Abstract
Type I interferons (IFNs) are pleiotropic cytokines well recognized for their role in the induction of a potent anti-viral gene program essential for host defense against viruses. They also modulate innate and adaptive immune responses. However, the role of type I IFNs in host defense against bacterial infections is enigmatic. Depending on the bacterium, they exert seemingly opposite and capricious functions. In this review we summarize the effect of type I IFNs on specific bacterial infections and highlight the effector mechanisms regulated by type I IFNs in an attempt to elucidate new avenues to understanding their role.
Introduction
The innate immune system is the first line of defense against invading bacteria. Germ line encoded pattern recognition receptors (PRRs) bind bacterial components and initiate an antibacterial inflammatory gene program that promotes immune cell recruitment and directs antibacterial activities. Engagement of select PRRs also leads to the induction of what is classically considered an antiviral gene program. The role of these antiviral genes in the context of bacterial infections is unclear.
Antiviral gene expression is directed by type I interferons (IFNs), a group of small, inducible cytokines that were discovered to ‘interfere’ with the ability of a virus to successfully replicate (Isaacs and Lindenmann, 1957). Type I IFNs are one of three families of IFNs. They include IFNβ, IFNω, IFNκ, IFNε, IFNζ, IFNδ, IFNτ, and 14 IFNα subtypes. Among them, IFNβ and the IFNαs are the most abundant and well-studied, thus all subsequent reference to type I IFNs herein will mainly refer to these two types. Type II IFN is composed of single IFNγ, while type III IFN includes IFNλ1 (IL-29), IFNλ2 (IL-28A), IFNλ3 (IL-28B), and IFNλ4. Unlike type II and type III IFNs, type I IFNs are broadly expressed. They signal through the heterodimeric, interferon α/β receptor (IFNAR) and induce over 300 interferon stimulated genes (ISGs). These ISGs directly inhibit key steps of the viral life cycle (Yan and Chen, 2012), stimulate host cell death, activate innate immune cells and promote the development of the adaptive immune response (Crouse et al., 2015).
Although early investigations focused on the antiviral properties of type I IFNs, several groups studying the intracellular bacteria Chlamydia observed that type I IFNs were induced by the bacterium (Sueltenfuss and Pollard, 1963) and that, in turn, they restricted bacterial growth (de la Maza et al., 1985). Much later, studies with another intracellular bacteria, Listeria monocytogenes (L. monocytogenes), substantiated a functional role for type I IFNs in directing the outcome of bacterial infections. Using mice deficient in IFNAR signaling, three groups revealed type I IFNs enhanced the susceptibility of mice to Listeria infection. Taken together, these seminal studies illustrated that type I IFN signaling plays a decisive role in Listeria infection by: (i) Reducing the efficiency of bacterial clearance; (ii) Decreasing the abundance of proinflammatory myeloid cells; (iii) Promoting the expression of proapoptotic genes; and (iv) Enhancing T cell sensitivity to apoptotic cell death (Auerbuch et al., 2004; Carrero et al., 2004; O'Connell et al., 2004).
These striking results encouraged further investigation into how type I IFNs modulated the outcome of other bacterial infections. Studies conducted for over more than a decade have revealed a paradoxical role for type I IFNs. They play an adverse role in certain bacterial infections, while in others they are critical for host defense. In this review we will focus on how type I IFNs function to direct disparate outcomes in a spectrum of bacterial infections.
Pathways of recognition and response
Expression of type I IFNs is driven by the interferon regulatory factor (IRF) family of transcription factors, namely IRF3 and IRF7. In most cells IRF3 is the dominant transcription factor during early type I IFN expression. Later, IRF7, which is also an ISG, is expressed and it amplifies type I IFN transcription (Honda et al., 2006). In specific cell types, however, other IRFs direct early expression of type I IFNs. For example, in plasmacytoid dendritic cells (pDCs) constitutive expression of IRF7 makes it the preferred IRF (Honda et al., 2006; Prakash et al., 2005). Interestingly, IRF5 appears to play a more dominant role in the induction of type I IFNs in response to bacterial pathogens (Bergstrom et al., 2015; Castiglia et al., 2016; Gratz et al., 2011; Pandey et al., 2009; Parker et al., 2014), and to a lesser extent against viruses (Lazear et al., 2013). Following PRR stimulation, IRFs are activated in a phosphorylation dependent manner. PRR ligation recruits a signaling adapter molecule which further recruits and activates the IRF kinases TBK1, IKKε, IRAK, IKKα. There are five classes of PRRs that detect bacterial components to activate IRFs. Toll like receptors (TLRs) are membrane bound PRRs, while RIG-I like receptors (RLRs), nucleotide-binding and oligomerizing domain (NOD) like receptors (NLRs), DNA sensors, and AIM-like receptors (ALRs) are found in the cytoplasm (Figure 1).
Figure 1. Signaling pathways leading to the induction of type I IFNs.
Recognition of components derived from bacteria occurs at the membrane and in the cytosol. Ligand binding of TLRs recruits the signaling adapter molecule MyD88 to TLR2/7/8/9/13 leading to activation of IRF kinases IRAK and IKKα. TLR4 recruits the adaptor TRIF and together they activate the IRF kinases TBK1 and IKKε. Engagement of cytosolic sensors leads to the recruitment of the signaling adapter molecule MAVS to the RLRs (RIG-I and MDA-5), STING to the DNA sensors (cGAS, DDX41 and IFI16), and RIP2 to the NLRs (NOD1 and NOD2). Like TRIF, cytosolic signaling adapters all activate TBK1 and IKKε. IRF kinases activate IRFs by phosphorylation dependent dimerization, allowing them to translocate into nucleus and drive type I FN expression.
TLRs are a family of 14 transmembrane receptors that are anchored in the cytoplasmic and endosomal membranes. Ligation of TLRs predominately induces proinflammatory and antibacterial genes, however a subset of TLRs also induce the expression of type I IFNs. Initially located at the plasma membrane, TLR4, and to a much lesser extent TLR2, induce type I IFNs following endocytosis, by ligating components derived from the bacterial cell surface (Barbalat et al., 2009; Kagan et al., 2008). In immune cells, such as dendritic cells (DCs), endosomal membrane anchored TLR9 is activated by bacterial DNA, whereas single stranded RNA is sensed by TLR7 (Mancuso et al., 2009), TLR8 (Eigenbrod et al., 2015), and TLR13 (Castiglia et al., 2016) to activate type I IFN expression. All TLRs, except TLR3, activate the downstream signaling adapter, MyD88. TLR4, however, activates both MyD88 and TRIF, but induces type I IFNs only through TRIF dependent signaling (Kawai and Akira, 2011).
In the cytoplasm, RNA is recognized by the RLRs, RIG-I and MDA-5. Ligand binding promotes association with the mitochondrial signaling adapter MAVS/Cardif/IPS/VISA (Kawai and Akira, 2011). Peptides derived from the bacterial cell wall elicit type I IFNs by engaging the NLRs, NOD1 (Watanabe et al., 2010) and NOD2; which in turn recruit the signaling adapter RIP2 (Pandey et al., 2009). Finally, DNA sensing is primarily carried out by cyclic GMP-AMP synthase (cGAS), an enzyme that has been recognized for its critical role in the catalyzing the formation of cyclic-GMP-AMP (cGAMP) (Sun et al., 2013). Other DNA sensors include the DExD/H box helicase, DDX41 which binds cyclic-di-GMP and cyclic-di-AMP, secondary metabolites unique to bacteria (Parvatiyar et al., 2012), and IFI16 (murine, IFI204), an ALR which binds double stranded DNA (Unterholzner et al., 2010). The adapter for the DNA sensors, as well as for cGAMP, is the Stimulator of Interferon Genes (STING), a transmembrane protein that resides in the endoplasmic reticulum (Ishikawa and Barber, 2008).
Unlike bacteria, viruses parasitize the host translation machinery to replicate and, as a consequence, the major pathways of recognition leading to type I IFNs are initiated by cytosolic PRRs. RLRs are engaged by viral RNAs, while viral DNA relies on STING dependent cGAS (Sun et al., 2013) and IFI16 (Unterholzner et al., 2010). Distinguishing viral and host nucleic acids is predicated on the detection of nucleotide sequence motifs and secondary structure formations unique to viruses (Kell et al., 2015; Sanchez et al., 2008). In the endosome, viral nucleic acids stimulate TLR3, TLR7, TLR8, and TLR9. TLR3 detects viral double stranded RNA, and like TLR4, signals through the TRIF signaling adapter. Lastly, contributing to a lesser extent, TLR2 and TLR4 ligation of viral proteins also triggers type I IFN induction (Kawai and Akira, 2011).
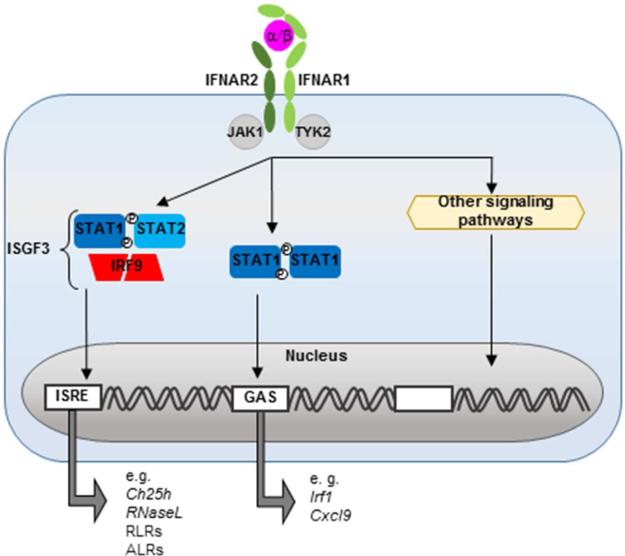
All type I IFNs signal in an autocrine and paracrine fashion through IFNAR, the heterodimeric transmembrane receptor composed of IFNAR1 and IFNAR2. Once crosslinked, the cytoplasmic tails of the IFNAR1/2 heterodimer activate Janus kinase 1 (Jak1) and tyrosine kinase 2 (Tyk2), which in turn phosphorylate members of the STAT family allowing them to dimerize and translocate to the nucleus. Formation of different transcription factor complexes is determined, in part, by the abundance and type of STAT produced by the cell (Miyagi et al., 2007), but also by positive and negative regulators (Ivashkiv and Donlin, 2014). Most cells express STAT1, STAT2 and IRF9, the canonical type I IFN transcription factors. STAT1/2 heterodimers recruit IRF9 to form the interferon stimulated gene factor 3 (ISGF3) complex. This complex binds to IFN-stimulated response elements (ISRE) located in the promoters of antiviral genes. In addition, both type I and type II IFNs can activate STAT1 homodimers, which bind γ-activated sequences that lead to the transcription of genes such as IRF1. STAT independent pathways are also activated by type I IFN and they contribute to the induction and expression of ISGs, reviewed elsewhere (Platanias, 2005) (Figure 2). Taken together, type I IFNs induce a diverse set of ISGs that extend beyond antiviral genes to include genes involved in modulating cellular activation and death pathways.
Figure 2. Type I IFNs induce ISGs expression through JAK/STAT signaling.
Binding of type I IFNs to the IFNAR1-IFNAR2 heterodimer activates Janus kinases, JAK1 and TYK2, to phosphorylate STAT transcription factors. Dimerization of activated STAT1 and STAT2 further recruits IRF9 and forms the ISGF3 complex, that promotes expression of genes containing an ISRE sequence in their promoter. Homodimerization of activated STAT1, drives the expression of genes containing a γ-activated sequence (GAS) in their promoter.
Disparate roles of type I IFNs during bacterial infection
How type I IFNs direct the outcome of bacterial infections hinges on many factors including, but not limited to, the bacterial replication strategy and virulence factors, as well as the route and site of the infection. These factors influence which type of host cell is activated, the magnitude of induction, the timing, and the duration of the response. Here we will present a vignette of different bacterial infections that, from the host perspective, are either negatively or positively affected by type I IFNs (also see Table 1).
Table 1.
Role of type I IFNs in different bacterial infections
| Pathogen | Gram reaction | Location | IFNα/β signaling | Key Mechanisms | Citations |
|---|---|---|---|---|---|
| Listeria monocytogenes | (+) | Intracellular | Detrimental | Promotion of apoptosis. Suppression of type II IFN and IL-17 | (Auerbuch et al., 2004; Carrero et al., 2004; Henry et al., 2010; O'Connell et al., 2004; Rayamajhi et al., 2010) |
| Francisella tularensis | (−) | Intracellular | Detrimental | Suppression of IL-17A | (Henry et al., 2007, 2010; Jones et al., 2010; Storek et al., 2015) |
| Salmonella enterica serovar Typhimurium | (−) | Intracellular | Detrimental | Promotion of necroptosis Suppression of innate cell recruitment and proinflammatory response. | (Perkins et al., 2015; Robinson et al., 2012; Schmolke et al., 2014) |
| Staphylococcus aureus | (+) | Extracellular | Detrimental | Suppression of innate cell recruitment and proinflammatory response. | (Martin et al., 2009; Parker et al., 2014) |
| Legionella pneumophilia | (−) | Intracellular | Protective | Promotion of IRG1 (itaconic acid) | (Lippmann et al., 2011; Naujoks et al., 2016; Plumlee et al., 2009) |
| Streptococcus pyogenes | (+) | Extracellular | Protective | Suppression of excessive proinflammatory response. | (Castiglia et al., 2016; Gratz et al., 2011; Mancuso et al., 2007, 2009) |
| Streptococcus pneumoniae | (+) | Extracellular | Protective | Promotion tissue integrity. | (Damjanovic et al., 2014; LeMessurier et al., 2013; Parker et al., 2011) |
| Helicobacter pylori | (+) | Extracellular | Protective | Promotion of Cxcl10 mediated cell recruitment. | (Flach et al., 2012; Watanabe et al., 2010) |
| Polymicrobial sepsis | (+) and (−) | Extracellular | Protective | Promotion of Cxcl10 mediated cell recruitment. | (Kelly-Scumpia et al., 2010) |
| Post influenza bacterial pneumonia | (+) or (−) | Extracellular | Detrimental | Suppression of proinflammatory response. | (Lee et al., 2015; Shahangian et al., 2009) |
| Mycobacterium tuberculosis | (+) | Intracellular | Detrimental | Promotion of IL-10 anti-inflammatory response. | (Berry et al., 2010; Manca et al., 2001; Mayer-Barber et al., 2014) |
| Mycobacterium leprae | (+) | Intracellular | Detrimental | Promotion of IL-10, IL-27 anti-inflammatory response. | (Liu et al., 2012; Teles et al., 2013, 2015) |
Type I IFN exacerbates bacterial infections
Widely used to study bacterial pathogenesis, Listeria, in humans, causes meningitis and sepsis in immunocompromised individuals and fetal infections in pregnant women. Infection by Listeria induces a strong type I IFN response that promotes host susceptibility, as deficiency in either IFNAR1 or IRF3 protects mice from Listeria infection (O'Connell et al., 2005). Listeriolysin O (LLO), a cytolysin secreted by Listeria to disrupt the integrity of the vacuole, allows the bacteria to escape into the cytoplasm. Escape is critical for bacterial replication, and it also exposes bacterial DNA to detection by cGAS, and to a lesser extent by IFI16 (Hansen et al., 2014) and bacterial RNA to RIG-I (Abdullah et al., 2012). Activation of these cytoplasmic sensors leads to type I IFN dependent induction of the proapoptotic genes such as DAXX (DAP6) and TRAIL, that promote macrophage (O'Connell et al., 2004) and lymphocyte (Carrero et al., 2004) apoptosis; ultimately promoting dissemination and proliferation of the bacteria. Consequently, Listeria is cleared more rapidly in the absence of IFNAR signaling (Auerbuch et al., 2004; Carrero et al., 2004; O'Connell et al., 2004). In addition, type I IFNs have been observed to attenuate expression of the IFNγ receptor (IFNRG1) (Rayamajhi et al., 2010), likely through the recruitment of gene silencing proteins by the early growth response transcription factor 3 (Erg3) in complex with NGFI-A binding protein 1 (Nab1), culminating in decreased IFNRG1 transcription (Kearney et al., 2013). Finally, type I IFNs inhibit the expression of IL-17A by γδ T cells, a type of innate immune lymphocyte, to suppress neutrophil recruitment (Henry et al., 2010).
Francisella tularensis, is responsible for tularemia, a highly contagious and life-threatening respiratory disease. Expression of genes in the Francisella pathogenicity island trigger the escape of the bacteria from the phagosome to the cytoplasm, and once in the cytoplasm the bacteria induce type I IFN through a cGAS- and IFI204-STING-IRF3 dependent pathways (Henry et al., 2007; Storek et al., 2015). Type I IFNs suppress the innate antibacterial response by inhibiting expression of IL-17A by γδ T cells. As in Listeria infections, elevated expression of IL-17A in IFNAR1 deficient mice enhances splenic neutrophil recruitment and is correlated with both improved bacterial clearance and survival (Henry et al., 2010). Interestingly, though deficiency in cGAS, STING, IFNAR1 or IRF3 renders mice resistant to infection by Francisella (Henry et al., 2007; Storek et al., 2015), deletion of AIM2, an ALR induced by type I IFNs, is detrimental to host defense (Jones et al., 2010; Rathinam et al., 2010). Yet, while type I IFNs potentiate AIM2 expression and indirectly its antibacterial activity, simultaneously AIM2 suppresses induction of type I IFNs by negatively regulating the cGAS-STING-IRF3 pathway (Corrales et al., 2016).
The Gram negative, intracellular bacterium, Salmonella enterica serovar Typhimurium causes acute gastroenteritis in humans, and, if uncontrolled, may disseminate and cause a more life-threatening disease. RIGI detection of Salmonella mRNA induces type I IFNs in non-phagocytic cells, whereas recognition of Salmonella LPS by TLR4 drives type I IFN induction in phagocytic cells (Schmolke et al., 2014). During systemic infection, IFNAR1 deficient mice accumulate less bacteria in the spleen and liver, and have markedly survival compared to wild type (WT) mice. Enhanced survival and defense noted in IFNAR1 and IFNβ deficient mice is associated with increased antibacterial proinflammatory responses (Perkins et al., 2015). Moreover, induction of type I IFNs during Salmonella infection promotes macrophage death by necroptosis (Henry et al., 2007; Robinson et al., 2012). Necroptosis is a type programmed cell death, that unlike TRAIL mediated apoptosis, proceeds in a caspase independent manner. Here IFNβ, not IFNα, promotes necroptosis (Robinson et al., 2012); illustrating an exclusive effector function for IFNβ.
Infections caused by the Gram positive, extracellular bacterium Staphylococcus aureus (S. aureus) are also exacerbated by type I IFNs. S. aureus is a common etiological agent of local skin infections, but it is also a primary cause of severe lung pneumonia and bloodstream infections. Type I IFNs are induced by protein A, a virulence factor, (Martin et al., 2009) and via TLR9-IRF1 or NOD2-IRF5 (Parker et al., 2014). In vivo, IFNAR1 deficiency confers resistance to lethal pneumonia. Survival is associated with increased CD11c+ DCs in the lungs and reduced TNFα in the bronchoalveolar lavage fluid (Martin et al., 2009). Interestingly, while IFNβ expression is inducible in cultured lung epithelial cells (Martin et al., 2009; Parker et al., 2014), transcript for IFNα is not detected (Martin et al., 2009). This suggests strain specific effects, as well as host specific capacities as differences between hematopoietic and non-hematopoietic cells are also noted (Parker et al., 2014).
Type I IFNs protect against bacterial infections
Legionella pneumophilia is responsible for Legionnaires’ disease, a severe lung pneumonia. Its type IV secretion system, Dot/Icm, is required for entry into and replication within the macrophage cytosol (Lippmann et al., 2011). It has been reported that the host detects Legionella in the cytosol by a STING dependent pathway (Lippmann et al., 2011), leading to IRF3 dependent type I IFN expression (Plumlee et al., 2009). While in a mouse model of pulmonary Legionella infection, IFNAR2 deficient mice did not reveal a role for type I IFNs (Ang et al., 2010), proliferation of Legionella in macrophages is inhibited by type I IFNs (Lippmann et al., 2011; Plumlee et al., 2009). Moreover, type I IFN, along with type II IFN, promote host defense likely through induction of cell intrinsic ISGs such as immune-responsive gene 1 (IRG1) (Naujoks et al., 2016).
Type I IFNs also fortify the host against infections caused by Gram positive Streptococci. Streptococcus pyogenes, the group A streptococcus, causes superficial and deep tissue infections that can develop into necrotizing fasciitis. Both group A streptococci and group B streptococci (Streptococcus agalactiae) activate the STING-TBK1-IRF3 pathway in macrophages (Gratz et al., 2011); while in cDCs, TLR7-Myd88-IRF5 and to a lesser extent IRF1, both play a role (Castiglia et al., 2016; Gratz et al., 2011; Mancuso et al., 2009). In a mouse model of S. pyogenes cellulitis, survival of WT mice is significantly greater than IFNAR1 deficient mice (Gratz et al., 2011). Enhanced survival is linked to the anti-inflammatory properties conferred by type I IFN signaling (Castiglia et al., 2016). Likewise, systemic infection of adult or neonatal mice with group B streptococci also requires IFNAR signaling to protect the host; and while IFNα4 was induced in WT group B streptococci infected mice, IFNβ plays a dominant role in conferring protection as IFNβ KO mice are more susceptible than WT mice (Mancuso et al., 2007).
Streptococcus pneumoniae causes life-threatening pneumonia. Host detection of S. pneumoniae is facilitated by the expression of autolysin and pneumolysin, two virulence factors that cooperate to introduce bacterial DNA into the cytosol. In epithelial cells, bacterial DNA activates STING to induce IFNβ (Parker et al., 2011). In two independent studies it was observed that in the absence of IFNAR1, even though immune cell recruitment was enhanced, more bacteria were found in the lungs (Parker et al., 2011) or escaping from the lungs into the bloodstream (LeMessurier et al., 2013). Exogenous IFNβ decreases transmigration into the bloodstream by promoting the expression of genes encoding tight junction proteins and, simultaneously, downregulating the expression of platelet activating receptor, the receptor by which the bacteria enter the cell (LeMessurier et al., 2013). Moreover, treatment of mice with an IFNα expressing adenoviral vector, led to decreased immunopathology and enhanced antibacterial activity in macrophages, resulting in an overall increase in survival (Damjanovic et al., 2014).
The Gram negative bacterium, Helicobacter pylori is the etiological agent of acute gastric infections, as well as chronic gastric ulcers and cancer. In non-hematopoietic cells, NOD1 sensing of a peptide derived from H. pylori peptidoglycan induces type I IFN expression in an IRF7 dependent manner. IFNAR1 or NOD1 deficient mice fail to restrict H. pylori proliferation. Concurrently, a significant reduction in expression of the chemotactic ISG, Cxcl10, suggests type I IFN induced Cxcl10 is critical for the control of H. pylori proliferation (Watanabe et al., 2010); and it is further supported by elevated Cxcl10 levels observed in vaccinated mice (Flach et al., 2012) and patients that are asymptomatic carriers (Jafarzadeh et al., 2013).
Type I IFN induced Cxcl10 also promotes host resistance during polymicrobial sepsis. Using a model of cecal ligation and puncture (CLP) in mice, investigators found the absence of IFNAR signaling led to increased mortality. Although the chemokine Cxcl1 was elevated in IFNAR1 deficient mice, decreased levels Cxcl10, were also noted. Administration of Cxcl10 to IFNAR1 deficient mice, restored hematopoietic cell recruitment and antibacterial activity which led to enhanced bacterial clearance and host resistance to polymicrobial sepsis (Kelly-Scumpia et al., 2010).
Type I IFNs are detrimental to secondary bacterial infections
High mortality rates associated with seasonal and pandemic influenza are driven by the development of secondary bacterial pneumonia caused by S. pneumoniae or S. aureus, and, at a lower incidence, other bacteria (McCullers, 2014). This enhanced susceptibility is transient, limited by the initiation and the duration of type I IFN expression. As discussed, type I IFNs promote host defense against S. pneumoniae infection, yet preceding expression of type I IFNs has detrimental effects. First, prior induction of type I IFNs attenuates the expression of neutrophil chemokines (Shahangian et al., 2009). Neutrophil recruitment is restored by deletion of either IFNAR1 (Shahangian et al., 2009) or the type I IFN responsive methyltransferase, Setdb2, which culminates in bacterial clearance (Schliehe et al., 2015; Shahangian et al., 2009). Second, prior expression of type I IFNs suppresses production of IL-17 by T cells (Cao et al., 2014; Kudva et al., 2011). In the absence of IL-17, diminished secretion of antibacterial peptides lipocalin 2 and BPIFA1 is associated with increased bacterial growth (Lee et al., 2015). Lastly, the duration of type I IFN expression dictates sensitivity to secondary bacterial infection, as superinfection do not develop if initiated 14 days after influenza infection (Lee et al., 2015).
Type I IFN promotes chronic bacterial infections
Mycobacteria comprises a group of highly contagious pathogens that establish chronic infections of the lungs (M. tuberculosis) or skin (M. leprae). The host relies on cGAS sensing of bacterial DNA (Collins et al., 2015; Wassermann et al., 2015; Watson et al., 2015), as well as NOD2 sensing of cell wall associated muramyl dipeptides (Pandey et al., 2009) to detect M. tuberculosis. Virulent strains provoke higher IFNα expression in the lungs compared to less virulent strains. IFNAR signaling suppresses IL-12 and IFNγ, thereby arresting the development of the antibacterial adaptive T cell (Th1) response and reducing host survival (Manca et al., 2001; Mayer-Barber et al., 2014). Similar results have been reported in human patients. Examination of the gene expression pattern of whole blood revealed a distinct gene expression signature defined by type I IFN and downstream ISGs in patients with active tuberculosis, that was not present in patients with latent tuberculosis (Berry et al., 2010). Likewise, ex vivo studies with M. leprae, demonstrate IFNβ induced anti-inflammatory cytokines drive the progression to chronic leprosy by inhibiting the development of Th1 immunity (Teles et al., 2013, 2015).
Disparate effector mechanisms of type I IFNs
Given their pleotropic nature, type I IFNs have the capacity to influence multiple host defense mechanisms. Productive host response to acute bacterial infection requires secretion of proinflammatory cytokines and chemokines that recruit and activate innate immune cells. Herein, we will explore the effect of type I IFN on key antibacterial mechanisms.
Suppression of Type II IFN responses by type I IFN
Like type I IFN, type II IFN is expressed during microbial infections and it exerts antiviral and immunomodulatory activities. Type II IFN, solely consisting of IFNγ, is induced by IL-12 and IL-18, with natural killer (NK) and T cells being the dominant producers. Type I and type II IFNs activate distinct and overlapping gene programs by signaling through their respective canonical JAK-STAT pathways. IFNγ preferentially induces phosphorylation and dimerization of STAT1 to promote expression of the γ-activated sequence-dependent genes. Type I IFNs favor the assembly of the ISGF3 transcription factor complex which drives ISRE controlled genes, but also activate STAT1 dimerization. (Manca et al., 2001). Intriguingly, NK cells express high basal levels of STAT4, which enables rapid induction of IFNγ following type I IFN stimulation. However, in the absence of IL-12 signaling, IFNγ expression is transient as STAT4 is displaced by STAT1, thus negatively regulating the production of type II IFN (Mack et al., 2011; Miyagi et al., 2007). IFNγ is critical for defense against many bacterial pathogens (Harty and Bevan, 1995; Lippmann et al., 2011; Teles et al., 2013). Despite numerous studies, it is still not clear how type I and type II IFN play opposite roles in host defense against certain bacteria and which downstream effector genes are responsible for such differences. In addition to regulating downstream effector genes, type I IFNs also suppress IFNγ expression by attenuating the transcription of its inducer, IL-12 (Berry et al., 2010; Carrero et al., 2004; Manca et al., 2001) and its receptor, IFNRG1 (Kearney et al., 2013; Rayamajhi et al., 2010) (Figure 3). Insufficient IFNγ expression can have drastic consequences. M. leprae infections that manifest as “self-healing” leprosy provoke an IFNγ driven Th1 response, while a type I IFN signature dominates during chronic, disseminated infections (Liu et al., 2012; Teles et al., 2013). Further, hypervirulent strains of M. tuberculosis promote the expression of type I IFNs, and concurrently attenuate IL-12 and IFNγ (Manca et al., 2001), further reinforcing that induction of type I IFNs is favorable for bacterial pathogenesis.
Figure 3. Effector mechanisms mediated by type I IFNs during bacterial infection.
Autocrine and paracrine signaling of type I IFNs suppress type II IFN signaling, proinflammatory responses and production of antimicrobial peptides to contribute to overall detrimental host outcomes. Modulation of metabolism by type I IFNs also exerts negative effects by either suppressing sterol biosynthesis and PGE2 or by upregulating CH25H, but may also promote host defense by inducing expression of IRG1. Tissue integrity is strengthened by type I IFNs through the induction of tight junction (TJs) proteins, claudins and occludins, yet suppressive effects on ILC2s and amphiregulin are also observed. See text for details.
Anti-inflammatory responses by type I IFN
Productive host antibacterial response relies on the coordination and balance of proinflammatory molecules, to clear bacteria, and anti-inflammatory molecules to limit tissue damage. Type I IFNs propagate the anti-inflammatory response by upregulating the expression of IL-10 and IL-27. IL-10 is a type I IFN inducible immunosuppressive and anti-inflammatory cytokine. Induction of IL-10 has been described to occur through both IL-27-dependent (Iyer et al., 2010) and -independent mechanisms (McNab et al., 2014). IL-10 inhibits the acute phase cytokines, TNFα and IL-1 (Bogdans et al., 1992), thereby attenuating expression of adhesion molecules and chemokines required for leukocyte recruitment (Yarilina et al., 2008). Downregulation of the proinflammatory response is essential to protect against tissue damage induced mortality as observed during Escherichia coli sepsis (Sewnath et al., 2001), but it is detrimental to the clearance of other bacterial infections (Auerbuch et al., 2004; Di Paolo et al., 2015; Mayer-Barber et al., 2014; McNab et al., 2014). Similarly, IL-27 protects against tissue damage during acute influenza infection (Liu et al., 2014), but compromises the host response to both secondary (Cao et al., 2014; Robinson et al., 2015) and chronic bacterial infections (Teles et al., 2015). The delicate balance between proinflammatory and anti-inflammatory responses is further illustrated by chemokine recruitment. Type I IFN mediated suppression of proinflammatory chemokines, Cxcl1 and Cxcl2, reduces myeloid recruitment and may protect against excessive tissue damage (Ellis et al., 2015), but may also compromise myeloid mediated bacterial clearance (Perkins et al., 2015; Schliehe et al., 2015; Shahangian et al., 2009) (Figure 3).
Inhibition of antimicrobial peptides by type I IFN
One consequence of the anti-inflammatory action mediated by type I IFNs is the suppression of several antimicrobial peptides (Figure 3). Antimicrobial peptides exert non-enzymatic activities that inhibit microbial functions by directly inhibiting the pathogen or host specific targets. Cathelicidin (CAMP) and beta-defensin 2 (DEFB4) are two antimicrobial peptides induced by IL-1β and cytochrome P450 family 27 subfamily B member 1 (CYP27B1), an enzyme in the vitamin D pathway. They are indirectly downregulated by IFNβ-IL-10 (Teles et al., 2013), through the induction of miR-21, an ISG that inhibits the translation of IL1B and CYP27B1 transcripts (Liu et al., 2012). Type I IFNs also negatively regulate the expression antimicrobial peptides induced by IL-22, such as lipocalin 2 and BPIFA1 (Lee et al., 2015). Downregulation of these antimicrobial peptides also occurs indirectly, as type I IFNs target IL-23, the cytokine that induces IL-22 (Kudva et al., 2011).
Regulation of metabolic pathways by type I IFN
There is a growing appreciation for the role of cellular metabolism in the coordination of immune response. TLR signaling and microbial infections have been observed to redirect the metabolic strategy of macrophage and dendritic cells. Type I IFNs downregulate sterol biosynthesis triggering a shift away from de novo cholesterol synthesis (Blanc et al., 2011) and towards cholesterol import. This shift, by an unknown mechanism, drives spontaneous STING-IRF3 dependent IFNβ expression (York et al., 2015). Type I IFNs also promote key antiviral responses such as upregulation of cholesterol-25 hydroxylase (Ch25h), an ISG that inhibits viral entry (Liu et al., 2013). During Listeria infection, however, Ch25h expression increases the susceptibility of mice to bacterial infection by negatively regulating capase-1 and IL-1β maturation (Reboldi et al., 2014). Other metabolic pathways are also influenced by type I IFNs. Prostaglandin E2 (PGE2), an arachidonic acid derived lipid mediator that promotes host defense against Mycobacterium, is negatively regulated by type I IFN suppression of IL-1β, but, at the same time, negatively regulates type I IFN expression (Mayer-Barber et al., 2014). Conversely, type I IFNs induce IRG1, an ISG that generates, itaconic acid, an antimicrobial metabolite that effectively restricts the growth of intracellular bacteria (Naujoks et al., 2016) (Figure 3).
Regulation of tissue integrity by type I IFN
Tissue integrity is maintained by the expression of tight junction proteins such as cadherins, claudins, and the peripheral scaffolding proteins such as occludins. The arrangement and density of these proteins determines the permeability of the epithelial barrier and act to confine local tissue infections (LeMessurier et al., 2013; Long et al., 2014). Type I IFNs, augmented by RNase-L, an ISG that mediates induction of type I IFNS, promote the expression of tight junction proteins (LeMessurier et al., 2013). RNase-L deficiency reduces IFNβ expression and, subsequently, diminishes tissue integrity (Long et al., 2013). This promotes host susceptibility to opportunistic (Long et al., 2013) and acquired bacterial infections (Li et al., 2008). In addition, group 2 innate lymphoid cells (ILC2s) have recently emerged as the key tissue resident cell type orchestrating tissue repair (Monticelli et al., 2011). ILC2s foster tissue integrity by upregulating expression of amphiregulin, an epithelial growth factor that promotes sustained signaling. Type I IFNs attenuate the expression of amphiregulin and stunt ILC2 proliferation (Monticelli et al., 2011) (Figure 3). This has been observed to confer long lasting deficiencies to the integrity of the mesenteric adipose tissue and the mesenteric lymph nodes (Fonseca et al., 2015).
Conclusion and Future Outlook
Type I IFNs direct a potent antiviral response through the induction a diverse set of ISGs. Some ISGs have evolved to exert broad antiviral activities, while others act to specifically target different classes of viruses. Over the past few decades, numerous studies have uncovered a pivotal role for type I IFNs in dictating host response to bacterial infections. Unlike their role in viral infections, type I IFNs play an unpredictable role in bacterial infections. Their pleiotropic nature leads not only to the induction of antiviral genes, but also to genes that modulate innate and adaptive immune responses. While some of the immunomodulatory genes are beneficial to host defense against bacterial infections, many are detrimental to the formation of an antibacterial inflammatory response.
Given the disparate role of type I IFNs among different bacterial infections, it is clear that the field will benefit from more comprehensive analyses of the type I IFN response. Crucially, the current state of our knowledge does not allow us to determine whether type I IFNs will be beneficial or detrimental based on the biology of the pathogen. Studies of the role of type I IFNs during viral infections have shown that a short, strong type I IFN response is generally beneficial to clearing the infection. However, sustained expression seems to be detrimental to the clearance of persistent viral infections (Wilson et al., 2013). Recently, it has been demonstrated that IFNβ and IFNα exert discreet functions during chronic viral infection; in early stages IFNβ promotes disorganization of the spleen, whereas IFNα limits viral dissemination (Ng et al., 2015). Thus, it is becoming increasingly apparent that type I IFNs are not redundant, but that they have overlapping and distinct functions.
Furthermore, functional differences may result not only from preferential expression of IFNα or IFNβ by different cell types (Honda et al., 2006; Prakash et al., 2005) or from their difference in affinity for IFNAR (Weerd et al., 2013), but also from the route of infection. Listeria administered by oral gavage enhances host resistance, whereas infection initiated by intraperitoneal injection leads to host susceptibility (Kernbauer et al., 2013). Both routes elicit type I IFN expression by inflammatory myeloid cells, however differences in hepatic colonization appear to shape the response (Kernbauer et al., 2013; Stockinger et al., 2009). Few have undertaken comprehensive studies of route even though it has been shown to play a role in both host response and pathogen biology (Fitzgeorge et al., 1983; Kernbauer et al., 2013; Martins et al., 2013).
Extending these observations to the role of type I IFNs on the outcome of bacterial infections, we suggest that more detailed examinations of the composition, magnitude, and duration of type I IFN expression will complement mechanistic investigations into the effector functions of specific type I IFNs and lead to more precise management of infections.
Acknowledgments
We thank Kislay Parvartiyar, Maxime Chapon and Elisa Deiru for helpful discussions and editing of this manuscript. We apologize, but due to space limitations, we were unable to cite all relevant studies. Work in the G.C. laboratory is supported by the National Institutes of Health grants AI056154 and AI069120.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- Abdullah Z, Schlee M, Roth S, Mraheil MA, Barchet W, Böttcher J, Hain T, et al. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. The EMBO Journal. 2012 Oct;:4153–4164. doi: 10.1038/emboj.2012.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang DKY, Oates CVL, Schuelein R, Kelly M, Sansom FM, Bourges D, Boon L, et al. Cutting edge: pulmonary Legionella pneumophila is controlled by plasmacytoid dendritic cells but not type I IFN. Journal of Immunology (Baltimore, Md. : 1950) 2010;184(10):5429–33. doi: 10.4049/jimmunol.1000128. [DOI] [PubMed] [Google Scholar]
- Auerbuch V, Brockstedt DG, Meyer-Morse N, O'Riordan M, Portnoy D. a. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. The Journal of Experimental Medicine. 2004;200(4):527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbalat R, Lau L, Locksley RM, Barton GM. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nature Immunology. 2009;10(11):1200–7. doi: 10.1038/ni.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom B, Aune MH, Awuh J. a., Kojen JF, Blix KJ, Ryan L, Flo TH, et al. TLR8 Senses Staphylococcus aureus RNA in Human Primary Monocytes and Macrophages and Induces IFN-Production via a TAK1-IKK -IRF5 Signaling Pathway. The Journal of Immunology. 2015;195(3) doi: 10.4049/jimmunol.1403176. doi:10.4049/jimmunol.1403176. [DOI] [PubMed] [Google Scholar]
- Berry MPR, Graham CM, McNab FW, Xu Z, Bloch S. a a, Oni T, Wilkinson K. a, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature, Nature Publishing Group. 2010;466(7309):973–7. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc M, Hsieh WY, Robertson K. a, Watterson S, Shui G, Lacaze P, Khondoker M, et al. Host defense against viral infection involves interferon mediated down-regulation of sterol biosynthesis. PLoS Biology, Vol. 2011;9(3):e1000598. doi: 10.1371/journal.pbio.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdans C, Vodovotz Y, Nathan C. Contrasting Mechanisms for Suppression of Macrophage Cytokine Release. The Journal of Biological Chemistry. 1992;267(32):23301–23308. [PubMed] [Google Scholar]
- Cao J, Wang D, Xu F, Gong Y, Wang H, Song Z, Li D, et al. Activation of IL-27 signalling promotes development of postinfluenza pneumococcal pneumonia. EMBO Molecular Medicine. 2014;6(1):120–140. doi: 10.1002/emmm.201302890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero J. a, Calderon B, Unanue ER. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. The Journal of Experimental Medicine. 2004;200(4):535–540. doi: 10.1084/jem.20040769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglia V, Piersigilli A, Ebner F, Janos M, Goldmann O, Damböck U, Kröger A, et al. Type I Interferon Signaling Prevents IL-1β-Driven Lethal Systemic Hyperinflammation during Invasive Bacterial Infection of Soft Tissue. Cell Host & Microbe. 2016;19(3):375–387. doi: 10.1016/j.chom.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Collins AC, Cai H, Li T, Franco LH, Li X-D, Nair VR, Scharn CR, et al. Cyclic GMP-AMP Synthase Is an Innate Immune DNA Sensor for Mycobacterium tuberculosis. Cell Host & Microbe, Elsevier Inc. 2015;17(6):820–828. doi: 10.1016/j.chom.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales L, Woo S-R, Williams JB, McWhirter SM, Dubensky TW, Gajewski TF. Antagonism of the STING Pathway via Activation of the AIM2 Inflammasome by Intracellular DNA. The Journal of Immunology. 2016 doi: 10.4049/jimmunol.1502538. available at: http://doi.org/10.4049/jimmunol.1502538. [DOI] [PMC free article] [PubMed]
- Crouse J, Kalinke U, Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nature Reviews Immunology, Nature Publishing Group. 2015;15(4):231–242. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- Damjanovic D, Khera A, Medina MF, Ennis J, Turner JD, Gauldie J, Xing Z. Type 1 interferon gene transfer enhances host defense against pulmonary Streptococcus pneumoniae infection via activating innate leukocytes. Molecular Therapy. Methods & Clinical Development. 2014 Jan;1:5. doi: 10.1038/mtm.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo NC, Shafiani S, Day T, Papayannoupoulou T, Russell DW, Iwakura Y, Sherman D, et al. Interdependence between Interleukin-1 and Tumor Necrosis Factor Regulates TNF-Dependent Control of Mycobacterium tuberculosis Infection. Immunity, Elsevier Inc. 2015;43(6):1125–1136. doi: 10.1016/j.immuni.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigenbrod T, Pelka K, Latz E, Kreikemeyer B, Dalpke AH. TLR8 Senses Bacterial RNA in Human Monocytes and Plays a Nonredundant Role for Recognition of Streptococcus pyogenes. Journal of Immunology (Baltimore, Md. : 1950) 2015;195(3):1092–9. doi: 10.4049/jimmunol.1403173. [DOI] [PubMed] [Google Scholar]
- Ellis GT, Davidson S, Crotta S, Branzk N, Papayannopoulos V, Wack A. TRAIL+ monocytes and monocyte-related cells cause lung damage and thereby increase susceptibility to influenza-Streptococcus pneumoniae coinfection. EMBO Reports. 2015;16(9):1203–18. doi: 10.15252/embr.201540473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgeorge RB, Baskerville a, Broster M, Hambleton P, Dennis PJ. Aerosol infection of animals with strains of Legionella pneumophila of different virulence: comparison with intraperitoneal and intranasal routes of infection. The Journal of Hygiene. 1983;90:81–89. doi: 10.1017/s0022172400063877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach C.-F.. c, Mozer M, Sundquist M.. b, Holmgren J.. c, Raghavan S.. b. Mucosal vaccination increases local chemokine production attracting immune cells to the stomach mucosa of Helicobacter pylori infected mice. Vaccine, Elsevier Ltd. 2012;30(9):1636–1643. doi: 10.1016/j.vaccine.2011.12.111. [DOI] [PubMed] [Google Scholar]
- Fonseca D.M. da, Hand TW, Han S-J, Gerner MY, Zaretsky AG, Byrd AL, Harrison OJ, et al. Microbiota-Dependent Sequelae of Acute Infection Compromise Tissue-Specific Immunity. Cell, Elsevier Inc. 2015;163(2):354–366. doi: 10.1016/j.cell.2015.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz N, Hartweger H, Matt U, Kratochvill F, Janos M, Sigel S, Drobits B, et al. Cheung A, editor. Type I Interferon Production Induced by Streptococcus pyogenes-Derived Nucleic Acids Is Required for Host Protection. PLoS Pathogens. 2011;7(5):e1001345. doi: 10.1371/journal.ppat.1001345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K, Prabakaran T, Laustsen A, Jørgensen SE, Rahbæk SH, Jensen SB, Nielsen R, et al. Listeria monocytogenes induces IFNβ expression through an IFI16-, cGAS- and STING-dependent pathway. The EMBO Journal. 2014;33(15):1654–66. doi: 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty JT, Bevan MJ. Specific immunity to Listeria monocytogenes in the absence of IFN gamma. Immunity. 1995;3(1):109–117. doi: 10.1016/1074-7613(95)90163-9. [DOI] [PubMed] [Google Scholar]
- Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. The Journal of Experimental Medicine. 2007;204(5):987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T, Kirimanjeswara GS, Ruby T, Jones JW, Peng K, Perret M, Ho L, et al. Type I IFN signaling constrains IL-17A/F secretion by gammadelta T cells during bacterial infections. J Immunol. 2010;184(7):3755–3767. doi: 10.4049/jimmunol.0902065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Takaoka A, Taniguchi T. Type I Inteferon Gene Induction by the Interferon Regulatory Factor Family of Transcription Factors. Immunity. 2006;25(3):349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Isaacs A, Lindenmann J. Proceedings of the Royal Society of London. Series B, Containing Papers of a Biological Character. Royal Society; Great Britain: 1957. Virus interference. I. The interferon. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv LB, Donlin LT. Nature Reviews. Immunology. 1. Vol. 14. Nature Publishing Group; 2014. Regulation of type I interferon responses. pp. 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer SS, Ghaffari AA, Cheng G. Lipopolysaccharide-mediated IL-10 transcriptional regulation requires sequential induction of type I IFNs and IL-27 in macrophages. Journal of Immunology (Baltimore, Md. : 1950) 2010;185(11):6599–607. doi: 10.4049/jimmunol.1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarzadeh A, Nemati M, Rezayati MT, Khoramdel H, Nabizadeh M, Hassanshahi G, Abdollahi H. Lower circulating levels of chemokine CXCl10 in Helicobacter pyloriinfected patients with peptic ulcer: Influence of the bacterial virulence factor CagA. Iranian Journal of Microbiology. 2013;5(1):28–35. [PMC free article] [PubMed] [Google Scholar]
- Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O'Rourke K, Chan S, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci U S A. 2010;107(21):9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nature Immunology. 2008;9(4):361–8. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34(5):637–50. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Kearney SJ, Delgado C, Eshleman EM, Hill KK, O'Connor BP, Lenz LL. Type I IFNs Downregulate Myeloid Cell IFN-γ Receptor by Inducing Recruitment of an Early Growth Response 3/NGFI-A Binding Protein 1 Complex That Silences ifngr1 Transcription. Journal of Immunology (Baltimore, Md. : 1950) 2013;191(6):3384–92. doi: 10.4049/jimmunol.1203510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell A, Stoddard M, Li H, Marcotrigiano J, Shaw GM, Gale M. Pathogen-Associated Molecular Pattern Recognition of Hepatitis C Virus Transmitted/Founder Variants by RIG-I Is Dependent on U-Core Length. Journal of Virology. 2015;89(21):11056–11068. doi: 10.1128/JVI.01964-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly-Scumpia KM, Scumpia PO, Delano MJ, Weinstein JS, Cuenca AG, Wynn JL, Moldawer LL. Type I interferon signaling in hematopoietic cells is required for survival in mouse polymicrobial sepsis by regulating CXCL10. The Journal of Experimental Medicine. 2010;207(2):319–26. doi: 10.1084/jem.20091959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernbauer E, Maier V, Rauch I, Müller M, Decker T. Route of Infection Determines the Impact of Type I Interferons on Innate Immunity to Listeria monocytogenes. PLoS ONE. 2013;8(6) doi: 10.1371/journal.pone.0065007. available at: http://doi.org/10.1371/journal.pone.0065007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudva A, Scheller EV, Robinson KM, Crowe CR, Choi SM, Slight SR, Khader SA, et al. Influenza A inhibits Th17-mediated host defense against bacterial pneumonia in mice. Journal of Immunology (Baltimore, Md. : 1950) 2011;186(3):1666–74. doi: 10.4049/jimmunol.1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Maza LM, Peterson EM, Goebel JM, Fennie CW, Czarniecki CW. Interferon-induced inhibition of Chlamydia trachomatis: dissociation from antiviral and antiproliferative effects. Infection and Immunity. 1985;47(3):719–22. doi: 10.1128/iai.47.3.719-722.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Lancaster A, Wilkins C, Suthar MS, Huang A, Vick SC, Clepper L, et al. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathogens. 2013;9(1):e1003118. doi: 10.1371/journal.ppat.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Robinson KM, McHugh KJ, Scheller EV, Mandalapu S, Chen C, Di YP, et al. Influenza-induced Type I Interferon Enhances Susceptibility to Gram-negative and Gram-positive Bacterial Pneumonia in Mice. American Journal of Physiology. Lung Cellular and Molecular Physiology. 2015:ajplung.00338.2014. doi: 10.1152/ajplung.00338.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMessurier KS, Häcker H, Chi L, Tuomanen E, Redecke V. Type I interferon protects against pneumococcal invasive disease by inhibiting bacterial transmigration across the lung. PLoS Pathogens. 2013;9(11):e1003727. doi: 10.1371/journal.ppat.1003727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-L, Ezelle HJ, Kang T-J, Zhang L, Shirey KA, Harro J, Hasday JD, et al. An essential role for the antiviral endoribonuclease, RNase-L, in antibacterial immunity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20816–20821. doi: 10.1073/pnas.0807265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann J, Müller HC, Naujoks J, Tabeling C, Shin S, Witzenrath M, Hellwig K, et al. Dissection of a type I interferon pathway in controlling bacterial intracellular infection in mice. Cellular Microbiology. 2011;13(11):1668–1682. doi: 10.1111/j.1462-5822.2011.01646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu FDM, Kenngott EE, Schröter MF, Kühl A, Jennrich S, Watzlawick R, Hoffmann U, et al. Timed Action of IL-27 Protects from Immunopathology while Preserving Defense in Influenza. PLoS Pathogens. 2014;10(5) doi: 10.1371/journal.ppat.1004110. available at: http://doi.org/10.1371/journal.ppat.1004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PT, Wheelwright M, Teles R, Komisopoulou E, Edfeldt K, Ferguson B, Mehta MD, et al. MicroRNA-21 targets the vitamin D–dependent antimicrobial pathway in leprosy. Nature Medicine, Nature Publishing Group. 2012;18(2):267–273. doi: 10.1038/nm.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S-Y, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK, Pernet O, et al. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity, Elsevier Inc. 2013;38(1):92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TM, Chakrabarti A, Ezelle HJ, Brennan-Laun SE, Raufman J, Polyakova I, Silverman RH, et al. RNase-L deficiency exacerbates experimental colitis and colitis-associated cancer. Inflammatory Bowel Diseases. 2013;19(6):1295–1305. doi: 10.1097/MIB.0b013e318281f2fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TM, Nisa S, Donnenberg MS, Hassel B. Enteropathogenic Escherichia coli inhibits type I interferon- and RNase L-mediated host defense to disrupt intestinal epithelial cell barrier function. Infection and Immunity. 2014;82(7):2802–14. doi: 10.1128/IAI.00105-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack EA, Kallal LE, Demers DA, Biron CA. Type 1 interferon induction of natural killer cell gamma interferon production for defense during lymphocytic choriomeningitis virus infection. mBio. 2011;2(4):e00169–11. doi: 10.1128/mBio.00169-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manca C, Tsenova L, Bergtold a, Freeman S, Tovey M, Musser JM, Barry CE, et al. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha /beta. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(10):5752–5757. doi: 10.1073/pnas.091096998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso G, Gambuzza M, Midiri A, Biondo C, Papasergi S, Akira S, Teti G, et al. Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nature Immunology. 2009;10(6):587–594. doi: 10.1038/ni.1733. [DOI] [PubMed] [Google Scholar]
- Mancuso G, Midiri a., Biondo C, Beninati C, Zummo S, Galbo R, Tomasello F, et al. Type I IFN Signaling Is Crucial for Host Resistance against Different Species of Pathogenic Bacteria. The Journal of Immunology. 2007;178(5):3126–3133. doi: 10.4049/jimmunol.178.5.3126. [DOI] [PubMed] [Google Scholar]
- Martin FJ, Gomez MI, Wetzel DM, Memmi G, O'Seaghdha M, Soong G, Schindler C, et al. Staphylococcus aureus activates type I IFN signaling in mice and humans through the Xr repeated sequences of protein A. Journal of Clinical Investigation. 2009;119(7):1931–1939. doi: 10.1172/JCI35879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins NE, Faria VG, Teixeira L, Magalhäes S, Sucena Ëlio. Host Adaptation Is Contingent upon the Infection Route Taken by Pathogens. PLoS Pathogens. 2013;9(9) doi: 10.1371/journal.ppat.1003601. available at: http://doi.org/10.1371/journal.ppat.1003601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer-Barber KD, Andrade BB, Oland SD, Amaral EP, Barber DL, Gonzales J, Derrick SC, et al. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature, Nature Publishing Group. 2014;511(7507):99–103. doi: 10.1038/nature13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA. Nature Reviews Microbiology. 4. Vol. 12. Nature Publishing Group; 2014. The co-pathogenesis of influenza viruses with bacteria in the lung; pp. 252–262. [DOI] [PubMed] [Google Scholar]
- McNab FW, Ewbank J, Howes A, Moreira-Teixeira L, Martirosyan A, Ghilardi N, Saraiva M, et al. Type I IFN induces IL-10 production in an IL-27-independent manner and blocks responsiveness to IFN-γ for production of IL-12 and bacterial killing in Mycobacterium tuberculosis-infected macrophages. Journal of Immunology (Baltimore, Md. : 1950) 2014;193(7):3600–12. doi: 10.4049/jimmunol.1401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi T, Gil MP, Wang X, Louten J, Chu W-M, Biron C. a. High basal STAT4 balanced by STAT1 induction to control type 1 interferon effects in natural killer cells. The Journal of Experimental Medicine. 2007;204(10):2383–2396. doi: 10.1084/jem.20070401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli L. a, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CGK, Doering T. a, Angelosanto JM, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nature Immunology. 2011;12(11):1045–54. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naujoks J, Tabeling C, Dill BD, Hoffmann C, Brown AS, Kunze M, Kempa S, et al. IFNs Modify the Proteome of Legionella-Containing Vacuoles and Restrict Infection Via IRG1-Derived Itaconic Acid. PLoS Pathogens. 2016;12(2):e1005408. doi: 10.1371/journal.ppat.1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CT, Sullivan BM, Teijaro JR, Lee AM, Welch M, Rice S, Sheehan KCF, et al. Cell Host & Microbe. 5. Vol. 17. Elsevier Inc.; 2015. Blockade of interferon Beta, but not interferon alpha, signaling controls persistent viral infection. pp. 653–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Saha SK, Vaidya S. a, Bruhn KW, Miranda G. a, Zarnegar B, Perry AK, et al. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. The Journal of Experimental Medicine. 2004;200(4):437–45. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Vaidya S. a., Perry AK, Saha SK, Dempsey PW, Cheng G. Immune activation of type I IFNs by Listeria monocytogenes occurs independently of TLR4, TLR2, and receptor interacting protein 2 but involves TNFR-associated NF kappa B kinase-binding kinase 1. Journal of Immunology (Baltimore, Md. : 1950) 2005;174(3):1602–7. doi: 10.4049/jimmunol.174.3.1602. [DOI] [PubMed] [Google Scholar]
- Pandey AK, Yang Y, Jiang Z, Fortune SM, Coulombe F, Behr MA, Fitzgerald KA, et al. NOD2, RIP2 and IRF5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PLoS Pathogens. 2009;5(7):e1000500. doi: 10.1371/journal.ppat.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D, Martin FJ, Soong G, Harfenist BS, Aguilar JL, Ratner AJ, Fitzgerald KA, et al. Streptococcus pneumoniae DNA initiates type I interferon signaling in the respiratory tract. mBio. 2011;2(3):e00016–11. doi: 10.1128/mBio.00016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D, Planet PJ, Soong G, Narechania A, Prince A. Induction of Type I Interferon Signaling Determines the Relative Pathogenicity of Staphylococcus aureus Strains. PLoS Pathogens. 2014;10(2) doi: 10.1371/journal.ppat.1003951. available at: http://doi.org/10.1371/journal.ppat.1003951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver S, et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nature Immunology. 2012;13(12) doi: 10.1038/ni.2460. available at: http://doi.org/10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DJ, Rajaiah R, Tennant SM, Ramachandran G, Higginson EE, Dyson TN, Vogel SN. Salmonella Typhimurium Co-Opts the Host Type I IFN System To Restrict Macrophage Innate Immune Transcriptional Responses Selectively. The Journal of Immunology. 2015 doi: 10.4049/jimmunol.1500105. available at: http://doi.org/10.4049/jimmunol.1500105. [DOI] [PMC free article] [PubMed]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nature Reviews. Immunology. 2005;5(5):375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Plumlee CR, Lee C, Beg A. a., Decker T, Shuman H. a., Schindler C. Interferons direct an effective innate response to Legionella pneumophila infection. Journal of Biological Chemistry. 2009;284(44):30058–30066. doi: 10.1074/jbc.M109.018283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash A, Smith E, Lee CK, Levy DE. Tissue-specific positive feedback requirements for production of type I interferon following virus infection. Journal of Biological Chemistry. 2005;280(19):18651–18657. doi: 10.1074/jbc.M501289200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam V. a K., Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, et al. Nature Immunology. 5. Vol. 11. Nature Publishing Group; 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. pp. 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi M, Humann J, Penheiter K, Andreasen K, Lenz LL. Induction of IFN-alphabeta enables Listeria monocytogenes to suppress macrophage activation by IFN-gamma. The Journal of Experimental Medicine. 2010;207(2):327–37. doi: 10.1084/jem.20091746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboldi A, Dang EV, McDonald JG, Liang G, Russell DW, Cyster JG. Inflammation. 25-Hydroxycholesterol suppresses interleukin-1-driven inflammation downstream of type I interferon. Science (New York, N.Y.) 2014;345(6197):679–84. doi: 10.1126/science.1254790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson KM, Lee B, Scheller EV, Mandalapu S, Enelow RI, Kolls JK, Alcorn JF. The role of IL-27 in susceptibility to post-influenza Staphylococcus aureus pneumonia. Respiratory Research. 2015;16(1):10. doi: 10.1186/s12931-015-0168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson N, McComb S, Mulligan R, Dudani R, Krishnan L, Sad S. Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nature Immunology. 2012;13(10):954–62. doi: 10.1038/ni.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez DJ, Miranda D, Arumugaswami V, Hwang S, Singer AE, Senaati A, Shahangian A, et al. A repetitive region of gammaherpesvirus genomic DNA is a ligand for induction of type I interferon. Journal of Virology. 2008;82(5):2208–17. doi: 10.1128/JVI.01718-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliehe C, Flynn EK, Vilagos B, Richson U, Swaminathan S, Bosnjak B, Bauer L, et al. The methyltransferase Setdb2 mediates virus-induced susceptibility to bacterial superinfection. Nature Immunology. 2015;16(1):67–74. doi: 10.1038/ni.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmolke M, Patel JR, de Castro E, Sánchez MTA, Uccellini MB, Miller JC, Manicassamy B, et al. RIG-I detects mRNA of intracellular Salmonella enterica serovar typhimurium during bacterial infection. mBio. 2014;5(2):1–9. doi: 10.1128/mBio.01006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewnath ME, Olszyna DP, Birjmohun R, ten Kate FJ, Gouma DJ, van Der Poll T. IL-10-deficient mice demonstrate multiple organ failure and increased mortality during Escherichia coli peritonitis despite an accelerated bacterial clearance. Journal of Immunology (Baltimore, Md. : 1950) 2001;166(10):6323–31. doi: 10.4049/jimmunol.166.10.6323. [DOI] [PubMed] [Google Scholar]
- Shahangian A, Chow EK, Tian X, Kang JR, Ghaffari A, Liu SY, Belperio JA, et al. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. The Journal of Clinical Investigation. 2009;119(7):1910–20. doi: 10.1172/JCI35412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger S, Kastner R, Kernbauer E, Pilz A, Westermayer S, Reutterer B, Soulat D, et al. Characterization of the interferon-producing cell in mice infected with Listeria monocytogenes. PLoS Pathogens. 2009;5(3):e1000355. doi: 10.1371/journal.ppat.1000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storek KM, Gertsvolf NA, Ohlson MB, Monack DM. cGAS and Ifi204 Cooperate To Produce Type I IFNs in Response to Francisella Infection. The Journal of Immunology. 2015;194(7):3236–3245. doi: 10.4049/jimmunol.1402764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueltenfuss EA, Pollard M. Cytochemical Assay of Interferon Produced by Duck Hepatitis Virus. Science (New York, N.Y.) 1963;139(3555):595–6. doi: 10.1126/science.139.3555.595. [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science (New York, N.Y.) 2013;339(6121):786–91. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles RMB, Graeber TG, Krutzik SR, Montoya D, Schenk M, Lee DJ, Komisopoulou E, et al. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science (New York, N.Y.) 2013;339(6126):1448–53. doi: 10.1126/science.1233665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles RMB, Kelly-Scumpia KM, Sarno EN, Rea TH, Ochoa MT, Cheng G, Modlin RL. The Journal of Investigative Dermatology. 10. Vol. 135. Nature Publishing Group; 2015. IL-27 Suppresses Antimicrobial Activity in Human Leprosy. pp. 2410–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, et al. Nature Immunology. 11. Vol. 11. Nature Publishing Group; 2010. IFI16 is an innate immune sensor for intracellular DNA. pp. 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassermann R, Gulen MF, Sala C, Perin SG, Lou Y, Rybniker J, Schmid-Burgk JL, et al. Cell Host & Microbe. 6. Vol. 17. Elsevier Inc.; 2015. Mycobacterium tuberculosis Differentially Activates cGAS- and Inflammasome-Dependent Intracellular Immune Responses through ESX-1; pp. 799–810. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Asano N, Fichtner-Feigl S, Gorelick PL, Tsuji Y, Matsumoto Y, Chiba T, et al. NOD1 contributes to mouse host defense against Helicobacter pylori via induction of type I IFN and activation of the ISGF3 signaling pathway. Journal of Clinical Investigation. 2010;120(5):1645–1662. doi: 10.1172/JCI39481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RO, Bell SL, MacDuff DA, Kimmey JM, Diner EJ, Olivas J, Vance RE, et al. Cell Host & Microbe. 6. Vol. 17. Elsevier Inc.; 2015. The Cytosolic Sensor cGAS Detects Mycobacterium tuberculosis DNA to Induce Type I Interferons and Activate Autophagy; pp. 811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerd N.A. De, Vivian JP, Nguyen TK, Mangan NE, Gould JA, Braniff S, Zaker-tabrizi L, et al. Nature Immunology. Nature Publishing Group; Jul, 2013. Structural basis of a unique interferon- b signaling axis mediated via the receptor IFNAR1; pp. 1–9. [DOI] [PubMed] [Google Scholar]
- Wilson EB, Yamada DH, Elsaesser H, Herskovitz J, Deng J, Cheng G, Aronow BJ, et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science (New York, N.Y.) 2013;340(6129):202–7. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Chen ZJ. Nature Immunology. 3. Vol. 13. Nature Publishing Group; 2012. Intrinsic antiviral immunity; pp. 214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarilina A, Park-Min K-H, Antoniv T, Hu X, Ivashkiv LB. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nature Immunology. 2008;9(4):378–87. doi: 10.1038/ni1576. [DOI] [PubMed] [Google Scholar]
- York AG, Williams KJ, Argus JP, Zhou QD, Brar G, Vergnes L, Gray EE, et al. Cell. 7. Vol. 163. Elsevier Inc.; 2015. Limiting Cholesterol Biosynthetic Flux Spontaneously Engages Type I IFN Signaling. pp. 1716–29. [DOI] [PMC free article] [PubMed] [Google Scholar]