Abstract
The transcription factor GaaR is needed for the expression of genes required for pectin degradation and transport and catabolism of the main degradation product, D-galacturonic acid (GA) in Aspergillus niger. In this study, we used the strong constitutive gpdA promoter of Aspergillus nidulans to overexpress gaaR in A. niger. Overexpression of gaaR resulted in an increased transcription of the genes encoding pectinases, (putative) GA transporters, and catabolic pathway enzymes even under non-inducing conditions, i.e., in the absence of GA. Exoproteome analysis of a strain overexpressing gaaR showed that this strain secretes highly elevated levels of pectinases when grown in fructose. The genes encoding exo-polygalacturonases were found to be subjected to CreA-mediated carbon catabolite repression, even in the presence of fructose. Deletion of creA in the strain overexpressing gaaR resulted in a further increase in pectinase production in fructose. We showed that GaaR localizes mainly in the nucleus regardless of the presence of an inducer, and that overexpression of gaaR leads to an increased concentration of GaaR in the nucleus.
Electronic supplementary material
The online version of this article (10.1007/s00253-018-8753-7) contains supplementary material, which is available to authorized users.
Keywords: Transcriptome, Exoproteome, Gene regulation, Transcription factor localization, GFP fluorescence, Transcription factor concentration
Introduction
Aspergillus niger is an important filamentous fungus for the industrial production of pectinases (Pedrolli et al. 2009). Pectinases are widely used in the food industry (Kashyap et al. 2001; Toushik et al. 2017; Khan et al. 2013) and are important enzymes in the utilization of pectin-rich feedstock in biofuel production (Edwards and Doran-Peterson 2012). Pectin is a complex plant cell wall polysaccharide and four substructures have been defined which include polygalacturonic acid (PGA), rhamnogalacturonan I, rhamnogalacturonan II, and xylogalacturonan. PGA is the most abundant pectic substructure and consists of D-galacturonic acid (GA) residues. GA is also present in the backbones of rhamnogalacturonan I, rhamnogalacturonan II, and xylogalacturonan (Caffall and Mohnen 2009).
A. niger contains a large number of enzymes potentially acting on pectin substructures (Martens-Uzunova and Schaap 2009; Coutinho et al. 2009; De Vries et al. 2017). In the presence of GA, the main sugar acid in pectin, the expression of the genes encoding pectinases (Martens-Uzunova and Schaap 2009; Alazi et al. 2016), the GA transporter GatA (Sloothaak et al. 2014), and the GA catabolic pathway enzymes GaaA, GaaB, GaaC, and GaaD (Martens-Uzunova and Schaap 2008; Alazi et al. 2016) are induced via the Zn2Cys6 type transcription factor GaaR (Alazi et al. 2016). Apart from the transcriptional activator (GaaR), the expression of GA-responsive genes is controlled by a repressor protein, GaaX. Loss of function of GaaX leads to constitutive and inducer-independent expression of pectinases (Niu et al. 2017). The repressor protein GaaX is postulated to inhibit the transcriptional activity of GaaR under non-inducing conditions, i.e., in the absence of GA. The presence of an inducer is suggested to inhibit the repressing activity of GaaX, thereby leading to the transcriptional induction of GA-responsive genes via GaaR (Niu et al. 2017). The GA catabolic pathway intermediate 2-keto-3-deoxy-L-galactonate has recently been identified as the physiological inducer of the GA-responsive genes (Alazi et al. 2017).
Overexpression of transcription factors has been shown to be an effective method to increase the expression of their target genes in Saccharomyces cerevisiae, even under conditions in which the transcription factors under consideration are normally not active (Chua et al. 2006). Similarly, overexpression of transcription factors involved in plant biomass degradation in filamentous fungi, such as xlnR (Noguchi et al. 2009) and manR (Ogawa et al. 2012) in Aspergillus oryzae and xyr1 in Trichoderma reesei (Jiang et al. 2016), was previously reported to result in elevated expression of their target genes in the presence of inducers. Inducer-independent production of cellulases was also observed in T. reesei strains overexpressing xyr1 (Lv et al. 2015; Wang et al. 2013).
In this study, we demonstrate that overexpression of gaaR results in constitutive transcription and secretion of pectinases under non-inducing conditions, probably by disturbing the stoichiometric balance of GaaR and GaaX in favor of GaaR. We further show that the effect of gaaR overexpression on pectinase production is sensitive to CreA-mediated carbon catabolite repression even when fructose, a less repressing carbon source compared to glucose, was used. A further increase in pectinase production on fructose upon gaaR overexpression was accomplished when the CreA-mediated carbon catabolite repression was inactivated via creA deletion.
Materials and methods
Strains, media, and growth conditions
All A. niger strains used in this study are listed in Online Resource 1. Media were prepared as described previously (Arentshorst et al. 2012). Radial growth assays of the strains were performed on minimal medium (MM) (pH 5.8) containing 1.5% (w/v) agar (Scharlau, Barcelona, Spain) and various carbon sources: 50 mM glucose (VWR International, Amsterdam, The Netherlands), fructose (Sigma-Aldrich, Zwijndrecht, The Netherlands), or GA (Chemodex, St Gallen, Switzerland), or 1% (w/v) PGA (Sigma, Zwijndrecht, The Netherlands) or apple pectin (AP) (Sigma-Aldrich, Zwijndrecht, The Netherlands). Plates were inoculated with 5 μL 0.9% NaCl containing 104 freshly harvested spores and cultivated at 30 °C for 7 days. MM (pH 5.8) containing 1.5% (w/v) agar, 10 mM acetamide (Sigma-Aldrich, Steinheim, Germany) as the sole nitrogen source, and acetate (Merck, Darmstadt, Germany), glucose, fructose, sorbitol (Roth, Karlsruhe, Germany) or GA as the carbon source was prepared as described previously (Arentshorst et al. 2012). Plates were inoculated with 5 μL 0.9% NaCl containing 5 × 104 freshly harvested spores. Filter sterilized carbon source solutions were added after autoclaving MM containing agar. PGA and AP were autoclaved together with the medium. All growth experiments were performed in duplicate.
For enzymatic analysis, 106 freshly harvested spores were inoculated per mL in 100 mL shake flasks that include 25 or 50 mL MM (pH 5.8) containing 50 mM glucose, fructose, sorbitol, or GA and were grown for 36 h in a rotary shaker at 30 °C and 250 rpm. Experiments were performed in duplicate.
For microscopic analysis of the co-localization of the nuclear specific SYTO59 dye (Invitrogen, Eugene, Oregon, USA) with the eGFP-tagged H2B protein, conidia of the MA26.1 strain were propagated on complete medium containing 1.5% (w/v) agar. 2 × 105 freshly harvested spores were placed on cover slips in a Petri dish with 20 mL MM containing 50 mM fructose and grown at 30 °C. After 16 h, the cover slips were rinsed twice with water and transferred to a new Petri dish with 20 mL MM containing 50 mM GA and growth was continued at 30 °C for 1.5 h. For microscopic analysis of the co-localization of the nuclear specific SYTO59 dye with the eGFP-tagged GaaX or GaaR proteins, conidia of the JN126.2, EA19.2, and EA20.10 strains were propagated on MM containing 1.5% (w/v) agar and 50 mM GA. 3 × 105 freshly harvested spores were inoculated on cover slips in Petri dishes that include 3 mL MM containing10 mM GA and 0.003% yeast extract and grown at 30 °C for approximately 22 h. For microscopic analysis of the fluorescence intensity, conidia of the EA19.2 and EA20.10 strains were propagated on complete medium containing 1.5% (w/v) agar. 2 × 105 freshly harvested spores were placed on cover slips in Petri dishes that include 20 mL MM containing 50 mM fructose, or 50 mM GA and 1 mM fructose, and grown at 30 °C for 17.5 h. For each condition, two biological replicates were performed.
Construction of strains overexpressing gaaR
Protoplast-mediated transformation of A. niger, purification of the transformants and extraction of the genomic DNA were performed as described by Arentshorst et al. (2012).
The plasmid pEA4 containing the PgpdA-gaaR-TtrpC construct was created as follows: the Aspergillus nidulans gpdA promoter was obtained from plasmid pAN52.1-NOTI (Punt et al. 1987) by restriction digestion with NotI and NcoI. The gaaR gene was amplified by PCR using the primer pairs listed in Online Resource 2 with A. niger N402 genomic DNA as template, ligated into pJET1.2/blunt cloning vector (Thermo Fisher Scientific, Carlsbad, CA) and amplified in Escherichia coli DH5α. Following plasmid isolation, gaaR was excised using restriction enzymes PscI and BglII. The NotI-NcoI fragment of PgpdA and the PscI-BglII fragment of gaaR were ligated into NotI-BamHI opened pAN52.1-NOTI. pEA4 was sequenced to ensure no PCR errors have occurred and proper ligation and orientation of the fragments. To create strains EA21.3, EA21.5, EA21.6, and EA21.8, pEA4 was co-transformed into strain JN36.1 together with the plasmid pMA357 containing the A. nidulans amdS gene behind the A. nidulans gpdA promoter (Alazi et al. 2016). Transformants were selected on plates containing acetamide as the sole nitrogen source. To create strain TK1.1, strain JN36.1 was co-transformed with pEA4 and the plasmid p3SR2 (Hynes et al. 1983). p3SR2 contains the A. nidulans amdS gene behind the endogenous amdS promoter (Hynes et al. 1983). Transformants were selected on plates containing acrylamide as the sole nitrogen source. Ectopic integration of the PgpdA-gaaR-TtrpC construct was confirmed via Southern blot analysis. Genomic DNA was restricted overnight with NcoI restriction enzyme. A 501 bp fragment containing the gaaR gene was PCR-amplified using the primer pairs listed in Online Resource 2 with N402 genomic DNA as template, and was used as a probe.
Construction of strains (over)expressing eGFP-gaaR
The gaaR and eGFP genes were amplified by PCR using the primer pairs listed in Online Resource 2 with N402 genomic DNA and the plasmid pFG029 (unpublished vector, containing PgpdA-eGFP-TtrpC) as template, respectively. eGFP and gaaR were combined by fusion PCR using primers eGFP_P1_NcoI and gaaR_comp_P2_BglII, and the eGFP-gaaR fusion product was ligated into pJET1.2/blunt cloning vector and amplified in E. coli DH5α. Following plasmid isolation, the eGFP-gaaR fusion product was excised in two parts using restriction enzymes NcoI and BglII, resulting in an NcoI-NcoI fragment and an NcoI-BglII fragment.
The plasmid pEA3 containing the PgpdA-eGFP-gaaR-TtrpC construct was created as follows: The NotI-NcoI fragment of PgpdA and the NcoI-BglII fragment of eGFP-gaaR were ligated into NotI-BamHI opened pAN52.1-NOTI. The resulting plasmid was digested with NcoI and ligated with the NcoI-NcoI fragment of eGFP-gaaR. pEA3 was sequenced to ensure no PCR errors and proper ligation and orientation of the fragments. Strain EA20.10 was created by co-transformation of strain JN36.1 with pEA3 together with the plasmid pMA357.
To construct plasmid pEA2 (PgaaR-eGFP-gaaR-TtrpC), the gaaR promoter was PCR-amplified using the primer pairs listed in Online Resource 2 with N402 genomic DNA as template, ligated into pJET1.2/blunt cloning vector and amplified in E. coli DH5α. Following plasmid isolation, PgaaR was excised using restriction enzymes NotI and NcoI. pEA2 was created in a similar way to pEA3, except that the NotI-NcoI fragment of PgaaR was used instead of PgpdA. pEA2 was sequenced to ensure no PCR errors and proper ligation and orientation of the fragments. Strain EA19.2 was created by co-transformation of strain JN36.1 with pEA2 together with the plasmid pMA357 and transformants were selected on plates containing acetamide as the sole nitrogen source. Ectopic integrations of the PgpdA-eGFP-gaaR-TtrpC and PgaaR-eGFP-gaaR-TtrpC constructs were confirmed by diagnostic PCRs (data not shown).
Construction of creA deletion strains
Loss of the pyrE gene in EA21.6 was mediated by counter selection on MM-5’-FOA plates (Arentshorst et al. 2012), resulting in the strain EA23.6. The split marker approach was employed in the deletion of the creA gene (Arentshorst et al. 2015). 5′ and 3′ flanks of creA were PCR-amplified using the primer pairs listed in Online Resource 2 with N402 genomic DNA as template. The A. nidulans pyrF gene (named pyrE in A. niger) was PCR-amplified as two fragments using the primer pairs listed in Online Resource 2 with A. nidulans strain A234genomic DNA as template. Split marker fragments with the pyrF selection marker were created by fusion PCR and used to transform the strain EA23.6, resulting in the strain TK2.1. Proper deletion of creA was confirmed by diagnostic PCR (data not shown).
MA342.2 was also constructed using the split marker approach (Arentshorst et al. 2015). 5′ and 3′ flanks of creA were PCR-amplified using the primer pairs listed in Online Resource 2 with N402 genomic DNA as template. The hygromycin resistance cassette was PCR-amplified using primers hygP3f and hygP4r and a derivative of pAN7.1 (Punt et al. 1987) as template. creA-hygR split marker fragments were created by fusion PCR and transformed to strain MA234.1, resulting in the ΔcreA strain MA342.2. Proper deletion of creA was confirmed by diagnostic PCR (data not shown).
Bioreactor cultivations and transcriptome and exoproteome analyses
Controlled bioreactor cultivations of MA234.1 (the reference strain) (in triplicate) and JN123.1 (ΔgaaX) (in duplicate) in MM containing 0.75% fructose and the subsequent transcriptome analyses were performed previously (Niu et al. 2017). Controlled bioreactor cultivations of the EA21.6strain (OEgaaR) (in duplicate) under exactly the same growth conditions and the subsequent RNA-seq analyses were performed as previously described by Niu et al. (2017). Both biomass accumulation (offline) and base addition (online) were determined to monitor exponential growth.
Broth samples were taken during exponential growth after every 4 mL of base addition. RNA isolated from exponentially growing cells at the sample point at which about 75–80% of the maximum biomass yield was reached was used for the RNA-seq experiment. RNA-seq data were submitted to the Sequence Read Archive under accession number SRP078485 for MA234.1 and JN123.1 (Niu et al. 2017) and accession number SRP114830 for EA21.6 (this study).
Supernatant samples from an exponentially growing culture of each strain at two successive sample points (based on base addition) following the RNA-seq sample point were withdrawn and filtered. The filtered supernatants were lyophilized, resuspended in 1 mL 50 mM citric acid buffer pH 5.0, and used for the exoproteome analysis. For each sample, proteins were precipitated with TCA (trichloroacetic acid), the pellet was washed twice with acetone and resuspended in 75 μL 200 mM ammonium bicarbonate and 0.1% AALS II. Protein concentrations were determined by the RCDC assay Kit (BioRad, Mississauga, Ontario). Three micrograms of total protein were loaded on 12% SDS-PAGE gel. The gel was colored with silver stain and developed for 3 min. Five micrograms of total protein were trypsin digested in solution overnight at 37 °C. Samples were desalted with C18 ziptips (Millipore, Billerica, MA), the eluate was dried and the peptides were resuspended in 50 μL 5% acetonitrile and 0.1% formic acid. Five microlitres of peptide digest were analyzed by LC-MS/MS on a Velos LTQ-Orbitrap. Extracted Ion Chromatogram (EIC) peak area values of proteins were calculated by averaging the top three most abundant peptide ion EIC values assigned to each protein as per the Proteome Discoverer 1.4 (Thermo Fisher, San Jose, CA) precursor peak area quantification workflow. Protein EIC area values were normalized using the determined value of a fixed spiked amount of trypsin-digested bovine serum albumin.
Enzymatic analysis
Supernatants from bioreactor or shake flask cultures were obtained by filtration through glass microfiber filters (Whatman, Buckinghamshire, UK) or sterile miracloth, and the filtrate was stored at − 80 °C. PGA plate assays were performed as described by Niu et al. (2017). Twenty-five microlitres of supernatant from each culture was spotted on plates containing 0.2% or 0.5% PGA, and plates were incubated at 37 °C for 16 or 20 h. PGA degradation was indicated by the formation of a clear zone of hydrolysis.
Microscopy
The coverslips with adherent germlings were placed upside down on glass slides and observed under a Zeiss Observer confocal laser scanning microscope (Zeiss, Jena, Germany). For nuclear staining, 0.5 mL of 25 μM SYTO59 dye solution was dropped on glass slides before placing the cover slips, and imaging was performed approximately after 2 h. The GFP and SYTO59 fluorescence were excited using 488 and 625 nm laser lines, respectively. Images were analyzed using the ImageJ software (Abramoff et al. 2004). To analyze the fluorescence intensity, 1–2 images were taken for each biological replicate. On each image, the exact same brightness and contrast adjustments were applied, 3–10 nuclear and 3–10 cytoplasmic fluorescence intensities in a defined area were measured, and calibrated for the background fluorescence.
Results
Expression of pectinase genes is upregulated in strains overexpressing gaaR
To create strains that overexpress the GA-responsive transcription factor gene gaaR (OEgaaR), the A. niger gaaR gene was fused with the strong constitutive A. nidulans gpdA promoter and transformed into a ΔgaaR strain. Southern blot analysis indicated that the gaaR overexpression construct was ectopically integrated in one or two copies in the genomes of the OEgaaR strains EA21.3, EA21.5, EA21.6, and EA21.8 (Fig. ESM_3.1 b). An additional multicopy OEgaaR strain, TK1.1, was obtained by using a more stringent selection method using acrylamide, and Southern blot analysis confirmed ectopic integration of at least four copies of the gaaR overexpression construct in its genome (Fig. ESM_3.1 c). We compared the radial growth of the OEgaaR strains on different monomeric and polymeric carbon sources (Fig. ESM_3.2). The ΔgaaR strain showed a strongly reduced growth on GA, PGA, and AP as previously shown (Alazi et al. 2016). Reintroducing the gaaR gene expressed from the gpdA promoter resulted in growth on GA, PGA, and AP in EA21.3, EA21.5, EA21.6, and EA21.8, indicating the presence of a functional GaaR. However, the OEgaaR strains showed partial and different levels of complementation of growth on GA-containing carbon sources (Fig. ESM_3.2 a). The TK1.1 strain, containing the highest copy number of the gaaR overexpression construct, showed a severely impaired growth on GA, PGA, and AP (data not shown and Fig. ESM_3.2 b). Growth of all OEgaaR strains on glucose or fructose was similar to the growth of the reference strain.
To assess the pectinase production capacity of the OEgaaR strains, the strains were grown in shake flasks in minimal medium containing non-inducing (50 mM glucose, 50 mM fructose, or 50 mM sorbitol) or inducing (50 mMGA) carbon sources, and the culture supernatants were spotted on PGA plates. As indicated by the clear zones of hydrolysis on PGA plates, the polygalacturonase activity in the culture supernatants of the OEgaaR strains EA21.3, EA21.5, EA21.6, and EA21.8 grown in glucose or fructose was higher compared to the reference strain (Fig. ESM_3.3 a). The culture supernatant of EA21.6 grown on sorbitol or GA displayed the highest polygalacturonase activity compared to EA21.3, EA21.5, EA21.8, and the reference strain (Fig. ESM_3.3 a and data not shown). EA21.6 was selected to be used in further experiments based on the growth profiles and pectinase production capacities of the OEgaaR strains. A further increased level of polygalacturonase activity was observed in the culture supernatant of TK1.1 grown on fructose compared to the reference strain as well as to EA21.6 (Fig. ESM_3.3 b).
Transcriptome analysis of the OEgaaR strain
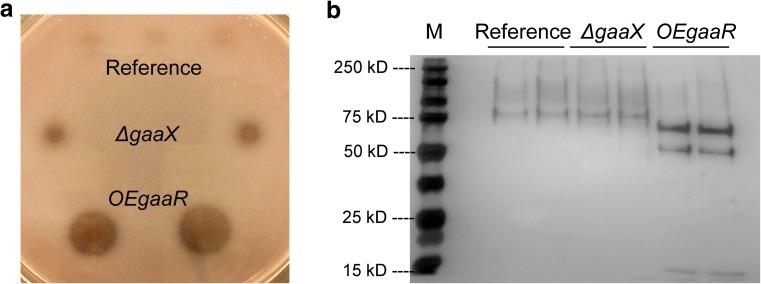
To investigate the expression of the multitude of genes involved in pectin degradation, GA transport, and catabolism in a strain overexpressing gaaR, we performed a genome-wide gene expression analysis using RNA-seq (Online Resource 4). The reference and OEgaaR (EA21.6) strains were grown in bioreactors on fructose, a carbon source that does not induce the expression of GA-responsive genes (Martens-Uzunova and Schaap 2008). Growth of the OEgaaR strain under the controlled bioreactor conditions (μmax 0.200 ± 0.001 g dry weight/kg/h, Ymax 4.117 ± 0.167 g dry weight/kg (n = 2)) was similar to the growth of the reference strain (μmax 0.214 ± 0.007 g dry weight/kg/h, Ymax 4.151 ± 0.134 g dry weight/kg (n = 3)). Analysis of RNA-seq data showed that the expression of gaaR was highly increased in the OEgaaR strain compared to the reference strain with a fold change of 63.8 (Online Resource 4 and Table 1). Overexpression of gaaR resulted in the upregulation (FC ≥ 4, FDR ≤ 0.001) of 19 of 48 genes encoding extracellular enzymes specifically assigned to the degradation of pectin according to de Vries et al. (2017) (see Table 1). Almost all of these genes (18 out of 19) belong to the GaaR/GaaX panregulon (Niu et al. 2017) and include several exo- and endo-polygalacturonases, pectin methylesterases, and pectin lyases, all acting on the PGA backbone of pectin, as well as the xylogalacturonase NRRL3_07469 acting on xylogalacturonan, and the arabinogalactanase encoded by gan53A and the α-L-rhamnosidase NRRL3_10558 acting on rhamnogalacturonan I. Nine of 19 pectinases that were upregulated in the OEgaaR strain were previously shown to be upregulated in ΔgaaX (JN123.1), the repressor deletion mutant, on fructose compared to the reference strain (FC ≥ 4, FDR ≤ 0.001) (Niu et al. 2017). The expression of these nine pectinase genes, except NRRL3_05252, was generally much lower in ΔgaaX than in the OEgaaR strain, and no additional pectinases were found to be upregulated in ΔgaaX (Table 1). Constitutive production of pectinases in the OEgaaR strain and to a lesser extent in ΔgaaX grown in bioreactors on fructose was also observed via a PGA plate assay (Fig. 1a). This analysis clearly indicates that overexpression of gaaR results in a more dramatic increase in the expression of several pectinases compared to deletion of the repressor gaaX.
Table 1.
Transcriptome and exoproteome analyses of predictive pectinases, GA transporters and catabolic pathway genes, gaaR and gaaX in the reference, OEgaaR and ΔgaaX strains. Transcript and extracellular protein levels are represented as TPM and normalized protein (EIC) area values, respectively. Transcript levels with a fold change≥4 and FDR≤0.001 compared to the reference strain are indicated with an asterisk. Genes belonging to the GaaR/GaaX panregulon (Niu et al. 2017) are written in bold. Transcriptome data for the reference and ΔgaaX strains were taken from Niu et al. 2017
| Gene ID NRRL3 |
Gene ID
CBS 513.88 |
Description | CAZy family | Transcript level (ave TPM) |
Extracellular protein level
(ave normalized protein (EIC) area) |
||||
|---|---|---|---|---|---|---|---|---|---|
| Reference | ΔgaaX | OEgaaR | Reference | ΔgaaX | OEgaaR | ||||
| NRRL3_00169 | An09g02160 | rhamnogalacturonan acetylesterase | CE12 | 0.455 | 0.680 | 0.822 | #N/A | #N/A | #N/A |
| NRRL3_07501 | An04g09360 | carbohydrate esterase family 12 protein | CE12 | 1.313 | 1.134 | 1.680 | 0.00 | 0.00 | 1.31 |
| NRRL3_08325 | An03g06310 | pectin methylesterase Pme8A | CE8 | 0.037 | 0.556* | 24.737* | 0.00 | 1.48 | 105.91 |
| NRRL3_07470 | An04g09690 | pectin methylesterase | CE8 | 0.755 | 5.722* | 57.977* | 0.55 | 7.49 | 45.73 |
| NRRL3_05252 | An02g12505 | pectin methylesterase-like protein | CE8 | 1.061 | 31.320* | 17.513* | 0.00 | 109.39 | 48.98 |
| NRRL3_00839 | An14g02920 | glycoside hydrolase family 105 protein | GH105 | 4.997 | 5.090 | 4.027 | #N/A | #N/A | #N/A |
| NRRL3_01038 | An14g05340 | glycoside hydrolase family 105 protein | GH105 | 0.330 | 0.367 | 0.289 | #N/A | #N/A | #N/A |
| NRRL3_06782 | An16g06990 | endo-polygalacturonase Pga28A | GH28 | 0.000 | 0.000 | 0.000 | 8.56 | 11.51 | 19.74 |
| NRRL3_05859 | An02g04900 | endo-polygalacturonase Pga28B | GH28 | 16.541 | 18.675 | 27.407 | 0.00 | 0.22 | 1.76 |
| NRRL3_08805 | An05g02440 | endo-polygalacturonase Pga28C | GH28 | 0.053 | 0.596 | 376.634* | 0.00 | 0.00 | 321.96 |
| NRRL3_00263 | An09g03260 | endo-polygalacturonase Pga28D | GH28 | 0.929 | 1.066 | 1.322 | 0.605 | 0 | 0 |
| NRRL3_02835 | An01g14670 | endo-polygalacturonase Pga28E | GH28 | 0.483 | 1.152 | 479.334* | 0.00 | 0.00 | 415.78 |
| NRRL3_02571 | An01g11520 | endo-polygalacturonase Pga28I | GH28 | 0.215 | 1.243 | 23.092* | 0.00 | 0.00 | 8.40 |
| NRRL3_04000 | An15g05370 | endo-polygalacturonase Pga28II | GH28 | 0.293 | 1.055 | 2.076* | 0.00 | 2.94 | 11.24 |
| NRRL3_03144 | An12g07500 | exo-polygalacturonase | GH28 | 1.361 | 51.633* | 65.839* | 0.00 | 23.17 | 46.86 |
| NRRL3_08281 | An03g06740 | exo-polygalacturonase Pgx28B | GH28 | 0.000 | 2.102* | 14.290* | 0.00 | 1.04 | 7.85 |
| NRRL3_05260 | An02g12450 | exo-polygalacturonase Pgx28C | GH28 | 0.955 | 16.768* | 45.050* | 0.00 | 10.00 | 27.24 |
| NRRL3_09810 | An11g04040 | exo-polygalacturonase | GH28 | 0.007 | 0.120 | 1.221* | #N/A | #N/A | #N/A |
| NRRL3_07469 | An04g09700 | xylogalacturonase | GH28 | 0.167 | 0.344 | 4.828* | 0.00 | 0.00 | 10.86 |
| NRRL3_09126 | An12g00950 | endo-rhamnogalacturonase Rhg28A | GH28 | 5.967 | 6.717 | 4.859 | #N/A | #N/A | #N/A |
| NRRL3_00953 | An14g04200 | endo-rhamnogalacturonase Rhg28B | GH28 | 0.000 | 0.009 | 0.000 | 0.69 | 2.31 | 0.00 |
| NRRL3_04303 | An07g01000 | endo-rhamnogalacturonase | GH28 | 0.183 | 0.249 | 0.256 | #N/A | #N/A | #N/A |
| NRRL3_09450 | An11g08700 | endo-rhamnogalacturonase | GH28 | 1.380 | 1.585 | 1.286 | #N/A | #N/A | #N/A |
| NRRL3_11790 | An06g02070 | endo-rhamnogalacturonase | GH28 | 0.217 | 0.061 | 0.034 | #N/A | #N/A | #N/A |
| NRRL3_09623 | An11g06320 | endo-rhamnogalacturonase | GH28 | 0.169 | 0.301 | 0.035 | #N/A | #N/A | #N/A |
| NRRL3_02832 | An01g14650 | glycoside hydrolase family 28 protein | GH28 | 3.472 | 5.049 | 3.482 | #N/A | #N/A | #N/A |
| NRRL3_08631 | An03g02080 | alpha-L-rhamnosidase Rha28B | GH28 | 0.000 | 0.021 | 0.159 | #N/A | #N/A | #N/A |
| NRRL3_10559 | An18g04810 | glycoside hydrolase family 28 protein | GH28 | 0.084 | 3.112* | 16.880* | 0.00 | 2.34 | 7.39 |
| NRRL3_10643 | An18g05940 | arabinogalactanase Gan53A | GH53 | 1.738 | 3.850 | 28.517* | 0.00 | 2.91 | 22.91 |
| NRRL3_02162 | An01g06620 | alpha-L-rhamnosidase | GH78 | 9.524 | 8.130 | 8.251 | #N/A | #N/A | #N/A |
| NRRL3_03279 | An12g05700 | alpha-L-rhamnosidase-like protein | GH78 | 1.326 | 1.329 | 0.921 | #N/A | #N/A | #N/A |
| NRRL3_03924 | An15g04530 | glycoside hydrolase family 78 protein | GH78 | 0.000 | 0.000 | 0.000 | #N/A | #N/A | #N/A |
| NRRL3_04245 | An07g00240 | alpha-L-rhamnosidase-like protein | GH78 | 0.070 | 0.077 | 0.043 | #N/A | #N/A | #N/A |
| NRRL3_06304 | An10g00290 | alpha-L-rhamnosidase-like protein | GH78 | 1.072 | 1.365 | 1.259 | #N/A | #N/A | #N/A |
| NRRL3_07520 | An04g09070 | alpha-L-rhamnosidase | GH78 | 2.228 | 2.178 | 1.749 | #N/A | #N/A | #N/A |
| NRRL3_11451 | An08g09140 | alpha-L-rhamnosidase-like protein | GH78 | 0.085 | 0.166 | 0.009 | #N/A | #N/A | #N/A |
| NRRL3_10558 | An18g04800 | alpha-L-rhamnosidase | GH78 | 0.350 | 3.847* | 17.470* | #N/A | #N/A | #N/A |
| NRRL3_01739 | An01g01340 | glycoside hydrolase family 88 protein | GH88 | 0.163 | 0.219 | 0.212 | #N/A | #N/A | #N/A |
| NRRL3_00824 | sialidase-like protein | GH93 | 149.296 | 144.208 | 132.943 | #N/A | #N/A | #N/A | |
| NRRL3_00965 | An14g04370 | pectin lyase Pel1A | PL1_4 | 1.657 | 3.255 | 332.933* | 2.21 | 2.57 | 653.22 |
| NRRL3_08767 | An03g00190 | pectin lyase Pel1B | PL1_4 | 5.072 | 5.031 | 2.944 | #N/A | #N/A | #N/A |
| NRRL3_04153 | An15g07160 | pectin lyase Pel1C | PL1_4 | 18.220 | 18.640 | 18.246 | #N/A | #N/A | #N/A |
| NRRL3_06269 | pectin lyase | PL1_4 | 0.015 | 0.033 | 0.083 | #N/A | #N/A | #N/A | |
| NRRL3_01237 | An19g00270 | pectin lyase | PL1_4 | 0.174 | 1.475* | 13.613* | 0.00 | 0.00 | 20.51 |
| NRRL3_09811 | An11g04030 | pectin lyase | PL1_4 | 0.000 | 0.011 | 0.503* | #N/A | #N/A | #N/A |
| NRRL3_06359 | An10g00870 | pectate lyase Ply1A | PL1_7 | 0.148 | 0.134 | 1.366 | #N/A | #N/A | #N/A |
| NRRL3_00684 | An14g01130 | rhamnogalacturonan lyase | PL4_1 | 0.021 | 0.052 | 0.320 | #N/A | #N/A | #N/A |
| NRRL3_10115 | An11g00390 | rhamnogalacturonan lyase | PL4_3 | 0.648 | 0.810 | 11.176 | #N/A | #N/A | #N/A |
| NRRL3_00958 | An14g04280 | MFS-type sugar/inositol transporter GatA | 3.472 | 524.952* | 125.820* | #N/A | #N/A | #N/A | |
| NRRL3_08663 | An03g01620 | MFS-type sugar/inositol transporter | 0.274 | 5.622* | 0.577 | #N/A | #N/A | #N/A | |
| NRRL3_04281 | An07g00780 | MFS-type transporter | 3.110 | 4.596 | 13.958* | #N/A | #N/A | #N/A | |
| NRRL3_05650 | An02g07710 | D-galacturonate reductase GaaA | 19.917 | 1515.440* | 135.009* | #N/A | #N/A | #N/A | |
| NRRL3_06890 | An16g05390 | L-galactonate dehydratase GaaB | 47.765 | 6256.695* | 782.361* | #N/A | 3.78 | #N/A | |
| NRRL3_05649 | An02g07720 | 2-keto-3-deoxy-L-galactonate aldolase GaaC | 12.536 | 2283.765* | 240.798* | #N/A | 2.23 | #N/A | |
| NRRL3_10050 | An11g01120 | L-glyceraldehyde reductase GaaD | 256.409 | 2732.370* | 570.508 | #N/A | 6.00 | 0.24 | |
| NRRL3_08195 | An04g00780 | D-galacturonic acid responsive transcription factor GaaR | 14.444 | 18.512 | 850.848* | #N/A | #N/A | #N/A | |
| NRRL3_08194 | An04g00790 | Repressor of D-galacturonic acid utilization GaaX | 15.968 | 0.000 | 352.560* | #N/A | #N/A | #N/A | |
Fig. 1.
Enzymatic analysis and secretome profiles of the A. niger reference (MA234.1), ∆gaaX (JN123.1) and OEgaaR (EA21.6) strains grown in bioreactors on 0.75% fructose. a PGA plate assay. Supernatant from each bioreactor culture at the sample point following the RNA-seq sample point was spotted on a PGA plate. b Silver stained SDS-PAGE patterns of secretomes from a bioreactor culture of each strain at two successive sample points following the RNA-seq sample point. Three micrograms of total protein were loaded in each lane. Marker (M) molecular weight in kD is indicated
Comparison of the expression of the genes encoding the (putative) GA transporters and the GA catabolic pathway enzymes (gaaA, gaaB, gaaC, and gaaD) between the OEgaaR, the reference and ΔgaaX strains revealed that gatA, the putative GA transporter NRRL3_04281, gaaA, gaaB, and gaaC were significantly upregulated (FC ≥ 4, FDR ≤ 0.001) in the OEgaaR strain (Table 1). The expression of gaaD was also significantly (FDR ≤ 0.001) increased in the OEgaaR strain compared to the reference strain with a fold change of 2.5 (Table 1). Interestingly, the expression of the genes encoding the GA catabolic pathway enzymes were moderately induced in the OEgaaR strain and expressed at much higher levels in the ΔgaaX strain. In contrast, many of the genes encoding the extracellular enzymes were expressed at higher levels in the OEgaaR strain compared to the ΔgaaX strain (see “Discussion”).
We also analyzed the effect of overexpression of gaaR on the expression of all 375 genes predicted to encode carbohydrate active enzymes (CAZymes) in A. niger strain NRRL3 (Online Resource 4). In addition to the above mentioned 19 pectinases (belonging to CAZy families CE8, GH28, GH53, GH78, PL1_4, and PL4_3), 20 CAZymes acting specifically on cellulose (AA9, GH5_5), starch (GH13_5, GH31), and xyloglucan (GH12, GH74, GH95) or acting on multiple substrates (CE16, GH18, GH3, GH35, GH43, GH51, GH54, GH79) (de Vries et al. 2017) were highly upregulated in the OEgaaR strain on fructose (Table 2).
Table 2.
Transcriptome and exoproteome analyses of 20 additional CAZymes upregulated in the OEgaaR strain compared to the reference strain. Transcript and extracellular protein levels are represented as TPM and normalized protein (EIC) area values, respectively. Transcript levels with a fold change≥4 and FDR≤0.001 compared to the reference strain are indicated with an asterisk. Genes belonging to the GaaR/GaaX panregulon (Niu et al. 2017) are written in bold. Transcriptome data for the reference and ΔgaaX strains were taken from Niu et al. 2017
| Gene ID NRRL3 |
Gene ID
CBS 513.88 |
Description | CAZy family | Transcript level (ave TPM) |
Extracellular protein level
(ave normalized protein (EIC) area) |
||||
|---|---|---|---|---|---|---|---|---|---|
| Reference | ΔgaaX | OEgaaR | Reference | ΔgaaX | OEgaaR | ||||
| NRRL3_00814 | An14g02670 | lytic polysaccharide monooxygenase | AA9 | 1.053 | 2.625 | 20.043* | #N/A | #N/A | #N/A |
| NRRL3_06379 | acetylesterase | CE16 | 0.042 | 0.142 | 24.511* | 0.00 | 0.00 | 0.84 | |
| NRRL3_06053 | An02g02540 | carbohydrate esterase family 16 protein | CE16 | 2.055 | 17.617* | 105.698* | 0.00 | 12.71 | 71.49 |
| NRRL3_01918 | An01g03340 | xyloglucanase Xeg12A | GH12 | 9.203 | 11.484 | 911.087* | #N/A | #N/A | #N/A |
| NRRL3_02746 | An01g13610 | glucan 1,4-alpha-maltohexaosidase | GH13_5 | 2.813 | 5.088 | 15.211* | #N/A | #N/A | #N/A |
| NRRL3_01212 | chitinase-like protein | GH18 | 8.261 | 25.549 | 43.967* | 0.00 | 0.00 | 0.34 | |
| NRRL3_06419 | An17g00300 | glycoside hydrolase family 3 protein | GH3 | 2.992 | 4.962 | 30.714* | 0.00 | 0.14 | 3.32 |
| NRRL3_00268 | An09g03300 | alpha-xylosidase Axl31A | GH31 | 1.211 | 3.397 | 8.676* | 0.00 | 0.00 | 0.00 |
| NRRL3_02630 | An01g12150 | beta-galactosidase Lac35B | GH35 | 1.589 | 2.203 | 33.505* | #N/A | #N/A | #N/A |
| NRRL3_02479 | An01g10350 | exo-beta-1,4-galactanase | GH35 | 3.480 | 22.294* | 24.938* | 0.00 | 1.39 | 0.00 |
| NRRL3_06244 | An02g00140 | glycoside hydrolase family 43 protein | GH43 | 0.812 | 22.185* | 22.908* | #N/A | #N/A | #N/A |
| NRRL3_04608 | An07g04930 | glycoside hydrolase family 43 protein | GH43 | 1.269 | 2.703 | 5.432* | #N/A | #N/A | #N/A |
| NRRL3_06791 | An16g06800 | glycoside hydrolase family 5 protein | GH5_5 | 22.647 | 68.809 | 665.632* | 24.31 | 128.89 | 700.71 |
| NRRL3_10865 | An08g01710 | alpha-arabinofuranosidase | GH51 | 0.795 | 11.209* | 10.280* | #N/A | #N/A | #N/A |
| NRRL3_03768 | An15g02300 | alpha-arabinofuranosidase Abf54B | GH54 | 0.807 | 3.673 | 10.070* | 0.38 | 6.53 | 2.49 |
| NRRL3_01787 | An01g01870 | xyloglucanase Xeg74C | GH74 | 0.055 | 0.174 | 4.919* | #N/A | #N/A | #N/A |
| NRRL3_00701 | An14g01330 | glycoside hydrolase family 79 protein | GH79 | 0.070 | 0.460 | 8.260* | #N/A | #N/A | #N/A |
| NRRL3_05305 | An02g11890 | beta-glucuronidase Gus79A | GH79 | 0.209 | 0.447 | 2.675* | 0.00 | 0.00 | 0.38 |
| NRRL3_07382 | An16g00540 | alpha-L-fucosidase-like protein | GH95 | 0.036 | 0.667* | 2.139* | 0.00 | 0.00 | 0.32 |
| NRRL3_07089 | An16g02760 | alpha-L-fucosidase | GH95 | 1.619 | 2.893 | 11.179* | 0.00 | 0.00 | 0.61 |
In total, 124 genes were significantly upregulated (FC ≥ 4, FDR ≤ 0.01) in the OEgaaR strain compared to the reference strain (Online Resource 4). The promoter regions of 110 upregulated genes for which the A. niger CBS 513.88 gene ID was available were screened for the presence of transcription factor binding sites using TFBSF (Meyer et al. 2009), and it was found that 69 genes contain the galacturonic acid-responsive element GARE (CCNCCAA) (Martens-Uzunova and Schaap 2008) required for GA-responsive gene induction (Niu et al. 2015) in their 1 kb upstream sequences. A gene ontology enrichment analysis via FetGOat (Nitsche et al. 2011) indicated that the genes upregulated in theOEgaaR strain were highly enriched with genes involved in carbohydrate (xyloglucan, pectin, lactose) metabolism. Out of 53 genes belonging to the GaaR/GaaX panregulon (Niu et al. 2017), 34 were upregulated in the OEgaaR strain, including gaaX with a fold change of 24.1 (Online Resource 4 and Table 1). Apart from the aforementioned genes, several genes with unknown relation to GA utilization were also upregulated in the OEgaaR strain. These include genes encoding hypothetical/uncharacterized proteins, proteins involved in diverse processes such as dehydrogenases and non-ribosomal peptide synthetases, MFS-type transporters and a Zn2Cys6 type transcription factor (NRRL3_11827) (Online Resource 4).
Exoproteome analysis of the OEgaaR strain
To support the observed transcriptional upregulation of CAZymes in the OEgaaR strain, we analyzed the exoproteome of the OEgaaR strain and compared it to the exoproteome of the reference and ΔgaaX strains grown in bioreactors on fructose (Online Resource 5). Mass-spectrometric analysis revealed 18 pectinases in the exoproteome of the OEgaaR strain. Seventeen of them were secreted at higher levels compared to the reference strain and the ΔgaaX strain. The protein level of the putative pectin methylesterase NRRL3_05252 was higher in ΔgaaX than in the OEgaaR strain similar to observed higher mRNA level of this gene in ΔgaaX (Table 1). Fifteen of these detected pectinases were also transcriptionally upregulated in the OEgaaR strain. In addition, eight genes encoding CAZymes that were expressed at higher levels in the OEgaaRstrain compared to the reference strain and ΔgaaX were found to accumulate at higher levels in the culture media of the OEgaaR strain (Table 2). With regard to the degradation of pectin, there is a good correspondence between the upregulated expression of genes and the increased extracellular accumulation of their encoding pectinolytic enzymes in the OEgaaR strain, for example the pectinases Pel1A, Pga28E, and Pga28C (Table 1). The distinct SDS-PAGE profile of the OEgaaR strain compared to the reference strain and ΔgaaX might represent the differences in abundance of the aforementioned extracellular proteins, such as NRRL3_06791 and Pel1A with predicted molecular weights of unglycosylated proteins of 54.8 and 39.7 kDa, respectively (Fig. 1b).
Nuclear concentration of GaaR is increased in the OEgaaR strain
Strains expressing an eGFP-tagged gaaR, directed by either the endogenous gaaR promoter (eGFP-gaaR) or the strong constitutive A. nidulans gpdA promoter (OEeGFP-gaaR), were constructed to investigate the subcellular localization of GaaR in the reference and OEgaaR strains, respectively. Expression of eGFP-tagged gaaR in a ∆gaaR background resulted in partial complementation of growth on GA and full complementation of growth on PGA and AP, in both eGFP-gaaR (EA19.2) and OEeGFP-gaaR (EA20.10) strains (Fig. ESM_3.2 a). The polygalacturonase activity in the culture supernatant of the OEeGFP-gaaR strain grown in fructose was higher compared to the eGFP-gaaR strain, resembling the polygalacturonase production capacities of the OEgaaR and reference strains, respectively (Fig. ESM_3.3 c). These results indicate that the eGFP-tagged GaaR is able to activate the transcription of the GA-responsive genes required for growth on GA-containing carbon sources, and that overexpression of the eGFP-tagged gaaR results in an increased accumulation of pectinases as in the overexpression of the untagged gaaR.
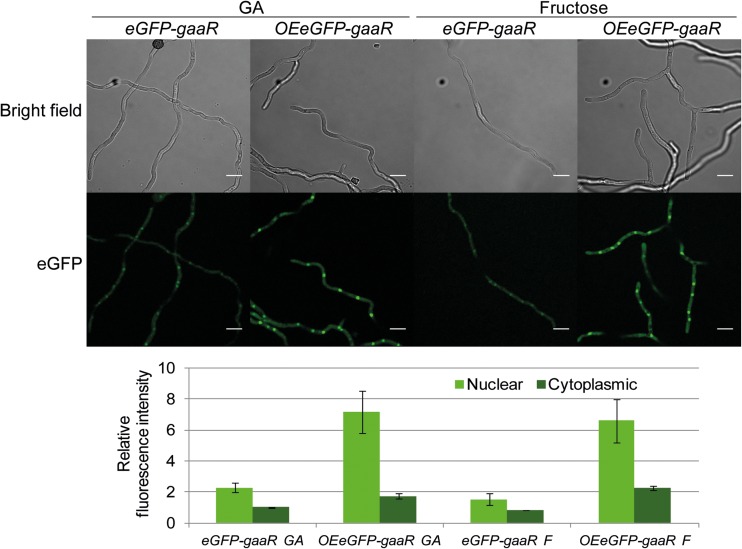
The subcellular localization of GaaR and GaaX was analyzed qualitatively using confocal laser scanning microscopy. As a nuclear marker suitable for co-localization experiments, the SYTO59 dye was used. The nuclear localization of the SYTO59 dye was confirmed in an A. niger strain harboring the eGFP-tagged H2B protein (MA26.1) (Fig. ESM_3.4 a). The eGFP-gaaR, OEeGFP-gaaR, and gaaX-eGFP (JN126.2) strains were grown in GA and nuclei were stained with SYTO59 (Fig. ESM_3.4 b). Both eGFP-GaaR and GaaX-eGFP were found to be present in the cytoplasm and nucleus, although we cannot exclude the possibility that eGFP was cleaved off from the fusion proteins and resulted in cytoplasmic or nuclear fluorescence signal. The co-localization experiment showed that eGFP-GaaR was mainly localized in the nucleus in both eGFP-gaaR and OEeGFP-gaaR strains. In the gaaX-eGFP strain grown in GA, GaaX-eGFP was present in both the cytoplasm and the nuclei at roughly the same level.
We next quantified the cytoplasmic and nuclear eGFP-GaaR intensity in the eGFP-gaaR and OEeGFP-gaaR strains grown in GA or fructose. As shown in Fig. 2, nuclear eGFP-GaaR fluorescence was higher than the cytoplasmic intensity regardless of the presence of an inducing carbon source or the promoter used to overexpress eGFP-tagged gaaR. The GFP signals in the eGFP-gaaR strain were low after growth in GA or fructose, confirming previous findings that gaaR is expressed at low levels on both GA and fructose (Alazi et al. 2016 and Table 1). Overexpression of eGFP-gaaR resulted in a much higher nuclear eGFP-GaaR concentration in the OEeGFP-gaaR strain than in the eGFP-gaaR strain, while only a slight increase in the cytoplasmic concentration was observed. This indicates that the excess eGFP-GaaR produced in the OEeGFP-gaaR strain localizes mainly in the nucleus. This result is in line with the observation that in a Botrytis cinerea strain overexpressing BcgaaR-eGFP, BcGaaR-eGFP mainly localizes in the nucleus under inducing or non-inducing conditions (Zhang et al. 2016).
Fig. 2.
Nuclear and cytoplasmic fluorescence intensity of the eGFP-tagged GaaR protein. The eGFP-gaaR (EA19.2) and OEeGFP-gaaR (EA20.10) strains were grown in MM containing 50 mM GA and 1 mM fructose, or 50 mM fructose (F) for 17.5 h. Example micrographs representing each condition are shown. Bars represent averages of two biological replicates and standard deviation is shown. Data is represented relative to the cytoplasmic fluorescence intensity in the eGFP-gaaR strain on GA. Scale bar 10 μm
Deletion of creA in the OEgaaR strain results in elevated production of pectinases
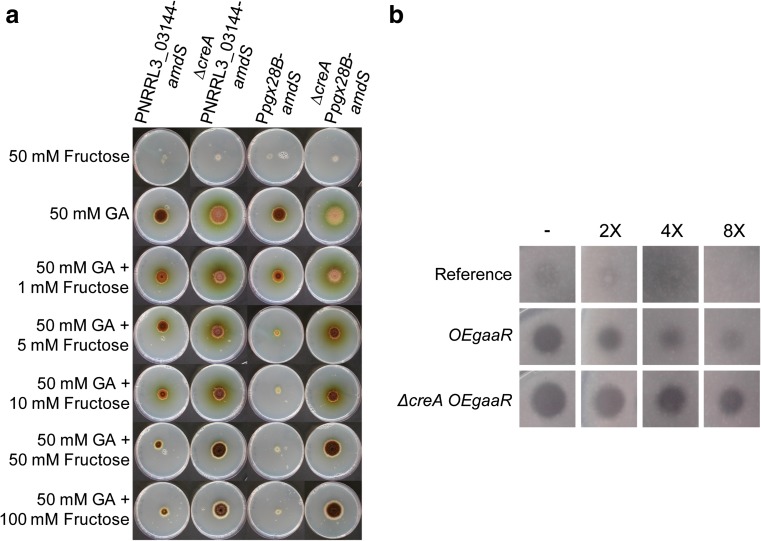
Carbon catabolite repression in the presence of glucose on several GA-responsive genes encoding exo-polygalacturonases, e.g., NRRL3_03144 (pgaX) and pgx28B (pgxB), was previously shown to be CreA-mediated (Niu et al. 2015). The OEgaaR strains produced more polygalacturonases during growth in fructose compared to glucose, indicating that fructose exerts less repression than glucose on pectinase gene expression (Fig. ESM_3.2 a). To investigate to which extent the presence of fructose affects CreA-mediated carbon catabolite repression on pectinase gene expression, we used promoter-reporter strains PNRRL3_03144-amdS and Ppgx28B-amdS, which are able to grow on acetamide as the sole nitrogen source only when the amdS gene encoding the acetamidase enzyme is expressed via the GA-responsive promoters of the pectinase genes NRRL3_03144 and pgx28B, respectively (Niu et al. 2015). Growth of the promoter-reporter strains on plates containing acetamide and GA decreased as the fructose concentration in the growth media increased (Fig. 3a), indicating that fructose also represses the expression of genes encoding those pectinases. In addition, we directly compared the repression power of glucose, fructose, sorbitol, and acetate on NRRL3_03144 expression in a single experiment (Fig.ESM_3.5). Radial growth assay confirmed that the expression NRRL3_03144 is repressed strongly by glucose and mildly by fructose. Sorbitol and acetate exerted negligible repression on NRRL3_03144 (Fig. ESM_3.5). Deletion of creA restored the growth of the promoter-reporter strains on fructose, showing that fructose-imposed carbon catabolite repression on pectinase gene expression is also mediated by CreA (Fig. 3a).
Fig. 3.
Analysis of CreA-mediated carbon catabolite repression on pectinase genes. a Growth phenotype of the PNRRL3_03144-amdS (JC1.5), ∆creA PNRRL3_03144-amdS (JN29.2), Ppgx28B-amdS (JC3.6) and ∆creA Ppgx28B-amdS (JN31.3) strains on solid MM containing 50 mM fructose, 50 mM GA, or 50 mM GA with increasing amounts of fructose after 7 days at 30 °C. All plates contain 10 mM acetamide as the sole nitrogen source. b PGA plate assay. The reference (MA234.1), OEgaaR (EA21.6), and ∆creAOEgaaR (TK2.1) strains were grown in MM containing 50 mM fructose for 36 h, and serial dilutions of culture supernatants were spotted on PGA plates. Dilution factors are indicated
All 124 genes that were upregulated in the OEgaaR strain in fructose and the promoter regions of which could be screened for the presence of transcription factor binding sites, contain at least one CreA binding motif (SYGGRG) (Cubero and Scazzocchio 1994) in their 1 kb upstream sequences (Online Resource 4), suggesting that carbon repression has a major effect on the expression on these GaaR target genes. Because the presence of fructose has a repressing effect on the expression of pectinase genes such as exo-polygalacturonases, NRRL3_03144 and pgx28B, we hypothesized that deletion of creA in the OEgaaR background would result in an elevated expression of pectinase genes on fructose. Therefore, the ∆creAOEgaaR strain (TK2.1) was created in the EA21.6 background to allow a direct comparison. Growth analysis on plates showed a reduced growth of the ∆creAOEgaaR strain on glucose, fructose, and AP, which was also observed in a control ∆creA strain (MA342.2) indicating that the reduced growth is caused by the creA deletion and not by gaaR overexpression (ESM_3.2 a).To assess the effect of creA deletion on pectinase production in combination with gaaR overexpression, the reference strain (MA234.1), OEgaaR (EA21.6) and ∆creAOEgaaR (TK2.1) strains were grown in fructose and the polygalacturonase activity in the culture supernatants was analyzed via a PGA plate assay. The culture supernatant of the ∆creAOEgaaR strain displayed the highest polygalacturonase activity, thereby providing additional evidence that fructose exerts repression on pectinase gene expression through CreA (Fig. 3b).
Discussion
The Zn2Cys6 transcriptional activator GaaR (Alazi et al. 2016) and the repressor protein GaaX (Niu et al. 2017) are the two important players in the transcriptional regulation of the GA-responsive genes in A. niger. Both GaaR and GaaX are highly conserved in filamentous fungi of the phylum ascomycetes. Therefore, the GaaR/GaaX module is expected to be the main regulatory mechanism in controlling GA-induced gene expression in filamentous fungi of ascomycetes. The combination of an activator (GaaR) and repressor (GaaX) protein to control gene expression represents a conserved mechanism which shows striking similarities with the regulation of genes involved in quinic acid utilization in A. nidulans (Lamb et al. 1996). In both the regulation of GA-responsive genes as well as in quinic acid-responsive genes, loss of function of the respective repressor proteins results in constitutive and inducer-independent expression of target genes. Importantly, the induced expression still requires the corresponding transcriptional activator (Grant et al. 1988; Niu et al. 2017). These observations suggest a model in which the transcriptional activator is kept inactive by its corresponding repressor protein under non-inducing conditions. Upon inducing conditions, an inducer molecule is expected to bind to the repressor thereby causing its dissociation from the transcriptional activator. Non-repressor bound activator is expected to be active as a transcription factor to induce the expression of target genes (Lamb et al. 1996; Niu et al. 2017).
In this study, we constructed several A. niger strains that overexpress gaaR via the A. nidulans gpdA promoter. The OEgaaR strains, carrying different copy numbers of the ectopically integrated gaaR overexpression construct, showed partial and different levels of complementation of growth on GA-containing carbon sources, whereas their growth on glucose or fructose was similar to the reference strain. While in the wild type high levels of pectinases are produced only under inducing conditions, the OEgaaR strains secreted high levels of polygalacturonases under both inducing and non-inducing conditions. These results imply that the OEgaaR strains possess a functional GaaR that is able to activate the expression of genes required for growth on GA and genes encoding polygalacturonases. Among all OEgaaR strains, EA21.6 and TK1.1 displayed the most impaired growth on GA, PGA, and AP and produced the highest levels of polygalacturonases in fructose. This might indicate a possible cofactor imbalance due to increased amounts of NAD(P)H-dependent GaaA and NADPH-dependent GaaD enzymes, or accumulation of a toxic GA catabolic pathway intermediate, when OEgaaR strains grow on GA-rich carbon sources.
The GA-responsive genes are transcriptionally induced by GaaR under inducing conditions (Alazi et al. 2016; Martens-Uzunova and Schaap 2008) and the transcriptional activity of GaaR is suggested to be controlled by GaaX, possibly via a protein-protein interaction, under non-inducing conditions (Niu et al. 2017). We showed that eGFP-GaaR is localized mainly in the nucleus under both inducing and non-inducing conditions, indicating that the transcriptional activity of GaaR is not regulated through nuclear translocation upon induction and the mechanism which keeps GaaR inactive under non-inducing conditions is likely to occur in the nucleus. Prediction of nuclear localization signals using the prediction tool NucPred (Brameier et al. 2007) indicated that GaaR likely localizes in the nucleus (score of 0.90) whereas GaaX (score of 0.27) is expected to spend less time in the nucleus. Nevertheless, GaaX-eGFP was found to be present in both cytoplasm and nucleus under inducing conditions, showing that it can enter the nucleus (Fig. ESM_3.4 b). These results imply that GaaX might inhibit the transcriptional activity of GaaR in the nucleus under non-inducing conditions.
Ectopic integration of gaaR in a ΔgaaR strain was previously shown to result in a full complementation of growth on GA (Alazi et al. 2016), whereas the eGFP-gaaR strain EA19.2 that was derived from a ΔgaaR strain and expresses N-terminally eGFP-tagged gaaR displayed a slightly reduced growth on GA compared to the reference strain. N-terminal eGFP-tagging might result in a minor decrease in GaaR transcription factor activity and therefore partial restoration of growth. As GaaR is expected to interact with GaaX, it was also assessed whether the N-terminal tagging of GaaR influenced its interaction with GaaX. The expression of eGFP-gaaR via the endogenous gaaR promoter did not result in a constitutive expression of the genes encoding polygalacturonases (Fig. ESM_3.3 c), indicating that eGFP-GaaR activity is properly controlled by GaaX under non-inducing conditions.
Overexpression of eGFP-GaaR driven by the A. nidulans gpdA promoter leads to a much higher nuclear eGFP-GaaR concentration under both inducing and non-inducing conditions compared to expression driven by the endogenous gaaR promoter. The increase in nuclear GaaR concentration was accompanied by transcriptional upregulation of GA-responsive genes and the increased accumulation of pectinases in the extracellular medium. The transcriptional activation of GA-responsive genes in the OEgaaR strain under non-inducing conditions can be explained by the possibility that the excess of GaaR titrates out the concentration of GaaX and escapes GaaX inhibition, even though gaaX is induced upon GaaR overexpression.
Genome-wide gene expression analysis in the reference strain grown in GA has been previously performed (Alazi et al. 2016). Direct comparison of the gene expression values between the study of Alazi et al. (2016), and this study needs careful interpretation due to different experimental setups (growth in shake flasks vs bioreactors) and representation of transcript levels (FPKM vs TPM). Notwithstanding, it can be observed that the expression level of the genes encoding pectinases are generally comparable between the reference and OEgaaR strains grown under inducing and non-inducing conditions, respectively. However, drastically higher expression of NRRL3_05252, NRRL3_03144, pgx28B, and gan53A in the reference strain and pga28C, pga28E, and pel1A in the OEgaaR strain was also observed.
Elimination of the repressing activity of GaaX by deleting gaaX was previously shown to be another way to activate the expression of GA-responsive genes under non-inducing conditions (Niu et al. 2017). The concentration of the nuclear GaaR in ΔgaaX is expected to be similar to wild type and much less compared to the OEgaaR strain. Only nine out of 48 genes encoding pectinases were upregulated in the ΔgaaX strain in fructose compared to the reference strain, and the transcript and extracellular protein levels of these pectinases were generally lower compared to the OEgaaR strain. This indicates that the nuclear concentration of active GaaR is indeed an important factor for transcriptional activation of GA-responsive genes. On the other hand, the genes encoding the (putative) GA transporters and catabolic pathway enzymes were expressed at higher levels in ΔgaaX compared to the OEgaaR strain, indicating that factors other than GaaR concentration might play a role in the regulation of these genes.
RNA-seq analysis showed that besides the genes encoding pectinases, 20 genes predicted to encode CAZymes involved in the degradation of multiple substrates or specifically of cellulose, starch, or xyloglucan were upregulated in the OEgaaR strain in fructose. This indicates that these enzymes might be involved or assist in enabling the degradation of pectin. Five of these CAZymes were shown to be upregulated in ΔgaaX, and therefore designated as part of the GaaR/GaaX panregulon (Niu et al. 2017). In addition, many of the 20 additional CAZymes were reported to be potentially regulated by transcription factors AraR and/or XlnR (Gruben et al. personal communication). However, the expression of the genes encoding AraR or XlnR were not significantly changed in OEgaaR, discounting the possibility that overexpression of GaaR caused transcriptional upregulation of the genes encoding CAZymes via their specific transcription factors.
Fructose was found to exert CreA-mediated repression of gene expression in case of the genes NRRL3_03144 (pgaX) and pgx28B (pgxB) encoding exo-polygalacturonases, which were previously shown to be strongly repressed in the presence of glucose (Niu et al. 2015). The repression power of fructose was lower than that of glucose (Fig. ESM_3.5). As shown by Niu et al. (2017), deletion of creA is not sufficient for an increased production of polygalacturonases under non-inducing conditions, showing that GA-responsive gene expression requires the presence of active GaaR relieved from GaaX inhibition. A similar phenomenon was previously observed in T. reesei, where high expression of the genes encoding cellulases in a Cre1-disrupted strain required the presence of the transcriptional activator Xyr1 under non-inducing conditions (Wang et al. 2013). The strain that lacks creA and overexpresses GaaR (TK2.1) secreted higher levels of polygalacturonases compared to the reference and OEgaaR strains, indicating that CreA substantially represses the expression of GA-responsive genes in the presence of fructose even when GaaR is abundant.
To conclude, genetic evidence suggests that the activity of the GaaR transcription factor is negated by the action of the GaaX repressor protein. Either deletion of GaaX or overexpression of GaaR results in a constitutive expression of GaaR/GaaX target genes. The simplest interpretation of these observations is that GaaX mediates its repressing activity by a direct interaction with GaaR. Loss of function of GaaX or overexpression of GaaR will affect the stoichiometry of GaaR-GaaX and lead to high levels of “repressor free” GaaR which is expected to act as an active transcription factor to induce expression of GA-responsive genes. We have shown that overexpression of GaaR leads to an increased level of pectinase production under non-inducing conditions, and that deletion of creA further increases the pectinase production capacity of A. niger. The ∆creAOEgaaR strain represents an interesting strain for applications in industry with its high pectinase production capacity in the absence of an inducing carbon source and in the presence of a repressing carbon source.
Electronic supplementary material
(PDF 977 kb)
(XLSX 1425 kb)
Funding
EA was supported by a grant from BE-Basic (Flagship 10). This work was in part supported by the NSERC IBN Strategic Network and Genome Canada.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s00253-018-8753-7) contains supplementary material, which is available to authorized users.
References
- Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with Image. J Biophoton Int. 2004;11(7):36–42. [Google Scholar]
- Alazi E, Niu J, Kowalczyk JE, Peng M, Aguilar Pontes MV, van Kan JAL, Visser J, de Vries RP, Ram AFJ. The transcriptional activator GaaR of Aspergillus niger is required for release and utilization of D-galacturonic acid from pectin. FEBS Lett. 2016;590:1804–1815. doi: 10.1002/1873-3468.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazi E, Khosravi C, Homan TG, du Pré S, Arentshorst M, Di Falco M, Pham TTM, Peng M, Aguilar-Pontes MV, Visser J, Tsang A, de Vries RP, Ram AFJ. The pathway intermediate 2-keto-3-deoxy-L-galactonate mediates the induction of genes involved in D-galacturonic acid utilization in Aspergillus niger. FEBS Lett. 2017;591:1408–1418. doi: 10.1002/1873-3468.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arentshorst M, Ram AFJ, Meyer V. Using non-homologous end-joining-deficient strains for functional gene analyses in filamentous fungi. Methods Mol Biol. 2012;835:133–150. doi: 10.1007/978-1-61779-501-5_9. [DOI] [PubMed] [Google Scholar]
- Arentshorst M, Niu J, Ram AFJ. Efficient generation of Aspergillus niger knock out strains by combining NHEJ mutants and a split marker approach. In: van den Berg MA, Maruthachalam K, editors. Genetic transformation systems in fungi. Cham: Springer International Publishing; 2015. pp. 263–272. [Google Scholar]
- Bos CJ, Debets AJ, Swart K, Huybers A, Kobus G, Slakhorst SM. Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr Genet. 1988;14:437–443. doi: 10.1007/BF00521266. [DOI] [PubMed] [Google Scholar]
- Brameier M, Krings A, MacCallum AM. NucPred—predicting nuclear localization of proteins. Bioinformatics. 2007;23(9):1159–1160. doi: 10.1093/bioinformatics/btm066. [DOI] [PubMed] [Google Scholar]
- Caffall KH, Mohnen D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res. 2009;344:1879–1900. doi: 10.1016/j.carres.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Chua G, Morris QD, Sopko R, Robinson MD, Ryan O, Chan ET, Frey BJ, Andrews BJ, Boone C, Hughes TR. Identifying transcription factor functions and targets by phenotypic activation. Proc Natl AcadSci U S A. 2006;103(32):12045–12050. doi: 10.1073/pnas.0605140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho PM, Andersen MR, Kolenova K, Vankuyk PA, Benoit I, Gruben BS, Trejo-Aguilar B, Visser H, van Solingen P, Pakula T, Seiboth B, Battaglia E, Aguilar-Osorio G, de Jong JF, Ohm RA, Aguilar M, Henrissat B, Nielsen J, Stalbrand H, de Vries RP. Post-genomic insights into the plant polysaccharide degradation potential of Aspergillus nidulans and comparison to Aspergillus niger and Aspergillus oryzae. Fungal Genet Biol. 2009;46(1):S161–S169. doi: 10.1016/j.fgb.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Cubero B, Scazzocchio C. Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 1994;13(2):407–415. doi: 10.1002/j.1460-2075.1994.tb06275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RP, Riley R, Wiebenga A, Aguilar-Osorio G, Amillis S, Uchima CA, Anderluh G, Asadollahi M, Askin M, Barry K, Battaglia E, Bayram Ö, Benocci T, Braus-Stromeyer SA, Caldana C, Cánovas D, Cerqueira GC, Chen F, Chen W, Choi C, Clum A, Dos Santos RA, Damásio AR, Diallinas G, Emri T, Fekete E, Flipphi M, Freyberg S, Gallo A, Gournas C, Habgood R, Hainaut M, Harispe ML, Henrissat B, Hildén KS, Hope R, Hossain A, Karabika E, Karaffa L, Karányi Z, Kraševec N, Kuo A, Kusch H, LaButti K, Lagendijk EL, Lapidus A, Levasseur A, Lindquist E, Lipzen A, Logrieco AF, MacCabe A, Mäkelä MR, Malavazi I, Melin P, Meyer V, Mielnichuk N, Miskei M, Molnár ÁP, Mulé G, Ngan CY, Orejas M, Orosz E, Ouedraogo JP, Overkamp KM, Park HS, Perrone G, Piumi F, Punt PJ, Ram AF, Ramón A, Rauscher S, Record E, Riaño-Pachón DM, Robert V, Röhrig J, Ruller R, Salamov A, Salih NS, Samson RA, Sándor E, Sanguinetti M, Schütze T, Sepčić K, Shelest E, Sherlock G, Sophianopoulou V, Squina FM, Sun H, Susca A, Todd RB, Tsang A, Unkles SE, van de Wiele N, van Rossen-Uffink D, Oliveira JV, Vesth TC, Visser J, Yu JH, Zhou M, Andersen MR, Archer DB, Baker SE, Benoit I, Brakhage AA, Braus GH, Fischer R, Frisvad JC, Goldman GH, Houbraken J, Oakley B, Pócsi I, Scazzocchio C, Seiboth B, vanKuyk PA, Wortman J, Dyer PS, Grigoriev IV. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017;18(1):28. doi: 10.1186/s13059-017-1151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MC, Doran-Peterson J. Pectin-rich biomass as feedstock for fuel ethanol production. Appl Microbiol Biotechnol. 2012;95:565–575. doi: 10.1007/s00253-012-4173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S, Roberts CF, Lamb H, Stout M, Hawkins AR. Genetic regulation of the quinic acid utilization (QUT) gene cluster in Aspergillus nidulans. J Gen Microbiol. 1988;134:347–358. doi: 10.1099/00221287-134-2-347. [DOI] [PubMed] [Google Scholar]
- Hynes MJ, Corrick CM, King JA. Isolation of genomic clones containing the amdS gene of Aspergillus nidulans and their use in the analysis of structural and regulatory mutations. Mol Cell Biol. 1983;3(8):1430–1439. doi: 10.1128/MCB.3.8.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Duarte AV, van den Brink J, Wiebenga A, Zou G, Wang C, de Vries RP, Zhou Z, Benoit I. Enhancing saccharification of wheat straw by mixing enzymes from genetically-modified Trichoderma reesei and Aspergillus niger. Biotechnol Lett. 2016;38(1):65–70. doi: 10.1007/s10529-015-1951-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap D, Vohra P, Chopra S, Tewari R. Applications of pectinases in the commercial sector: a review. Bioresour Technol. 2001;77:215–227. doi: 10.1016/S0960-8524(00)00118-8. [DOI] [PubMed] [Google Scholar]
- Khan M, Nakkeeran E, Umesh-Kumar S. Potential application of pectinase in developing functional foods. Annu Rev Food SciTechnol. 2013;4:21–34. doi: 10.1146/annurev-food-030212-182525. [DOI] [PubMed] [Google Scholar]
- Lamb HK, Newton GH, Levett LJ, Cairns E, Roberts CF, Hawkins AR (1996) The QUTA activator and QUTR repressor proteins of Aspergillus nidulans interact to regulate transcription of the quinate utilization pathway genes. Microbiology 142:1477–1490 [DOI] [PubMed]
- Lv X, Zheng F, Li C, Zhang W, Chen G, Liu W. Characterization of a copper responsive promoter and its mediated overexpression of the xylanase regulator 1 results in an induction-independent production of cellulases in Trichoderma reesei. Biotechnol Biofuels. 2015;8:67. doi: 10.1186/s13068-015-0249-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens-Uzunova ES, Schaap PJ. An evolutionary conserved D-galacturonic acid metabolic pathway operates across filamentous fungi capable of pectin degradation. Fungal Genet Biol. 2008;45(11):1449–1457. doi: 10.1016/j.fgb.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Martens-Uzunova ES, Schaap PJ (2009) Assessment of the pectin degrading enzyme network of Aspergillus niger by functional genomics. Fungal Genet Biol 46:S170–S179. [DOI] [PubMed]
- Meyer V, Arentshorst M, Flitter SJ, Nitsche BM, Kwon MJ, Reynaga-Peña CG, Bartnicki-Garcia S, van den Hondel CAMJJ, Ram AFJ. Reconstruction of signaling networks regulating fungal morphogenesis by transcriptomics. Eukaryot Cell. 2009;8(11):1677–1691. doi: 10.1128/EC.00050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsche BM, Crabtree J, Cerqueira GC, Meyer V, Ram AFJ, Wortman JR. New resources for functional analysis of omics data for the genus Aspergillus. BMC Genomics. 2011;12(1):486. doi: 10.1186/1471-2164-12-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J, Homan TG, Arentshorst M, de Vries RP, Visser J, Ram AF. The interaction of induction and repression mechanisms in the regulation of galacturonic acid-induced genes in Aspergillus niger. Fungal Genet Biol. 2015;82:32–42. doi: 10.1016/j.fgb.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Niu J, Alazi E, Reid ID, Arentshorst M, Punt PJ, Visser J, Tsang A, Ram AFJ. An evolutionarily conserved transcriptional activator-repressor module controls expression of genes for D-galacturonic acid utilization in Aspergillus niger. Genetics. 2017;205:169–183. doi: 10.1534/genetics.116.194050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi Y, Sano M, Kanamaru K, Ko T, Takeuchi M, Kato M, Kobayashi T. Genes regulated by AoXlnR, the xylanolytic and cellulolytic transcriptional regulator, in Aspergillus oryzae. Appl Microbiol Biotechnol. 2009;85(1):141–154. doi: 10.1007/s00253-009-2236-9. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Kobayashi T, Koyama Y. ManR, a novel Zn(II)2Cys6 transcriptional activator, controls the β-mannan utilization system in Aspergillus oryzae. Fungal Genet Biol. 2012;49:987–995. doi: 10.1016/j.fgb.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Pedrolli DB, Monteiro AC, Gomes E, Carmona EC. Pectin and pectinases: production, characterization and industrial application of microbial pectinolytic enzymes. Open Biotechnol J. 2009;3:9–18. doi: 10.2174/1874070700903010009. [DOI] [Google Scholar]
- Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel CA. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene. 1987;56(1):117–124. doi: 10.1016/0378-1119(87)90164-8. [DOI] [PubMed] [Google Scholar]
- Sloothaak J, Schilders M, Schaap PJ, de Graaff LH. Overexpression of the Aspergillus niger GatA transporter leads to preferential use of D-galacturonic acid over D-xylose. AMB Express. 2014;4(66):1–9. doi: 10.1186/s13568-014-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toushik SH, Lee K, Lee J, Kim K. Functional applications of lignocellulolytic enzymes in the fruit and vegetable processing industries. J Food Sci. 2017;88(3):583–595. doi: 10.1111/1750-3841.13636. [DOI] [PubMed] [Google Scholar]
- Vinck A (2007) Hyphal differentiation in the fungal mycelium. Dissertation, Utrecht University
- Wang S, Liu G, Wang J, Yu J, Huang B, Xing M. Enhancing cellulase production in Trichoderma reesei RUT C30 through combined manipulation of activating and repressing genes. J Ind Microbiol Biotechnol. 2013;40(6):633–641. doi: 10.1007/s10295-013-1253-y. [DOI] [PubMed] [Google Scholar]
- Yuan XL, Goosen C, Kools H, van den Maarel MJEC, van den Hondel CAMJJ, Dijkhuizen L, Ram AFJ. Database mining and transcriptional analysis of genes encoding inulin-modifying enzymes of Aspergillus niger. Microbiology. 2006;152(10):3061–3073. doi: 10.1099/mic.0.29051-0. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lubbers RJM, Simon A, Stassen JHM, Ribera PRV, Viaud M, van Kan JAL. A novel Zn2Cys6 transcription factor BcGaaR regulates D-galacturonic acid utilization in Botrytis cinerea. Mol Microbiol. 2016;100(2):247–262. doi: 10.1111/mmi.13314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 977 kb)
(XLSX 1425 kb)