with STAR+ stem cell activity differs between different stages of tumor development (Figure 4B). Previous studies (Drost et al., 2015, Fumagalli et al., 2017) indicated successful engraftment of the Triple, i.e., APC, KRAS, and TP53, and the Quadruple mutant lines. In agreement with those studies, our Triple mutant TPO remains a tumor in situ while the Quadruple mutant TPO showed irregular structured epithelium, a feature of invasive carcinomas (Figure S3A). In agreement with recent observations of LGR5 expression patterns in human tumors (Shimokawa et al., 2017), STAR+ cancer cells were detected at the outer regions of the tumors. This was particularly pronounced in the Quadruple mutant TPO (Figure 4B). Subsequent quantification of STAR+ cancer cells by FACS analysis revealed that the invasive Quadruple mutant tumor had an increased proportion of cancer stem cells as compared with the Triple mutant tumor (Figure 4C). Next, we performed mRNA expression profiling of the STAR+ cells that were derived from the Quadruple mutant tumor in the mouse versus STAR+ cells from cultured Quadruple mutant TPO line. Reassuringly, the cellular identities of STAR+ cells from both sources are very similar in overall expression profile (R2 of 0.94) (Figure 4D). Moreover, when specifically analyzing previously determined ISC-enriched genes (Muñoz et al., 2012), we noticed that the ISC signature in STAR+ cells is very robust and seems largely independent of environmental conditions (R2 of 0.93) (Figure 4D).
STAR+ Cancer Cells Contain Stem Cell Activity
In order to define a comprehensive gene expression signature for STAR+ stem cells while they evolve along the adenoma-carcinoma sequence of CRC development, we performed RNA sequencing on sorted STAR+ and STARneg cancer cells from multiple human TPO subtypes (Figures S3B and S4A), including the original normal organoids. We consistently identified 638 genes with significant differences in expression levels between the two populations. Among the most upregulated genes in STAR+ cancer cells is ASCL2, the master regulator of stem cell fate and the inducer of STAR activity (Figure 5A). In addition, known stem cell genes such as RGMB, SP5, EPHB3, and LRIG1 were consistently upregulated. In contrast, cancer cells without stem cell activity had consistent expression of differentiation markers such as REG4, VILLIN, and KRT7. To assess the gene expression profile of STAR+ cancer stem cells in an unbiased manner, we performed a GSEA comparing our data with available gene signatures of ISCs (Muñoz et al., 2012). The gene expression signature that is common among STAR+ cells of the different TPO subtypes resembles Lgr5+ ISCs to a large extent (Figure 5B), implying that the STAR+ cells in human tumor organoids represent the genuine LGR5 stem cell lineage. In agreement with the concept that common cancer mutations eliminate the dependency of niche signals rather than inducing de novo phenotypes (Fujii et al., 2016, Fumagalli et al., 2017), we observed that the general stem cell signature of STAR+ cells stays similar during tumor progression along the adenoma-carcinoma sequence.
Figure 5.
Characterization of STAR+ Cancer Stem Cells during Tumor Progression
(A) Gene expression profiling of STAR+ versus STARneg cells from various TPO subtypes. Shown are the 638 transcripts whose average over five samples show >1.25-fold difference. Mann-Whitney U test, p value < 0.005.
(B) Top: GSEA on gene expression dynamics of STAR+ versus STARneg cancer cells with the previously published gene set that is specific for Lgr5+ ISCs. The strong ISC signature in STAR+ cells confirms that the STAR minigene labels cancer cells with stem cell activity in human tumor organoids that is independent of mutational landscapes. Bottom: GSEA with the specific gene list for DCS cells shows strong DCS cell signature in STARneg cells, suggesting the presence of DCS-like-cancer cells within the non-stem cell population.
(C) Colony-forming efficiency of single STAR+ or STARneg cells scored at days 5 and 10 after plating. Normal human colon and APC-deficient cells were supplemented with Rock-inhibitor Y-27632 to prevent anoikis at the single-cell stage. N = 3 independent experiments. Data are represented as mean ± SEM.
(D) Cellular differentiation and fate plasticity create cellular heterogeneity of STAR expression within developing organoids. Three categories were assigned for an organoid phenotype: 100% positive for STAR expression (red), complete absence of STAR expression (white), or everything in between (mixed). Phenotypes were scored at days 0, 5, and 10 after plating. N = 3 independent experiments. Data are represented as mean ± SEM. See also Figure S4.
Normally, colon stem cells and DCS cells are both positioned at the entire base of a crypt. Since the STAR minigene marks stem cell activity in a heterogeneous fashion in all TPO subtypes, we wondered whether the STARneg population of cancer cells include cells that resemble the DCS cells of the colon. From previously performed gene expression analysis of Reg4+ DCS cells (Sasaki et al., 2016), we generated a Reg4+ DCS cell-specific gene signature consisting of 208 genes that are >2-fold upregulated with a p value of <0.05. Although multiple cellular lineages are likely present within the STARneg cell population, the STARneg tumor cell population from different TPO subtypes shows resemblance to the expression signatures of differentiated DCS cells (Figure 5B), indicating that at least a subset of tumor cells are DCS-like cells.
To study evolving characteristics of STAR+ stem cells while tumors progress from benign lesions to invasive carcinomas, we studied outgrowth potential of FACS-sorted STAR+ and STARneg cells of multiple TPO subtypes. The growth course of individual organoids was examined over time using brightfield imaging and fluorescent imaging of the nuclei (H2B-mNeonGreen) and STAR-labeled stem cell activity. Independent of TPO subtype, we observed three modes of outgrowth for STAR+ cells. First, the developing tumor organoids remain 100% positive for STAR activity. Alternatively, they lose STAR activity completely. In the third scenario, STAR+ cancer cells both self-renew into STAR+ daughter cells, but also generate STARneg progeny by means of differentiation, yielding organoids with a mixed composition of STAR+ and STARneg cells. Profiting from this “mixed” outgrowth scenario, we isolated STAR+ and STARneg cells from organoids that developed in 2 weeks from the FACS-sorted STAR+ cell population for quantitative follow-up experiments. Importantly, this experimental strategy excludes silencing of the minigene as a cause of absence for STAR activity (Figure S4A).
We first set out to further define colony-forming efficiencies of STAR+ and STARneg cells isolated from the various TPO tumor subtypes. Normal colon and the more benign tumor organoid lines, i.e., with single mutations in APC or KRAS, require inhibition of anoikis-mediated apoptosis in order to grow out (Figure 5C). Anoikis inhibition can be omitted for TPO tumor variants containing two or more cancer mutations. Although the TPO line with APC and TP53 deficiency shows modest but significant colony formation without anoikis inhibition, combining APC-deficiency with oncogenic KRAS shows increased efficiency of organoid formation. Further addition of a third (TP53) and/or fourth (SMAD4) mutation show similar efficiencies (Figures 5C and S4B).
Intriguingly, the colony-forming efficiencies for STARneg cells revealed almost identical behavior as STAR+ stem cells, albeit with slightly lower efficiencies (Figure 5C). In a parallel experiment, anoikis was inhibited for all TPO subtypes (Figures S4B and S4C), equalizing the overall colony formation efficiencies to similar levels among all TPO tumor lines. Also in this experiment, outgrowth from STAR+ and STARneg cells gave strikingly similar results.
Next, we decided to study the outgrowth of STARneg cells in closer detail. At day 0, FACS-sorted cells were confirmed to be nearly purely STARneg as assessed under the confocal microscope. However, when the same organoids were imaged at later time points (day 5 and 10), an increasing number of organoids reobtained STAR+ cells that resulted in organoids of mixed composition (Figure 5D, bottom panels), indicative of cell fate plasticity. In identical experiments, the STAR+ cells transformed from a pure population (day 0) into a population of organoids where an increasing number of organoids showed a mixed phenotype (Figure 5D, upper panels), indicative of STAR+ stem cells that not only self-renew but also generate STARneg progeny. Besides mixed phenotypes, some organoids lost all STAR+ stem cell activity, while a small but significant fraction of organoids remained completely composed of STAR+ stem cells (Figure 5D, top panels).
In addition to the presence of robust de-differentiation potential of progenitor cells within normal crypt compartments in mice (Buczacki et al., 2013, Metcalfe et al., 2014, Tetteh et al., 2016, Tian et al., 2011, van Es et al., 2012), we now reveal similar presence of de-differentiation capacity in normal human colon. Moreover, the capacity for fate plasticity of STARneg progenitors does not seem to be dramatically enhanced after acquisition of common cancer mutations.
Cancer stem cells have previously been identified in CRC samples based on their high level of Wnt signaling using WNT/β-catenin reporters (i.e., TOP reporters) (Vermeulen et al., 2010). As expected, most prominent WNT-responsive cells in tumor organoids are also labeled with STAR (Figure S4D). Moreover, reflecting the binary expression pattern of ASCL2 in intestinal and colon stem cells as observed in normal crypts, we noticed a more restrictive expression pattern of STAR in comparison to the wider WNT gradient in tumor organoids. Therefore, in addition to simple labeling of stem cells, the stem cell-exclusive expression pattern that is imposed by STAR also provides the unique opportunity to specifically manipulate cellular processes that occur in stem cells by overexpression of any protein-of-interest, e.g., dominant-negative or active effectors of signaling pathways.
STAR Minigene Labels Cancer Stem Cells in Patient-Derived CRC Organoids
Next, we introduced STAR minigenes in three different patient-derived CRC organoid lines from a well-characterized multi-patient organoid biobank (van de Wetering et al., 2015). Each of the three lines, i.e., P9T, P16T, and hypermutated P19bT, contain different mutational landscapes (Figure 6A). In clonal outgrowths of these patient-derived CRC organoids, thereby ensuring genetic homogeneity, different levels of STAR-labeled stem cell activity were observed (Figure 6A). Thus, heterogeneous patterning of STAR reported stem cell activity seems to be a common theme among organoids from healthy, early tumorigenic (TPOs) or end-stage CRC tissue.
Figure 6.
STAR Minigene Labels Cancer Stem Cells in Patient-Derived CRC Organoids
(A) Patient-derived CRC organoids P9T, P16T, and P19bT with stable integration of the STAR minigene. Top: Bright-field images. Middle: Heterogeneous pattern of STAR activity (white). Bottom: Merge of STAR expression (red) with H2B-Neon (green). Dashed lines indicate individual STAR+ cells. Panel at the right indicates overview of mutations in frequently mutated genes in cancer. Panel is reproduced from van de Wetering et al. (2015). Scale bars: 50 μm.
(B) GSEA on gene expression dynamics of STAR+ versus STARneg cancer cells from TPOs and patient-derived CRC organoids shows again a strong ISC-specific gene signature in STAR+ cancer cells.
(C) Heatmap of STAR-specific gene expression dynamics (Mann-Whitney U test, p value < 0.005) in multiple patient-derived CRC organoids and TPO subtypes reveals stem cell and differentiation markers in STAR+ and STARneg cells, respectively.
(D and E) Like Figure 5C (D) and Figure 5D (E), but now for indicated patient-derived CRC organoids. See also Figure S5.
Next, we performed mRNA expression profiling of the STAR+ and STARneg cells. As expected, also in these patient-derived CRC organoids, the STAR minigene marks cancer cells with an expression pattern that closely associates with the stem cell signature of well-characterized Lgr5+ ISCs from mouse (Figure 6B). Subsequently, we compared the STAR-specific expression profiles of patient-derived CRC organoids with the RNA expression data from STAR+ stem cells of the TPO subtypes. Genes consistently upregulated in STAR+ stem cells when compared to STARneg cancer cells include well-known stem cell marker genes such as ASCL2, EPHB3, NOTCH1, SOX4, and AXIN2 (Figure 6C). In contrast, among the most differential genes upregulated in the STARneg cancer cells is REG4, the marker gene of DCS cells, the Paneth cell equivalent cell in colon epithelium that supports stem cell function (Figure 6C).
In addition, we addressed the functional characteristics of the cancer cells with or without STAR+ stem cell activity. In line with the observations with the TPO subtypes, we observed near-identical colony-forming efficiencies for STAR+ and STARneg cancer cells and observed, again, significant fate plasticity (Figures 6D, 6E, and S5A). The degree of plasticity varied per patient-derived tumor line, potentially reflecting the differences in the mutational landscape.
Discussion
Organoid technology is a rapidly expanding research field. Its unique resemblance to in vivo epithelia not only draws attention from adult stem cell biologist, but a far wider public that ranges from tissue engineers and biophysicist for understanding self-organizational properties of tissues to physicians that strive to optimize disease modeling (Clevers, 2016). Considering this widespread mode of applications, user-friendly technology that enables straightforward monitoring and manipulation of specific cell types is of great interest. Especially labeling stem cells in human intestinal organoids is challenging and has so far only been demonstrated in colon organoids of cancerous origin using CRISPR knockin strategies that are both technically challenging and time consuming (de Sousa e Melo et al., 2017, Shimokawa et al., 2017).
Here, we report the STAR minigene as an alternative strategy for ISC labeling. The STAR minigene reports transcriptional activity of ASCL2, the master regulator of ISC fate (van der Flier et al., 2009) and as a result generates an expression pattern that is specific for stem cells of the gut. Importantly, due to the small size of the enhancer/promoter element (∼0.5 kb), STAR is compatible with standard transposon-based and lentiviral infection protocols for genomic integration. Therefore, in contrast to CRISPR-mediated strategies, STAR provides a simple and flexible strategy to fluorescently label and/or drive all sorts of proteins-of-interest in ISCs. Illustrative for its user friendliness is the successful labeling of stem cells in human colon organoids of normal origin using lentiviral infection.
In line with previous reports, we confirm that synthetic promoters with a repeating number of minimal ASCL2-optimal binding motifs do not provide transcriptional activity in an ASCL2-dependent manner (Schuijers et al., 2015). In contrast, multiple 41-bp STAR repeats do generate strong transcriptional activity. It will be interesting to identify the exact molecular mechanism how ASCL2 initiates transcriptional activity from a STAR repeat. Most importantly, all our data indicate that STAR activity is critically dependent on the presence of ASCL2 and hence is specific for stem cells of the gut.
Recently, the functional relevance of LGR5+ cancer stem cells has been reported for primary tumor growth in CRC (Cortina et al., 2017, de Sousa e Melo et al., 2017, Shimokawa et al., 2017). We confirm their findings of extensive plasticity in different patients CRCs, the presence of which seems virtually independent of the mutational landscape and that will likely impact the potential of cancer stem cells as a therapeutic target. In addition, using TPO subtypes representing the different stages of the adenoma-carcinoma sequence in CRC, we now show that the capacity for cell fate plasticity is present during the entire process of tumorigenesis in human colon. Moreover, in line with mouse studies (Buczacki et al., 2013, Metcalfe et al., 2014, Tetteh et al., 2016, Tian et al., 2011, van Es et al., 2012), we now confirm for human colon that the capacity of cancer cells to de-differentiate is independent of the acquisition of multiple cancer mutations, but predominantly a feature inherited from normal cells. In contrast, the acquisition of cancer mutations, especially the addition of oncogenic KRAS on top of APC deficiency, does have a significant impact on the efficiency of individual cancer cells to form organoids. However, this is independent of stem cell activity, but merely reflects the capacity of individual cancer cells to survive at the single-cell stage since plating efficiency could be rescued in normal and benign tumor organoids by inhibition of anoikis.
Besides stem cell-specific expression in small intestine and colon, expression of ASCL2 has recently been documented in cancerous tissues of among others lung, stomach, and breast (Wang et al., 2017). Although gene silencing has been reported for ASCL2 in a number of individual CRCs, potentially excluding such samples for STAR applications (Jubb et al., 2006), it will be of great interest to verify whether ASCL2 also marks stem cell activity in tumorigenic lesions of other organs, as well as in their corresponding normal epithelia. Libraries of patient-derived colorectal organoids have recently been generated that also include benign tumor samples and the organoid cultures of the normal epithelia (Fujii et al., 2016, van de Wetering et al., 2015). CRISPR/Cas9-mediated knockins have been used to label stem cell markers in human organoids, but their feasibility is technically challenging and has so far only been shown in CRC organoids of advanced stages (Cortina et al., 2017, Shimokawa et al., 2017). The STAR minigene is an alternative user-friendly strategy to label stem cells in intestinal organoids that has the potential to empower easy accessible stem cell research using primary human colon tissues from multiple different patients and pathogenic conditions.
Experimental Procedures
For a detailed description of experimental procedures, please see Supplemental Experimental Procedures.
Organoid Cultures
Mouse organoids were established and maintained as described previously (Sato et al., 2009). Patient-derived colon organoids were maintained as described previously (van de Wetering et al., 2015). The TPO series were maintained as described previously (Drost et al., 2015). For maturation of normal colon organoids that include true crypt and villus-like structures, organoids are grown in 15% Wnt15 culture medium (CM) and 7.5% R-spondin CM without SB202190 and nicotinamide. To confirm correct sample identity in the laboratory, used organoid lines were regularly tested by SNP analysis.
Genomic Integration Minigene
Organoid transfection using Tol2 transposase was performed with following reporter plasmids: Tol2 KLHDC4-min.pLGR5-tagRFPt_PGK-puro (in SIM Bl6 organoids); Tol2 8xSTAR-min.pLGR5-mNeonGreen_PGK-puro (in SIM Bl6 organoids); Tol2 8xSTAR-min.pLGR5-tagRFPt_PGK-puro (in SIM Lgr5-DTR-GFP organoids); Tol2 5xTOP-tagRFPt_PGK-Puro (in SIM Bl6 organoids and in human TripleAPC/ KRAS/ TP53 and Quadruple mutant STAR colon organoids); Tol2 5XAscl2-5xTOP-tagRFPt_PGK-Puro (in SIM Bl6 organoids) (genomic location of KLHDC4 enhancer [chr16: 87732692–87733464]).
Alternatively, we infected TPOs and patient-derived CRC organoids P16T and P19bT with a lentiviral vector containing 4xSTAR repeats driving TagBFP2 expression linked with an IRES sequence to a blasticidin selection cassette (pLV-4xSTAR-min.pLGR5-TagBFP2-IRES-Blast.) and a pLV-H2B-mNeonGreen-IRES-puro. P9T was infected with pLV-4xSTAR-min.pLGR5-TagRFPt::PGK-Puro-P2A-H2B-mNeonGreen. To exclude silencing of the minigenes during culture, the STAR-infected organoids were sorted by FACSAria III (BD Biosciences) for STARhigh cells 2 weeks before each experiment.
Microscopy, Live-Cell Imaging, and Colony-Forming Efficiency
Images were captured with a Leica SP8X microscope. For immunofluorescence analyses, the mouse organoids were washed with PBS for 1 hr, permeabilized with PBD0.2T (48.5 mL of PBS, 1 mL of 10% Triton, 0.5 mL of DMSO, and 0.5 g of BSA), and incubated with antibodies at 4°C overnight.
For live-cell analysis, sorted STAR+ and STARneg cells were plated on a glass-bottom 384-well plate (Corning 4581) and mounted onto a Leica SP8X microscope. The plate was held at 37°C in a microscope box and equipped with a culture chamber for humidity and 6.4% CO2 overflow. Organoids were imaged in XYZ(T)-mode using a water 25× objective (HCX IRAPO L; numerical aperture [N.A.], 0.95) with a tunable white light laser. For higher magnification and identification of STAR-positive cells, we used a water 40× objective (HC PL APO CS2; N.A., 1.1). Post-acquisition analyses of phenotypes was performed manually using ImageJ.
For the colony-forming efficiency assays, 3,000–5,000 viable single cells were collected via FACS and plated over five wells with a density of 50–300 cells per well of a glass-bottom 384-well plate. Using bright-field and live-fluorescence imaging, single cells were monitored at day 0 and at several days of culture organoid to quantify organoid formation (colony-forming efficiency %).
Cell Transfection and Dual-Luciferase Assay
The generated reporters were constructed in a pGL4.10 backbone and transfected with Xtremegene9 (Roche) in all cell lines, with Renilla luciferase as a transfection control. The dual-luciferase assay system of Promega was used to measure reporter activity. Empty pGL4.10 or pGL4.10-min.hLGR5 served as negative controls. All luciferase activities were normalized to Renilla. Reporter activities were normalized to pGL4.10-min.hLGR5 promoter.
STAR Interaction Studies
Nuclear extracts were prepared from LS174T cells. DNA oligos of the 4× STAR and 4× mSTAR repeats were amplified using biotinylated primers. LS174T nuclear extract was combined with protein incubation buffer and added to the beads. Pull-downs were performed in duplicate. Afterward on bead digestion was used to digest the proteins in tryptic peptides. The label-free ASCL2-flag pull-down was performed in triplicate using 20 μL of anti-FLAG M2 affinity gel (Sigma) per pull-down. Afterward on bead digestion was used to digest the affinity purified proteins in tryptic peptides. Finally, the tryptic peptides were acidified and desalted using C18 Stagetips prior to mass spec analysis. For liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of the DNA pull-down and the label-free FLAG pull-down, the tryptic peptides were separated by an Easy-nLC 1000 (Thermo Fisher) connected online to an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific). MaxQuant (version 1.5.1.0) was used to analyze the raw MS spectra by searching against the Uniprot curated human proteome (downloaded December 2015).
Orthotopic Xenotransplantation of Organoids
The orthotopic transplantation of organoids was executed as previously described (Fumagalli et al., 2017).
RNA Sequencing
RNA from the FACS-sorted organoids was extracted using the RNeasy RNA extraction kit (QIAGEN) with DNaseI treatment. A maximum of 250 ng of RNA per sample was used for ribosomal RNA depletion using the Ribo-Zero Gold rRNA removal kit (Illumina). A maximum of 2 ng of cDNA per sample was used for library preparation using the Kapa Hyper Prep Kit (KAPA Biosystems). Sequencing was performed using a Illumina HiSeq 2000 machine and generated 50-bp paired-end reads.
GSEA
For GSEA, we used the ISC signature from Muñoz et al. as ISC gene set. To define a REG4+ DCS gene set, we compared available RNA sequencing data of Lgr5+ and Reg4+ DCS cells from Sasaki et al., and considered the genes with at least a 2-fold upregulation and a p value < 0.05 (by Student’s t test) as a REG4+ DCS-specific gene signature. Human orthologs of both gene sets were identified with bioDBnet.
Acknowledgments
We thank all members of the H.J.G.S. and Bos laboratories for fruitful discussions and critical reading of the manuscript; F. de Sauvage (Genentech) for providing organoid cultures of Lgr5-DTR-GFP knockin mice; and Jeroen van Velzen and Pien van der Burght of the UMCU flow cytometry facility (Laboratory for Translational Immunology). H.J.G.S. is supported by a KWF fellowship from the Dutch Cancer Society (UU 2013-6070); general support came from the Netherlands Organization for Scientific Research Gravitation Program: consortium is Cancer Genomics Netherlands (J.v.R., M.V., and H.J.G.S.), NWO-VICI grant 91815604 (M.M.M.), and ERC Consolidator Grant 648804 (J.v.R.).
Author Contributions
K.C.O. and H.J.G.S. conceived the study. K.C.O., B.M.T.B., J.v.R., M.V., M.M.M., and H.J.G.S designed experiments, and K.C.O. performed most of the experiments. K.C.O. M.J.R.-C., M.C.H., and I.V.-K. performed culture experiments and integration of minigenes. K.C.O. and J.S. performed luciferase assays. A.F. and J.v.R. performed and analyzed in vivo transplantation experiments. L.v.V., R.G.H.L., and M.V. performed mass spectrometry experiments and RNA sequencing, as well as their analyses. M.O. performed smFISH experiments. K.C.O. and H.J.G.S. wrote the manuscript, which was reviewed by all authors.
Declaration of Interests
The authors declare no competing interests.
Published: February 6, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and five figures and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.01.033.
Data and Software Availability
The accession number for the RNA sequencing data reported in this paper is GEO: GSE99133. The project identifier for the mass spectrometric data reported in this paper is PRIDE (ProteomeXchange Consortium via the PRIDE repository): PXD006575.
Supplemental Information
References
- Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D., Dick J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Buczacki S.J.A., Zecchini H.I., Nicholson A.M., Russell R., Vermeulen L., Kemp R., Winton D.J. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- Cortina C., Turon G., Stork D., Hernando-Momblona X., Sevillano M., Aguilera M., Tosi S., Merlos-Suárez A., Stephan-Otto Attolini C., Sancho E., Batlle E. A genome editing approach to study cancer stem cells in human tumors. EMBO Mol. Med. 2017;9:869–879. doi: 10.15252/emmm.201707550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P., Kalisky T., Sahoo D., Rajendran P.S., Rothenberg M.E., Leyrat A.A., Sim S., Okamoto J., Johnston D.M., Qian D. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat. Biotechnol. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa e Melo F., Kurtova A.V., Harnoss J.M., Kljavin N., Hoeck J.D., Hung J., Anderson J.E., Storm E.E., Modrusan Z., Koeppen H. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature. 2017;543:676–680. doi: 10.1038/nature21713. [DOI] [PubMed] [Google Scholar]
- Drost J., van Jaarsveld R.H., Ponsioen B., Zimberlin C., van Boxtel R., Buijs A., Sachs N., Overmeer R.M., Offerhaus G.J., Begthel H. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- Farin H.F., Van Es J.H., Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology. 2012;143:1518–1529.e7. doi: 10.1053/j.gastro.2012.08.031. [DOI] [PubMed] [Google Scholar]
- Fujii M., Shimokawa M., Date S., Takano A., Matano M., Nanki K., Ohta Y., Toshimitsu K., Nakazato Y., Kawasaki K. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18:827–838. doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Fumagalli A., Drost J., Suijkerbuijk S.J., van Boxtel R., de Ligt J., Offerhaus G.J., Begthel H., Beerling E., Tan E.H., Sansom O.J. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc. Natl. Acad. Sci. USA. 2017;114:E2357–E2364. doi: 10.1073/pnas.1701219114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.E., Birren S.J., Saito T., Anderson D.J. DNA binding and transcriptional regulatory activity of mammalian achaete-scute homologous (MASH) proteins revealed by interaction with a muscle-specific enhancer. Proc. Natl. Acad. Sci. USA. 1992;89:3596–3600. doi: 10.1073/pnas.89.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubb A.M., Chalasani S., Frantz G.D., Smits R., Grabsch H.I., Kavi V., Maughan N.J., Hillan K.J., Quirke P., Koeppen H. Achaete-scute like 2 (ascl2) is a target of Wnt signalling and is upregulated in intestinal neoplasia. Oncogene. 2006;25:3445–3457. doi: 10.1038/sj.onc.1209382. [DOI] [PubMed] [Google Scholar]
- Kreso A., O’Brien C.A., van Galen P., Gan O.I., Notta F., Brown A.M., Ng K., Ma J., Wienholds E., Dunant C. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:543–548. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., Minden M., Paterson B., Caligiuri M.A., Dick J.E. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Metcalfe C., Kljavin N.M., Ybarra R., de Sauvage F.J. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 2014;14:149–159. doi: 10.1016/j.stem.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Muñoz J., Stange D.E., Schepers A.G., van de Wetering M., Koo B.K., Itzkovitz S., Volckmann R., Kung K.S., Koster J., Radulescu S. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent “+4” cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien C.A., Pollett A., Gallinger S., Dick J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- Prasetyanti P.R., Zimberlin C.D., Bots M., Vermeulen L., Melo Fde.S., Medema J.P. Regulation of stem cell self-renewal and differentiation by Wnt and Notch are conserved throughout the adenoma-carcinoma sequence in the colon. Mol. Cancer. 2013;12:126. doi: 10.1186/1476-4598-12-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci-Vitiani L., Lombardi D.G., Pilozzi E., Biffoni M., Todaro M., Peschle C., De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- Rothenberg M.E., Nusse Y., Kalisky T., Lee J.J., Dalerba P., Scheeren F., Lobo N., Kulkarni S., Sim S., Qian D. Identification of a cKit+ colonic crypt base secretory cell that supports Lgr5+ stem cells in mice. Gastroenterology. 2012;142:1195–1205.e6. doi: 10.1053/j.gastro.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki N., Sachs N., Wiebrands K., Ellenbroek S.I., Fumagalli A., Lyubimova A., Begthel H., van den Born M., van Es J.H., Karthaus W.R. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc. Natl. Acad. Sci. USA. 2016;113:E5399–E5407. doi: 10.1073/pnas.1607327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Sato T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van de Wetering M., Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers A.G., Snippert H.J., Stange D.E., van den Born M., van Es J.H., van de Wetering M., Clevers H. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- Schuijers J., Junker J.P., Mokry M., Hatzis P., Koo B.K., Sasselli V., van der Flier L.G., Cuppen E., van Oudenaarden A., Clevers H. Ascl2 acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell. 2015;16:158–170. doi: 10.1016/j.stem.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Scott I.C., Anson-Cartwright L., Riley P., Reda D., Cross J.C. The HAND1 basic helix-loop-helix transcription factor regulates trophoblast differentiation via multiple mechanisms. Mol. Cell. Biol. 2000;20:530–541. doi: 10.1128/mcb.20.2.530-541.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa M., Ohta Y., Nishikori S., Matano M., Takano A., Fujii M., Date S., Sugimoto S., Kanai T., Sato T. Visualization and targeting of LGR5+ human colon cancer stem cells. Nature. 2017;545:187–192. doi: 10.1038/nature22081. [DOI] [PubMed] [Google Scholar]
- Tetteh P.W., Basak O., Farin H.F., Wiebrands K., Kretzschmar K., Begthel H., van den Born M., Korving J., de Sauvage F., van Es J.H. Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell. 2016;18:203–213. doi: 10.1016/j.stem.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Tian H., Biehs B., Warming S., Leong K.G., Rangell L., Klein O.D., de Sauvage F.J. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M., Sancho E., Verweij C., de Lau W., Oving I., Hurlstone A., van der Horn K., Batlle E., Coudreuse D., Haramis A.P. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- van de Wetering M., Francies H.E., Francis J.M., Bounova G., Iorio F., Pronk A., van Houdt W., van Gorp J., Taylor-Weiner A., Kester L. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Flier L.G., Sabates-Bellver J., Oving I., Haegebarth A., De Palo M., Anti M., Van Gijn M.E., Suijkerbuijk S., Van de Wetering M., Marra G., Clevers H. The intestinal Wnt/TCF signature. Gastroenterology. 2007;132:628–632. doi: 10.1053/j.gastro.2006.08.039. [DOI] [PubMed] [Google Scholar]
- van der Flier L.G., van Gijn M.E., Hatzis P., Kujala P., Haegebarth A., Stange D.E., Begthel H., van den Born M., Guryev V., Oving I. Transcription factor Achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- van Es J.H., Jay P., Gregorieff A., van Gijn M.E., Jonkheer S., Hatzis P., Thiele A., van den Born M., Begthel H., Brabletz T. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat. Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- van Es J.H., Sato T., van de Wetering M., Lyubimova A., Yee Nee A.N., Gregorieff A., Sasaki N., Zeinstra L., van den Born M., Korving J. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verissimo C.S., Overmeer R.M., Ponsioen B., Drost J., Mertens S., Verlaan-Klink I., Gerwen B.V., van der Ven M., Wetering M.V., Egan D.A. Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. eLife. 2016;5:e18489. doi: 10.7554/eLife.18489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen L., De Sousa E Melo F., van der Heijden M., Cameron K., de Jong J.H., Borovski T., Tuynman J.B., Todaro M., Merz C., Rodermond H. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- Wang C.Y., Shahi P., Huang J.T., Phan N.N., Sun Z., Lin Y.C., Lai M.D., Werb Z. Systematic analysis of the Achaete-scute complex-like gene signature in clinical cancer patients. Mol. Clin. Oncol. 2017;6:7–18. doi: 10.3892/mco.2016.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.