Abstract
Nisin is a complex lanthipeptide that has broad spectrum antibacterial activity. In efforts to broaden the structural diversity of this ribosomally synthesized lantibiotic, we now report the recombinant expression of Nisin variants that incorporate non-canonical amino acids (ncAAs) at discrete positions. This is achieved by expressing the nisA structural gene, cyclase (nisC) and dehydratase (nisB), together with an orthogonal nonsense suppressor tRNA/aminoacyl-tRNA synthetase pair in E.coli. A number of ncAAs with novel chemical reactivity were genetically incorporated into NisA, including an α-chloroacetamide-containing ncAA which allowed for the expression of Nisin variants with novel macrocyclic topologies. This methodology should allow for the exploration of lanthipeptide variants with new or enhanced activities.
Graphical abstract
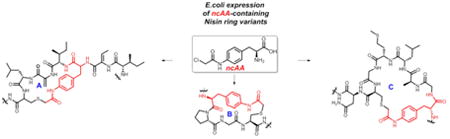
The lanthipeptides are a subclass of a large family of ribosomally synthesized and post-translationally modified macrocyclic peptides (RiPPs).1 Members that possess antibacterial activity are known as lantibiotics,2 and are defined by characteristic lanthionine or methyllantionine thioether bridges that impart backbone rigidity. The most studied member of the lanthipeptides, Nisin A, depicted as its prepeptide or mNisA (Figure 1), has been used as a food preservative for the past fifty years.3 It features multiple dehydrated amino acids arising from the activity of a dehydratase (NisB) on serine (Ser) and threonine (Thr) residues, and five thioether crosslinks (rings A, B, C and D/E) formed by a cyclase (NisC) that catalyzes thia-Michael reactions.4 Nisin exerts its antimicrobial activity through its N-terminal domain that recognizes and sequesters the pyrophosphate moiety of Lipid II, and the C-terminal domain which inserts into the outer membrane.5-6 This dual mode of action, directed towards non-proteinaceous bacterial targets, has slowed the development of resistant strains.7 However, its poor aqueous solubility and pH sensitivity have delayed its use as a human therapeutic.8
Figure 1.
Wild type mNisA. Highlighted thioether rings and key residues for this study (residues 30-34 omitted for simplicity).
To further explore the potential utility of this complex natural product, various efforts have been undertaken to generate variants of Nisin and other lantibiotics,9 including the use of solid phase synthesis,10-12 biomimetic approaches,13 the in vitro biosynthesis of lanthionine-containing peptides14-15 and an in vitro mutasynthetic approach.16
To expand the number of amino acids building blocks that can be biosynthetically incorporated into lantipeptides, we now report that orthogonal nonsense suppressor tRNA/aminoacyl-tRNA synthetase pairs (tRNA/aaRS) can be used to site-specifically insert ncAAs into Nisin in bacteria.17
To date, two examples of ncAA incorporation into lanthipeptides using nonsense codon suppression have been reported by van der Donk.18-19 Kuipers also reported the incorporation of tryptophan analogues into Nisin using an auxotrophic strain.20 With the goal of creating Nisin variants through biomimetic cyclization reactions (Figure 2), we built upon the former system18 and began by recombinantly expressing wild type (WT) NisA in bacterial strains in which we have previously genetically encoded a large number of ncAAs. The nisA structural gene together with the dehydratase (nisB), were inserted into a pRSF-1 vector to afford pRSF-NisA. The cyclase, nisC, was inserted into a pACYC backbone to afford pACYC-NisC. The Ser codon at position 5 of nisA was then mutated to the amber stop codon TAG (Ser5TAG), and the resulting plasmid (pZC1) was co-expressed, along with pACYC-NisC, and a pULTRA plasmid harboring either a polyspecific M.jannaschii tRNATyr/TyrRS or M.barkeri pyrrolysine tRNAPyl/PylRS.21 We then attempted to substitute a number of ncAAs containing diverse side chains including keto, azide and acetylene groups for Ser5 (Figure 3A). SDS-PAGE and MS analysis (Figure S1-S3) confirmed incorporation of pAcF, pAzF and ProcK at position 5 in yields ranging from 3 to 5 mg/L (Figure 3B). Although promising, in our hands the expression system afforded incomplete dehydration of Ser and Thr residues, giving rise to a distribution of products.
Figure 2.
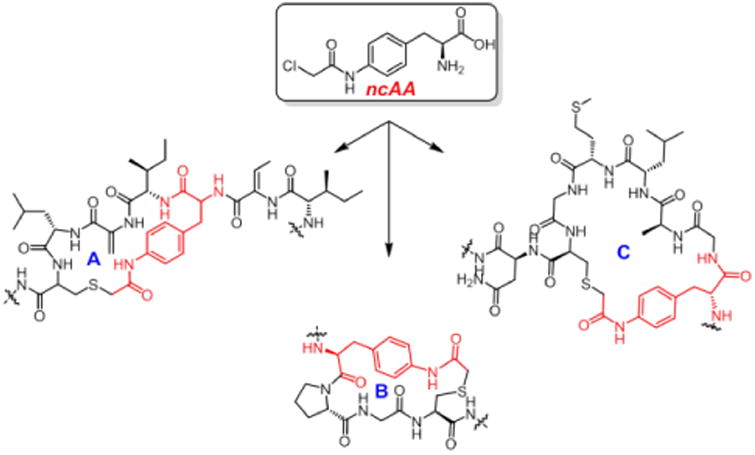
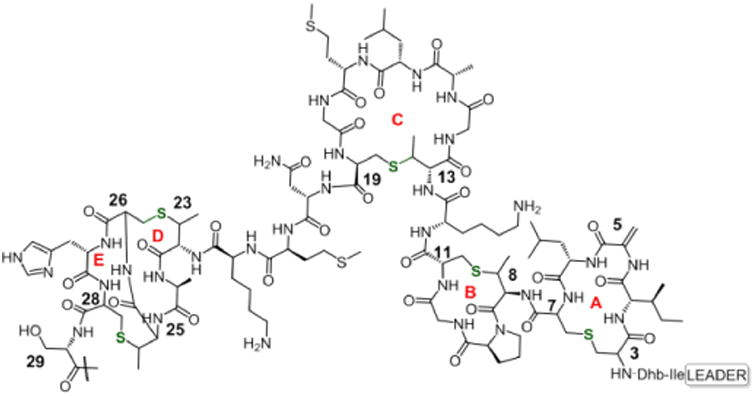
Altered ring structures of Nisin created by ncAA incorporation.
Figure 3.
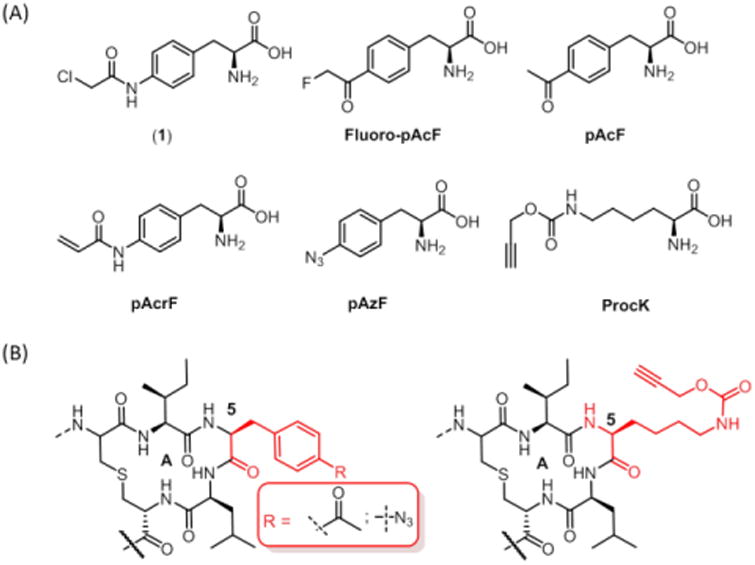
(A) ncAAs used in this study (B) Biorthogonal chemical handles incorporated into Ser5TAG NisA.
To solve this issue, we capitalized on the recent finding that the dehydratase NisB requires glutamylated tRNA (tRNAGlu) in order to acylate the Ser/Thr side chains of NisA with glutamate prior to dehydration.22 We reasoned that overexpressing tRNAGlu could improve the dehydration efficiency. To test this notion, we constructed a plasmid (pZC2) bearing E.coli tRNAGlu/GluRS downstream of a proK constitutive promoter encoded on pACYC-NisC. Co-expression of pZC2 with pRSF-NisAWT, showed a substantial increase in dehydration efficiency (WT mNisA = 7299 Da), confirming our hypothesis (Figure S4, S5).
Given this optimized system, we next explored the possibility of taking advantage of electrophilic ncAAs and the innate reactivity of cysteine residues present in NisA, to generate Nisin variants with altered ring structures. We began by incorporating mildly reactive ncAAs23 in place of Ser/Thr residues at lanthionine ring junctions, starting with the ring A Ser3TAG mutant (pZC3). Fluoro-pAcF (Figure 3A) was incorporated into NisA at residue 3 in a yield of 2 mg/L. MS analysis showed a mass (7418 Da) corresponding to the loss of fluoride (Figure S6). This result, although encouraging, was somewhat cryptic due to the similar molecular weight of fluorine and water. Because water acts as a leaving group during precursor peptide maturation, an extra post-translational dehydration would result in a product with a similar mass as that of the desired, cyclized peptide. This led us to reconsider the ncAA to be used.
We reasoned that a chloroacetamide ncAA24 (Figure 3A, 1) would be the best compromise in terms of reactivity and leaving group MW for mass analysis. Gratifyingly, when 1 was incorporated into ring A of mNisA (Ser3TAG), MS analysis showed a mass of 7433 Da [M/z]+1, consistent with a fully dehydrated and cyclized peptide containing a novel ring A thioether architecture. We next investigated whether this approach could be extended to other ring junctions as a general strategy towards lanthipeptides diversification. To this end, the remaining dehydratable residues (Ser/Thr) involved in ring cyclization were individually point mutated to TAG and the resulting constructs co-transformed with pULTRA and pZC2. Upon expression and isolation of the prepeptides, SDS-PAGE (Figure 4A) and mass analysis revealed cyclization of thioether rings A, B, and C, corresponding to the Ser3TAG, Thr8TAG and Thr13TAG mutations, respectively, in yields of 1.5 to 3 mg/L (Figure 4B, C and D, Figure S7 to S9). Efforts to create intertwined thioether ring D/E (Thr23TAG and Thr25TAG), were troublesome giving rise to a mixture of desired and truncated products at Lys22.
Figure 4.
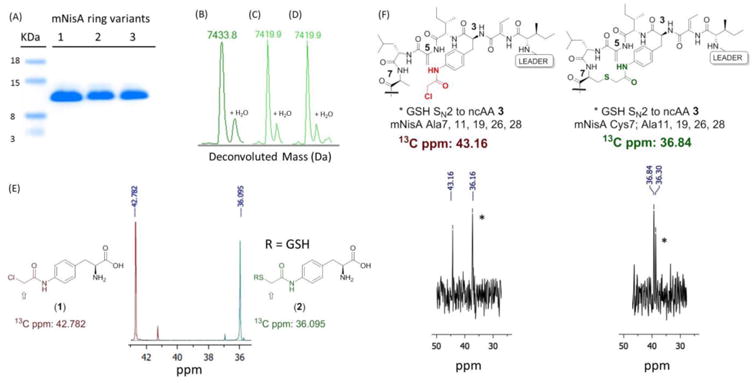
(A) SDS-PAGE mNisA ring variants incorporating 1. Lane 1: ring A, lane 2: ring B, lane 3: ring C. (B-D) QTOF-ESI for mNisA ring variants incorporating 1: (B) Ser3TAG, ring A. (C) Thr8TAG, ring B. (D) Thr13TAG, ring C. (E)13C-NMR of labeled 1 (red) and 2 (green). (F) 13C-NMR mNisA Cys to Ala mutants incorporating labeled 1, non-cyclized (red) and cyclized (green) peptides. Asterisks indicate GSH addition products.
Our next goal was to further characterize the newly formed bond in the Ser3 to 1 mutant. To simplify analysis, we decided to generate a monocyclic Nisin. To this end, we constructed two plasmids emanating from the common vector, NisA Ser3TAG (pZC3). In the first construct (pZC4), we sequentially mutated all cysteine residues to Ala (Cys7, 11, 19, 26, 28Ala). The second construct (pZC5) retained Cys7 and all the remaining Cys were mutated to Ala (Cys11, 19, 26, 28Ala). Each construct was individually co-transformed with the accessory plasmids (pULTRA, pZC2), expressed in the presence of 1, and the mature peptides were isolated. MS analysis confirmed cyclization only for the peptide bearing reactive Cys7 (Figure S11). In contrast, the fully mutated Cys to Ala construct showed a MS spectrum for a peptide carrying the unreacted chloroacetamide. Interestingly, this non-cyclized peptide showed an extra dehydration event that was ablated upon Ser29 to Ala mutation in the core peptide (Figure S10). This observation is in accordance with the hypothesis that dehydration and cyclization events in Nisin biosynthesis are coupled, and that precise PTM control depends upon conformationally restricted substrates.25-26
To obtain NMR evidence for the novel thioether linkage, we synthesized isotopically-labeled ncAA (1) bearing a 13C at the reaction center (2-chloroacetamide position) using chloroacetyl chloride-2-13C as a starting material. Incorporation of 13C-1 into Nisin mutants should afford chemical shift differences between cyclized and non-cyclized product. As a control experiment, we recorded the 13C-NMR spectrum of labeled 1, and then treated it with excess glutathione (GSH) to form a thioether bond as a standard. A substantial chemical shift difference between the starting ncAA-chloroacetamide (42.78 ppm) and the GSH-displaced product 2 (36.09 ppm), was observed (Figure 4E). We then expressed and HPLC purified mutants Ala7, Ala11, Ala19, Ala26, Ala28 and Cys7, Ala11, Ala19, Ala26, Ala28 encoded on pZC4 and pZC5, respectively, incorporating 13C-(1). The peptide lacking all cysteines showed 13C NMR peaks consistent with the presence of a chloroacetamide (43.16 ppm) and its GSH adduct. On the other hand, the peptide harboring reactive Cys7, showed a 13C resonance (36.84 ppm) consistent with the standard thioether 2, confirming the macrocyclization event (Figure 4F). The additional thioether resonance (36.30 ppm) is likely due either to GSH addition to 1, with Cys7 cyclizing to dehydroalanine 5 (Dha5), or GSH addition to Dha5 of the expanded cyclic peptide. Upon trypsin-mediated cleavage of the leader peptide, Nisin analogues bearing novel thioeter linkages were tested for antibacterial activity. Unfortunately, while a halo assay against M. luteus indicated that this Nisin ring variants are devoid of antibacterial activity (while WT Nisin retained activity) (Figure S19), the possibility of constructing non-natural lanthipeptides with positional precision should allow for the interrogation of broader chemical space.
In summary, we genetically incorporated diverse ncAAs containing either biorthogonal handles or mildly reactive functional groups into Nisin A. NMR and tandem MS spectroscopy confirmed the formation of a novel ring A thioether bond for one of the mutants. We are currently probing various ring topologies with the aim to restore biologically active conformations. The ability to diversify lanthipeptide macrocycles at the ribosomal stage may lead to improvements in their pharmacological properties.
Supplementary Material
Acknowledgments
Funding Sources: NIH 5R01 GM062159
Kristen Williams for assistance in manuscript preparation, Dr. Laura Pasternak for NMR analysis, Dr. Michael J. Bollong and Dr. Sean A. Reed for helpful discussions.
Footnotes
Supporting Information. Mass spectra (ESI-QTOF, LC MS/MS), NMR data, cloning and synthetic protocols are described in the supporting information. The Supporting Information is available free of charge on the ACS Publications website.
References
- 1.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJ, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Sussmuth RD, Tagg JR, Tang GL, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Nat Prod Rep. 2013;30(1):108–60. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bierbaum G, Sahl HG. Curr Pharm Biotechnol. 2009;10(1):2–18. doi: 10.2174/138920109787048616. [DOI] [PubMed] [Google Scholar]
- 3.Rogers LA. Journal of bacteriology. 1928;16(5):321–5. doi: 10.1128/jb.16.5.321-325.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koponen O, Tolonen M, Qiao M, Wahlstrom G, Helin J, Saris PE. Microbiology. 2002;148(Pt 11):3561–8. doi: 10.1099/00221287-148-11-3561. [DOI] [PubMed] [Google Scholar]
- 5.Hsu ST, Breukink E, Tischenko E, Lutters MA, de Kruijff B, Kaptein R, Bonvin AM. Nat Struct Mol Biol. 2004;11(10):963–7. doi: 10.1038/nsmb830. [DOI] [PubMed] [Google Scholar]
- 6.Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl HG, de Kruijff B. Science. 1999;286(5448):2361–4. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 7.Delves-Broughton J, Blackburn P, Evans RJ, Hugenholtz J. Antonie Van Leeuwenhoek. 1996;69(2):193–202. doi: 10.1007/BF00399424. [DOI] [PubMed] [Google Scholar]
- 8.van Heel AJ, Montalban-Lopez M, Kuipers OP. Expert Opin Drug Metab Toxicol. 2011;7(6):675–80. doi: 10.1517/17425255.2011.573478. [DOI] [PubMed] [Google Scholar]
- 9.Cotter PD, Hill C, Ross RP. Curr Protein Pept Sci. 2005;6(1):61–75. doi: 10.2174/1389203053027584. [DOI] [PubMed] [Google Scholar]
- 10.Okeley NM, Zhu Y, van Der Donk WA. Org Lett. 2000;2(23):3603–6. doi: 10.1021/ol006485d. [DOI] [PubMed] [Google Scholar]
- 11.Bregant S, Tabor AB. J Org Chem. 2005;70(7):2430–8. doi: 10.1021/jo048222t. [DOI] [PubMed] [Google Scholar]
- 12.Pattabiraman VR, McKinnie SM, Vederas JC. Angew Chem Int Ed Engl. 2008;47(49):9472–5. doi: 10.1002/anie.200802919. [DOI] [PubMed] [Google Scholar]
- 13.Burrage S, Raynham T, Williams G, Essex JW, Allen C, Cardno M, Swali V, Bradley M. Chemistry. 2000;6(8):1455–66. doi: 10.1002/(sici)1521-3765(20000417)6:8<1455::aid-chem1455>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 14.Goto Y, Iwasaki K, Torikai K, Murakami H, Suga H. Chem Commun (Camb) 2009;(23):3419–21. doi: 10.1039/b904314d. [DOI] [PubMed] [Google Scholar]
- 15.Seebeck FP, Ricardo A, Szostak JW. Chem Commun (Camb) 2011;47(21):6141–3. doi: 10.1039/c0cc05663d. [DOI] [PubMed] [Google Scholar]
- 16.Levengood MR, Knerr PJ, Oman TJ, van der Donk WA. J Am Chem Soc. 2009;131(34):12024–5. doi: 10.1021/ja903239s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie J, Schultz PG. Curr Opin Chem Biol. 2005;9(6):548–54. doi: 10.1016/j.cbpa.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Shi Y, Yang X, Garg N, van der Donk WA. J Am Chem Soc. 2011;133(8):2338–41. doi: 10.1021/ja109044r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bindman NA, Bobeica SC, Liu WR, van der Donk WA. J Am Chem Soc. 2015;137(22):6975–8. doi: 10.1021/jacs.5b04681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou L, Shao JF, Li Q, van Heel AJ, de Vries MP, Broos J, Kuipers OP. Amino Acids. 2016;48(5):1309–1318. doi: 10.1007/s00726-016-2186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatterjee A, Sun SB, Furman JL, Xiao H, Schultz PG. Biochemistry. 2013;52(10):1828–37. doi: 10.1021/bi4000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortega MA, Hao Y, Zhang Q, Walker MC, van der Donk WA, Nair SK. Nature. 2015;517(7535):509–12. doi: 10.1038/nature13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiang Z, Ren H, Hu YS, Coin I, Wei J, Cang H, Wang L. Nat Methods. 2013;10(9):885–8. doi: 10.1038/nmeth.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamamoto T, Sisido M, Ohtsuki T, Taki M. Chem Commun (Camb) 2011;47(32):9116–8. doi: 10.1039/c1cc12196k. [DOI] [PubMed] [Google Scholar]
- 25.Lubelski J, Khusainov R, Kuipers OP. J Biol Chem. 2009;284(38):25962–72. doi: 10.1074/jbc.M109.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Repka LM, Chekan JR, Nair SK, van der Donk WA. Chem Rev. 2017 doi: 10.1021/acs.chemrev.6b00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


