In vivo cells are reported to self-assemble at the microscale into complex functional configurations with coded spatial and temporal organization embedded in three-dimensional (3-D) microenvironments consisting of extracellular matrix (ECM). Tissue functionality originates, and it is regulated, based on the arrangement of these components and their relative positions.[1] Conventional two-dimensional (2-D) cultures fail to recreate complex heterogeneous architecture, cell-cell, and cell-ECM interactions that exist in vivo.[2] In contrast, three-dimensional (3-D) cultures have been shown to recapitulate in vivo native microenvironment in terms of cellular communication and ECM development. However, precise 3-D coding of living materials is challenging due to their architectural complexity and spatiotemporal heterogeneity.
3-D self-assembly is a promising approach for manufacturing and coding functional bio-architectures from individual building blocks.[3, 4] Self-assembly based on different physical principles, including magnetism,[5] fluidic force,[6] surface energy,[7] or gravity,[8] has been developed for several applications.[9] However, these methods offer limited control over uniformity, throughput, and spatiotemporal 3-D organization, especially when applied to cells, spheroids or organoids.[10] A detailed description of the advantages and disadvantages of most common cell assembly techniques is presented in the Supporting Information (SI) Table 1. In general, geometrical and functional heterogeneity of cellular constructs and organoid cultures represent a challenge to the accurate modeling of physiological and pathological behavior.[11, 12] Therefore, there is an unmet need for an effective assembly method with deterministic control on the biomanufacturing and reconfigurability of functional living systems, which could also enable the study of transient biological mechanisms.
A 3-D assembly method exploiting magnetic forces could enable a versatile and scalable approach for the biofabrication and coding of complex living constructs and organoids. Magnetic micro/nano-particles have been previously used for 3-D cell magnetic assembly,[4, 13] but they present concerns due to their release and subsequent systemic cytotoxicity.[14] Therefore, alternative biocompatible non-invasive strategies need to be developed to meet the requirement for biomedical and clinical applications. Although the use of paramagnetic ions in self-assembly has been investigated,[5] such a method has not been broadly applied for guided assembly and coding of 3-D living materials.
Here, we report a versatile, efficient, and inexpensive strategy that uses the principles of magnetic levitation to rapidly assemble individual cells or cellular constructs into functional 3-D architectures with controlled geometry and cellular organization. The developed 3-D assembly strategy presents multiple advantages, such as: (i) broad applicability to different cell types, such as cancer cells, stem cells, and neuronal cells; (ii) removable cell magnetization by washing after assembly; (iii) no requirements for magnetic nanoparticles that need to be physically removed from the constructs; (iv) selective assembly of live cells; (v) substrate-/scaffold-/biomaterial-free approach, allowing direct 3-D cell-cell interactions; (vi) continuous in situ real-time imaging capabilities; (vii) ability to maintain viability and functional properties; (viii) spatiotemporal control of cell interaction (ix) 3-D coding capabilities; and (x) rapid and high throughput assembly potential.
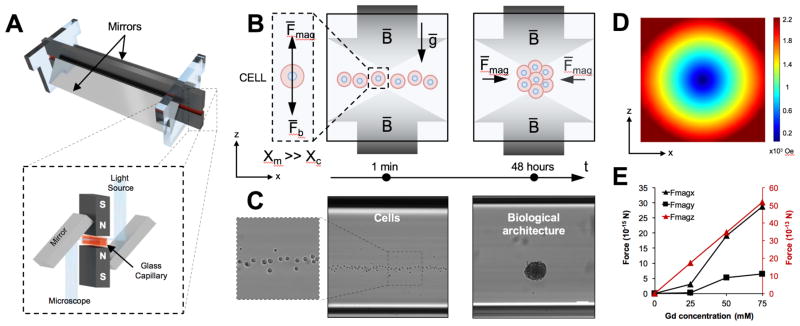
To precisely assemble 3-D cellular architectures, we developed a biocompatible magnetic levitation assembly method (Figure 1). The system is composed of two magnets with same poles facing each other with a glass microcapillary positioned between them (Figure 1A). Mirrors are added to the sides to allow imaging and real-time monitoring. In this system, a gadolinium-based, non-ionic paramagnetic agent was dissolved in the cellular medium and used in conjunction with the magnetic field to levitate cells. Gadolinium is an FDA approved contrast agent used for decades in magnetic resonance imaging (MRI).[15] The difference (Δx= Xc − Xm) between magnetic susceptibility of the medium (Xm) and cells (Xc) allows cellular units to levitate in an equilibrium plane where magnetic forces (Fmag) and buoyancy forces (Fb) equilibrate in opposite directions.[5, 16] The magnetic force directs the cellular units towards the minimum of the magnetic field, inducing them to interact (Figure 1B and 1C) and assemble into stable aggregates. 3-D cellular aggregates start to consolidate after 8 hours in levitation (Supporting Information, Figure 1), reaching a uniform size after 24–48 hours (Figure 2B and Supporting Information, Figure 1). As we previously reported,[16] only viable cells with an intact cellular membrane, exhibit a magnetic susceptibility difference with the surrounding media and can be levitated and assembled with magnetic levitation. On the other hand, dying cells and cellular debris sink to the bottom of the capillary and are excluded from the levitation induced assembly. This method can offer a unique advantage over existing technologies since dead cells and cellular debris, which can affect spheroid characteristics such as shape and size,[17] as well as cellular behavior, are removed.
Figure 1. Magnetically guided self-assembly and numerical simulation.
(A) Illustration of magnetic levitation system. N52-grade neodymium magnets are positioned at 1mm distance with same poles facing each other. Two mirrors are placed at 45° to allow microscope imaging. The components are held together with laser-cut plastic parts. (B) Schematic of the aggregation process in z-x (D) plane. The difference between magnetic susceptibility of the medium (Xm) and cells (Xc) allows cells to levitate in an equilibrium plane where magnetic force (Fmag) and buoyancy force (Fb) are equal and opposite in directions. During levitation, cells can be guided and assembled into stable 3-D biological architecture with controlled diameters and sizes. (C) Microscopy images of the cells levitation, and resulting biological architecture are presented. Scale bar, 100 μm. (D) 3-D simulation of the magnetic field norm for the cross section of the capillary (z-x plane). (E) Simulation of the magnetic force perceived by the cell in different spatial direction and different Gd3+ concentrations.
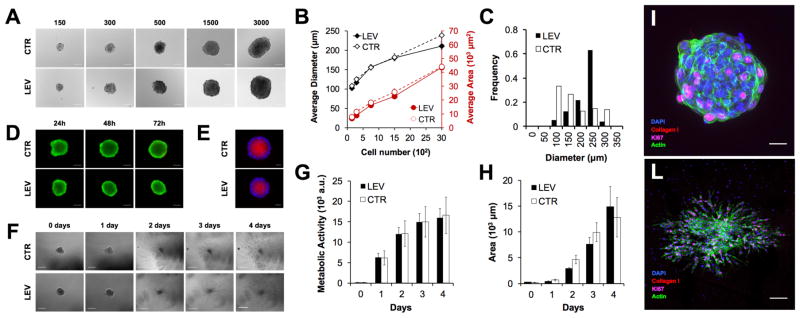
Figure 2. Assessment of size, viability, hypoxia, metabolism, and function of assembled cells by magnetic levitation.
(A) Micrographs showing cells assembled for 48 hours with magnetic levitation (LEV, 50 mM Gd3+) at different cell numbers (i.e., 150, 300, 750, 1500 and 3000 cells). Average diameters and area assembled by LEV at various cell numbers (B) Cells were also assembled using the hanging drop method, as a control (CTR). The data are presented as a mean ± standard error (SEM) (n≥6). (C) Histograms of average diameter for LEV and CTR are presented. (D) Fluorescent images showing live/dead assay (calcein/ethidium homodimer-1, green: live cell, red: dead cell) during LEV and CTR assembly (24, 48 and 72 hours). (E) Fluorescent images of hypoxia (Image-IT® Hypoxia, red) and nuclei (DAPI, blue) of LEV and CTR. Assembled cells (750 cells, 50 mM Gd3+, 48 hours) were cultured for 4 days in growth media and fibrin gel (F). Scale bar is 200 μm for days 0 to 2 and 500 μm for days 3 and 4 (F). Cell metabolism in growth media (G) and invasion area in fibrin ECM (H) were measured over time (i.e., day 0 to 4). The data are presented as a mean ± standard deviation (SD) (n≥4). Cell Immunostaining after levitation assembly (I) and growth in gel (4 days) (L). Cell proliferation (Ki-67, magenta), collagen I (red) secretion, nuclei (DAPI, blue) and Actin filaments (green). Scale bar is 50 μm for (I) and 200 μm for (L).
To investigate the magnetic characteristic of the system, we developed a quantitative model of the device (Figure 1D and Supporting Information, Figure 2). The computational model showed that the lowest value point of magnetic field is at the centroid of the channel. In addition, the magnetic field is symmetric with the channel center at x–y, y–z, and x–z sections (Supporting Information, Figure 2 A–C). The model was also used to explore how the units react under the influence of the magnetic field (Supporting Information, Figure 2 D–I). In particular, we evaluated the magnetic forces applied to a cell in x, y, and z directions and the time required to reach levitation equilibrium at different Gd3+ concentrations (0, 25, 50 and 75 mM) (Figure 1E). The simulations show that the force in y-direction is weaker than the force in x and z directions due to the preferred magnetization orientation and the time to reach equilibrium is directly proportional to the magnetic force and the Gd3+ concentration.
To investigate the ability of the system to assemble living cellular units we evaluated various paramagnetic conditions. First, we examined the viability of NIH 3T3 cells under different Gd3+ concentrations, ranging from 0 to 100mM over 72 hours (Supporting Information, Figure 3) using conventional tissue culture plates (2-D). Our results showed no effect of Gd3+ on cell viability up to 100mM over 3 days in culture (Supporting Information, Figure 3). However, resazurin-based metabolism assay results revealed that Gd3+ concentrations ≥75mM reduced cell proliferation (Supporting Information, Figure 3). We observed that this effect is temporary and can be reversed, for concentrations ≤75mM, by removing Gd3+ from the media. (Supporting Information, Figure 3). Based on the reported results, we selected the Gd3+ concentration of 50 mM to achieve rapid 3-D assembly while preserving cell viability and proliferation.
Dimensions of the cellular architectures assembled by the magnetic levitation method can be easily tuned by changing the number of cells (i.e., 150, 300, 750, 1500, 3000 cells per device; Figure 2A). Biological architectures assembled with magnetic levitation showed no significant difference in terms of average diameter and area compared to the controls (Figure 2A and 2B). Moreover, an evaluation of the size distribution revealed that the magnetic levitation method produces more uniform populations compared to the control (Figure 2C). Nevertheless, imperfect cell aggregation, leading to the formation of separate cell aggregates inside a single levitation device, was observed, similarly to the control (Supporting Information, Figure 1B). Our results showed that the magnetic levitation assembly platform preserves viability and functional activity of the NIH 3T3 cells assembled in 3-D architectures, while the effect of levitation and Gd3+ on cell viability during the assembly process was previously evaluated with a live/dead assay showing no significant difference compared to controls (Figure 2D and Supporting Information, Figure 4).
We demonstrated the presence of hypoxic regions both for magnetic levitation assembly and control (Figure 2E). The presence of gradients of oxygen and nutrient is due to the diffusion barriers imposed by the 3D cultures and it is critical to mimic the in vivo microenvironment where gradients orchestrate cellular and tissue behavior. We also evaluated the metabolic activity and growth of recovered cell architectures (Figure 2F–H). Our results showed that there is no significant difference in the metabolic activity of biological aggregates generated using magnetic levitation with respect to the control (Figure 2G). Moreover, to evaluate the invasion characteristics of the assembled architectures, they were embedded in fibrin gel and monitored as they invaded the surrounding ECM (Figure 2F). NIH 3T3 cells freely grew in the fibrin gel and their area of invasion increased similarly to the control (Figure 2H). Cell proliferation (i.e., Ki-67) and ECM deposition (i.e., collagen type I) markers immunostaining confirmed that the presented method allows the assembly of vital biological architectures, which can express normal behavior and phenotypic characteristics (Figure 2I–L).
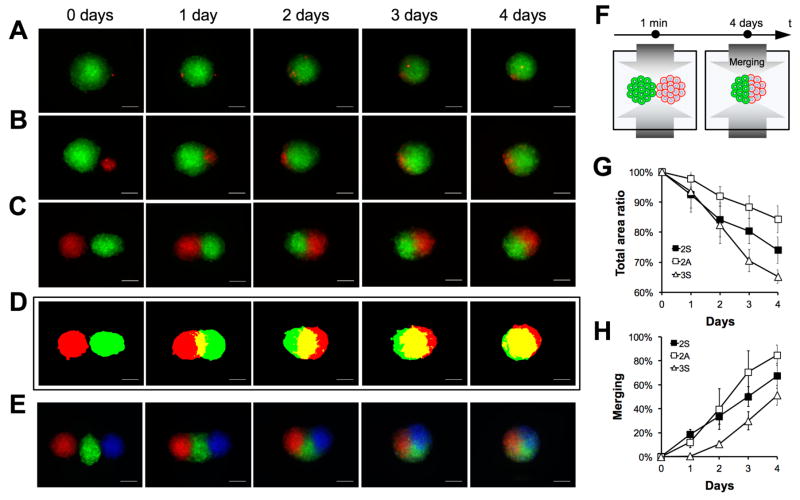
Cell-to-cell and tissue-to-tissue interactions are currently studied on conventional 2-D surfaces of petri dishes or flasks. However, the true nature of these interactions relies more on mere cellular contact rather than the ability to expand or grow over a surface, whose chemico-physical properties can significantly affect cell behavior. Using the magnetic levitation platform, we demonstrated the control over the interaction of biological units at various complexities, such as single-cell-to-spheroid (Figure 3A) and homo-/hetero-cellular spheroid-to-spheroid of varying sizes or numbers (Figure 3A–E), without the use of a 2-D substrate. We observed that smaller homo-cellular clusters tend to merge with larger units asymmetrically (Figure 3B), while spheroids that are equal in size fuse with a symmetric pattern (Figure 3C). This phenomenon was also observed when spheroids’ triplets interacted with each other (Figure 3E). We also built the representative 2-D topographic maps for complex interactive patterns (Figure 3C) and quantified the merging processes in real-time (Figure 3E–F). These results demonstrated the unique ability of magnetic levitation platform to monitor and guide the re-configurability of living materials in a predetermined and ordered manner, down to a single-cell level. This capability can be used to study and modulate the interaction and migration of cells and organoids, eliminating the effects associated with the surface, expanding the development of novel physiological and pathological in vitro models.
Figure 3. Biological units interaction and merging.
Single cell (A) and spheroid with different (B) or similar (C) sizes were magnetically guided to interact in pairs. The interaction was monitored in situ for 4 days using bright field and fluorescent microscopy. Representative topographical maps of similar size spheroids doublets are presented for each time point of the merging (D). Images of spheroid triplets merging over time are also presented (E). Scale bar, 200μm. Illustration of the merging process (F). Total fluorescence area ratio (G) and merging percentage (H) were measured and plotted as a function of time for symmetric (2S) and asymmetric duplets (2A), and symmetric triplets (3S). The data are presented as a mean ± SD (n=8 for 2S, n=3 for 2A and n=5 for 3S)
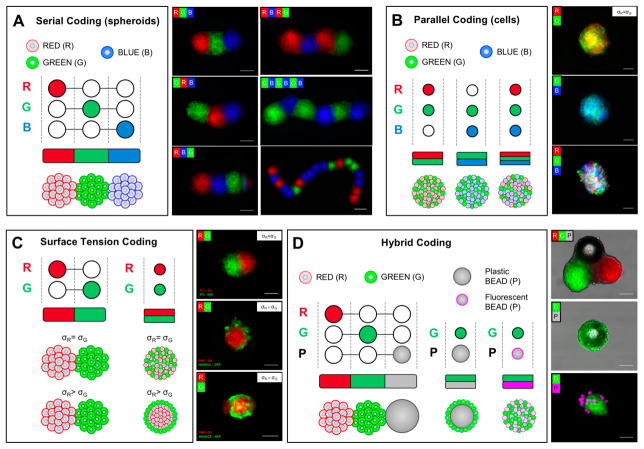
The developed platform was extended to code living architectures with 3-D organization and function arising from imposed location, specific biological properties, and functional compositions. To code different biological compositions, biological units were inserted into the device and allowed to self-assemble. In Figure 4, we present the schemes for controlling the steps of the units’ injection for living tissue fabrication and fluorescent micrographs, both for serial and parallel coding (Figure 4A–B). During serial coding, the controlled injection of different-colored biological units produced tissue strings that were color-coded along their length. With the current platform, we were able to fabricate tissue strings with more than 15 repeating units, but the number of units per string can be increased. Figure 4B shows different-colored cell populations coded in parallel inside a single biological unit. Although a unit with three colors is presented, the number, proportions and size of the parallel coded units are controllable and tunable.
Figure 4. Coding of spatially controlled cellular architectures with heterogeneous compositions.
Different biological units were cultured separately and then inserted into the levitation device with or without beads, as shown in the provided schematic for each coding type. The biological units were then magnetically guided to form different compositions that consolidated during culture. Schematic for serial (A) and parallel (B) coding with different cellular units (spheroids and cells, R, red; G, green; and B, blue). Schematic for surface tension (C) and hybrid (D) coding with different cellular units and plastic beads (P, gray or magenta). Fluorescence images are presented. Scale bars, 100μm.
The effect of cell composition and surface tension on 3-D arrangement was evaluated for recreating biofidelic architectures. It has been previously reported that surface tension (σ) is a known mechanism that directs cell and tissue organization.[18] Figure 4B shows that cells with the same surface tension (σ cell 1 = σ cell 2) tend to randomly distribute inside the 3-D biological architecture (Figure 4B). However, cells with different surface tensions (σcell 1 ≠ σcell 2) reorganized in layered structures. Further, cells with lower σ, such as endothelial cells, tend to envelop cells that exhibit higher surface tension, such as smooth muscle cells (SMCs). Our results are in agreement with the literature.[19] We have also observed that homogeneous biological units composed of same cell type fuse with a symmetric pattern (Figure 4C). However, when two different biological units consists of cells with different surface tensions (SMCs versus human umbilical vein endothelial cells (HUVECs)), the less cohesive unit dissociates and envelops the more cohesive one (Figure 4C). One of the advantages of the developed platform is that it can generate living materials with controllable composition and topography and can predict their behavior as they merge with a simple, measurable, physical parameter of surface tension. This aspect enables spatiotemporal control of cell co-culturing to form biofidelic architectures with 3-D organization arising from their specific surface tension properties.
To generate heterogeneous composite structures of living and non-living materials, plastic spheres were injected in series or in parallel to assemble multiple hybrid compositions (Figure 4D). We present the fluorescence micrographs of these hybrid units with variations in morphology, modulated by the size of the cellular units, beads, and in chemical composition. These results demonstrated control over the position of biological units to incorporate 3-D non-living objects into pre-organized 3-D structures. Hybrid-coding could be of potential benefit in broader applications to provide the spatiotemporal release of biochemical factors at the microscale to the assembled cells and dynamically change cellular microenvironments, which is challenging to achieve using existing technologies once the cellular constructs/organoids are assembled.
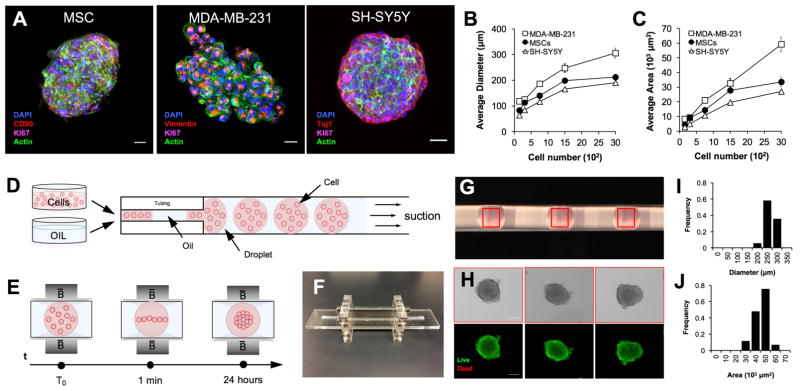
To show the broad biological applicability of the system, we assembled 3-D architectures of different cell types (i.e., human stem cells (MSC), brain cancer cells (SH-SY5Y), and breast cancer cells (MDA-MB-231)) (Figure 5A). The average diameter (Figure 5B) and area (Figure 5C) for different cell numbers are presented for each cell type. These results demonstrated that MSCs and SH-SY5Y generate uniform spheroids, while MDA-MB-231 aggregates appeared larger with uneven edges. Immunostaining assay of proliferation (i.e., Ki67) and characteristic markers (i.e., Cluster of Differentiation 90 (CD90), Vimentin, and Neuron-specific Class III β-tubulin (TuJ1)) confirmed that the cells assembled using magnetic levitation proliferate and present typical phenotypic markers (Figure 5A).
Figure 5. Scaling up the magnetically guided self-assembly of living architectures.
(A) To show broad applicability of the magnetic levitation platform to multiple cell types, human mesenchymal stem cells (MSCs), human breast cancer cells (i.e., MDA-MB-231), and human neuroblastic cancer cells (i.e., SH-SY5Y) were assembled (48 hours, 50mM Gd3+) and stained for Ki-67, CD90, Vimentin, Tuj1, nuclei and actin. Scale bar is 25μm. (B) Average diameter and (C) area at different cell numbers (i.e., 150, 300, 750, 1500, and 3000 cells) are shown for each cell type. The data are presented as a mean ± SEM (MSCs n≥6, MDA-MB-231 n≥6, SH-SY5Y n≥4). (D) Schematic representation of the droplet generation method. The alternate oil phase and aqueous phase containing cells were loaded through controlled suction in a tubing. (E) Schematic of magnetic levitation system for multiple biological unit assembly. During magnetic levitation, cells that are compartmentalized in the droplet levitate and self-assemble in a high-throughput fashion. (F) Photograph of the levitation device and (G) magnification of the droplet inside the tubing. (H) Bright field and fluorescent micrographs of cells assembled for 24 hours (3000 cells, 50mM Gd3+). Viability was assessed with live/dead assay (calcein/ethidium homodimer-1, green: live cell, red: dead cell). Scale bars, 100 μm. Histograms of average diameter (I) and area (J) are presented (n=12).
High-throughput of uniform cellular constructs and organoid cultures is critical for the biofabrication of reliable in vitro model for basic and preclinical research, [12, 20] and for bottom-up tissue engineering. Thus, to further demonstrate the feasibility of upscaling the method, we developed a droplet-based magnetic levitation assembly device to generate a larger number of uniform biological units (Figure 5). In this configuration, the cells that are compartmentalized in the water-in-oil droplets levitate and assemble into individual bio-architectures within 24 hours (Figure 5D–E). Given that reducing the volume of the media in the droplets reduces the time of aggregation, a minimum droplet volume of ~5μl was selected. Live/dead results showed that the media volume was sufficient to maintain a uniform viability of the biological units while the assembly was taking place (Figure 5H). This design choice improved the speed of aggregation and shape uniformity, reducing the event of imperfect cellular assembly, as showed in Figure 5I–J. Overall, these results confirmed that the magnetic levitation platform can achieve high throughput biomanufacturing of soft living systems.
Earlier reports have identified long term effects of magnetic fields and forces on cells using larger scale systems, such as super conducting high-tesla magnets up to 12T.[21] Here, we use approx. 0.4T magnets, significantly lowering the overall magnetic force for needed for cell levitation. This comes at the expense of using a paramagnetic medium based on gadolinium. We evaluated the long-term effects of various Gd3+ concentrations to ensure the toxicity levels would be safe at up to 75mM over a 3-day period.
Here, for the first time, we have presented the ability to control the spatiotemporal assembly of biological and non-biological units into heterogeneous materials using magnetic levitation, without employing nanoparticles or ferrofluids. This platform would allow for new capabilities for the engineering of living materials with complex 3-D organizations and functions, which are arising from the imposed location, predefined biological properties, and artificial functional components. Further, the method brings in a new direction to code soft living systems and bio-manufacture organoids and tumoroids in a high throughput droplet format. The developed system also provides an innovative approach to create, guide, and control the reconfigurability of living materials, which can broadly be used to study transient biological mechanisms (e.g., cell-to-cell, single-cell-to-organoid and organoid-organoid interactions, cell migration, cancer invasion, drug resistance and tissue formation). The surfaces-/scaffold-free nature of the presented approach has the potential to eliminate the effects associated with stiffness, roughness or chemical composition of these substrates that can uncontrollably affect cell behavior and phenotype. The biofidelic assembly process developed here can be applied in numerous fields, such as 3-D bio-printing and bottom-up tissue engineering, as well as drug discovery, developmental biology, neuroscience, and cancer research.
Experimental Section
Cell culture
NIH 3T3 murine fibroblasts and MDA-MB-231 were cultured in DMEM supplemented with 10% FBS and 1% Penicillin/Streptomycin. Bone Marrow derived stem cells were cultured in MEM alpha (Gibco) supplemented with 1% Penicillin/Streptomycin. SH-SY5Y human neuroblastoma cells were cultured in Advanced DMEM/F-12 (Gibco) supplemented with 1% Penicillin/Streptomycin. GFP-expressing Human Umbilical Vein Endothelial Cells (GFP-HUVECs) were cultured in EGM-2 (Lonza). AoSMC-Aortic Smooth Muscle Cells were cultured in SmGM-2 (Lonza). The cells were grown at 37 °C and 5% CO2 in a humidified atmosphere.
Levitation device fabrication
The levitation devices were fabricated following the previously reported procedure.[16] In brief, N52-grade neodymium magnets (NdFeB) (l × b × h: 50mm × 2mm × 5mm, K&J Magnetics) are positioned at 1mm distance with same poles facing each other. Two mirrors were placed at 45° to allow imaging using an inverted microscope. The components of the levitation system are held together with laser-cut plastic parts (VLS 2.30 Versa Laser).
Modelling and simulation
The simulation of the magnetic field was performed using COMSOL Multiphysics v.4.4 software. Theoretical modelling of the force distribution, levitation, and equilibrium of the cells were performed by balancing the magnetophoretic force with the buoyancy force, using the equations reported in our previous work.[16]
Magnetic levitation cell assembly method
Non-ionic paramagnetic agent (Gadavist, Gd3+) was dissolved in the cell suspension at the desired concentration. The paramagnetic cell suspension was injected into a squared-section glass capillary (1mm ID, Vitrocom), then carefully inserted between the magnets. The levitation device was enclosed in 60mm petri dish (Thermo Fisher Scientific) to maintain sterility and placed in a 100 × 100mm petri dish (Thermo Fisher Scientific) with ~15 ml of water to prevent excessive evaporation. Cells were assembled at 37 °C and 5% CO2 in a humidified atmosphere. Gd3+ concentration, media volume, cell number and levitation time, were varied according to different experiments.
Hanging drop method
Spheroids were fabricated with hanging drop method following reported protocol. In brief, the cell suspension (30 μl) was pipetted into each well of Perfecta3D® Hanging Drop Plate (3D Biomatrix). PBS(1×) (Life Technologies) was added to the reservoirs located on plate and tray perimeter to prevent evaporation during incubation culture. Spheroids were maintained in growth media and monitored.
Geometric measurements
Size and area were measured with ImageJ Fiji software. The diameter was calculated as the average of width and height of bounding rectangle.
Live Dead assay
3T3 NIH cells were exposed for different times (24, 48 and 72 hours) to different concentration of Gd3+ (0 and 50 mM) during magnetic levitation (LEV) and hanging drop (CTR) assembly. Cell viability was assessed with live/dead assay (calcein/ethidium homodimer-1).
Hypoxia assay
LEV and CTR spheroid were incubated in 10mM Image-IT® Hypoxia Reagent in DMEM for 24 hours. Hoechst 33342 (Thermo Fisher Scientific) was added to the media at the concentration of 1 μg/ml for 5–10 minutes incubation. Sample were washed with fresh DMEM before imaging with the microscope. Fluorescent images were processed using ImageJ Fiji and stack focuser tool.
Metabolism assay
NIH 3T3 cells were previously assembled using magnetic levitation (LEV) and hanging drop (CTR) method (48 hours, 50 or 0mM Gd3+). The fabricated biological architectures were then placed in non-adhesive 96-well plate (Ultra-Low Attachment surface microplates (ULA) Corning) and cultured for 48 hours in PrestoBlue solution (Thermo Fisher Scientific, 10% in DMEM). Changes in the fluorescence of the samples were measured as described in the supplementary information.
Invasion study size
NIH 3T3 spheroids assembled with magnetic levitation (LEV) and hanging drop method (CTR) (48 hours, 750 cells, 0 or 50 mM Gd3+). Similar size spheroids were selected and then placed in poly-HEMA coated 96-well plate, embedded in-situ within fibrin gel (~10 mg/ml) and cultured for 4 days. Spheroids invasion area in fibrin gel were manually measured with ImageJ Fiji software and plotted.
Cell interaction and merging assay
DIO, DII and DID-stained NIH 3T3 cells were previously fabricated in ULA microplates. Spheroids were then placed in duplets or triplets in one levitation device that was temporarily tilted to ensure initial spheroid-to-spheroid contact. Afterwards, spheroids interaction was monitored at regular intervals for 4 days with an inverted fluorescence microscope. For long term experiments, cellular media was changed by gently pipetting it out from one side of the capillary and injecting fresh one. Total area and merging percentage were quantified with ImageJ Fiji. In particular, the merging percentage was calculated as the ratio of pixels number of the fluorescence channels intersection and the total pixels number of the combined fluorescence. For quantification purposes, the fluorescence reflection on the glass capillary and the air bubble, if present, was manually removed.
Magnetic levitation coding
In the serial coding method, DIO, DII and DID-stained NIH 3T3 ULA spheroids (~1500 cells total) were inserted into the capillary, following the schematic in Figure 4A. The levitation device was temporarily tilted for approx. 5 min to ensure initial spheroid-to-spheroid contact. Cells were then normally assembled (24 hours, 60mM Gd3+). In the parallel coding approach, different combination of DIO, DII and DID-stained 3T3 cells were simultaneously injected in the capillary, following the schematic in Figure 4B, and assembled (~1500 cells total, 48–72 hours, 50mM Gd3+). In the surface tension-based coding method, DII-stained SMCs and GFP-HUVECs were inserted into the capillary both in serial and parallel fashion, following the schematic in Figure 4C, and assembled (~1500 cells total, 72 hours, 60mM Gd3+). In the parallel hybrid coding approach, plastic spheres were inserted into the capillary together with DIO-stained NIH 3T3 cells following the schematic in Figure 4D, and assembled for 24 hours (~900 cells total, 50mM Gd3+). Synthemax II microcarriers (Corning) and fluorescent violet polyethylene microspheres (1.06g/cc, 10–90μm, Cospheric), coated with gelatin, were used. In the serial hybrid coding DIO, DII-stained NIH 3T3 ULA spheroids (~1500 cells total) were inserted into the capillary together with Synthemax II microcarriers, following the schematic in Figure 4D, and assembled for 72 hours (50mM Gd3+).
Immunostaining
cells were fixed with 4% paraformaldehyde for 30 min, then blocked and permeabilized with 5% BSA and 0.3 % TritonX in PBS for 1 hour at 37°C. Samples were incubated with primary antibodies (collagen I (Abcam, 90395), ki67 (Abcam, 15580), CD90 (BD Pharmingen, 555593), Vimentin (Novus Biologicals, NB300-223), Tuj1 (R&S systems, MAB1195)) overnight at 4°C. After washing, secondary antibodies (anti-mouse for collagen I, CD90 Vimentin and Tuj1; anti-rabbit for ki67) mixed with phalloidin (Life Technologies, A12379) were applied for 2 hours at room temperature. DAPI solution was applied for 5 min. Washing steps were performed in PBS for 4 times for 5 minutes. Results were analyzed with the confocal microscope (Zeiss LSM710).
High-throughput droplet magnetic levitation method
High-throughput droplet magnetic levitation device was fabricated using a non-lithographic technique as reported previously. In brief, the levitation chip device was assembled using laser-cut poly(methyl methacrylate) (PMMA) (McMaster-Carr, Atlanta, GA) with a 3mm tubing housing and two magnets (l × b × h: 60mm × 4mm × 10mm, K&J Magnetics) with two magnets with the same pole facing each other. A vinyl tubing (2.8mm OD, 2.2mm ID and 70mm Length, Cole-Parmer) was connected to smaller silicone tubing (2.2mm OD, 0.7 mm ID and 0.8mm Length, Cole-Parmer) on one side and a 200 μL pipette (Eppendorf) on the other side. Multiple droplets (~10 per device) were generated by gentle alternate aspiration of mineral oil and NIH 3T3 cell suspension (~5 μL in volume, 3000 cells, 50mM Gd3+). The vinyl tube enclosing the droplet was then inserted in PMMA housing between the magnets and then placed in the incubator for assembly (37 °C and 5% CO2 in a humidified atmosphere). Cells were removed after 24 hours by gently flowing media inside the tubing.
Statistical information
Sample size, mean values, standard deviations, and standard errors were calculated and indicated on each figure.
Supplementary Material
Acknowledgments
The authors would like to thank all the members of BAMM Labs for their assistance. We would also like to thank Raphael Faustino Canadas for technical support and scientific discussion and Bera Hasan Coskun for contributing to the magnetic levitation device illustration. The research reported in this publication was partially supported by the National Institutes of Health under Award Number R15HL115556 and by the National Science Foundation under Award Number 1461602 and EAGER 1547791. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or National Science Foundation.
Footnotes
Author contributions
A.T, G.D., and U.D. developed the idea; A.T. and U.D designed the experimental approach; A.T., G.D., K.S., V.M. and B.C. performed the experiments; A.T., G.D., K.S., B.C., R.E. and U.D. analyzed the data; A.T., R.E. and U.D. wrote the manuscript.
References
- 1.Sasai Y. Nature. 2013;493:318. doi: 10.1038/nature11859. [DOI] [PubMed] [Google Scholar]; Griffith LG, Swartz MA. Nature reviews Molecular cell biology. 2006;7:211. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]; Davenport RJ. American Association for the Advancement of Science. 2005 [Google Scholar]
- 2.Asghar W, El Assal R, Shafiee H, Pitteri S, Paulmurugan R, Demirci U. Materials Today. 2015;18:539. doi: 10.1016/j.mattod.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Asghar W, Shafiee H, Chen P, Tasoglu S, Guven S, Gurkan UA, Demirci U. Cancer Targeted Drug Delivery. Springer; 2013. p. 635. [Google Scholar]
- 3.Whitesides GM, Grzybowski B. Science. 2002;295:2418. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]; Chen P, Luo Z, Güven S, Tasoglu S, Ganesan AV, Weng A, Demirci U. Advanced materials. 2014;26:5936. doi: 10.1002/adma.201402079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tasoglu S, Yu C, Gungordu H, Guven S, Vural T, Demirci U. Nature communications. 2014:5. doi: 10.1038/ncomms5702. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xu F, Wu CaM, Rengarajan V, Finley TD, Keles HO, Sung Y, Li B, Gurkan UA, Demirci U. Advanced Materials. 2011;23:4254. doi: 10.1002/adma.201101962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tasoglu S, Kavaz D, Gurkan UA, Guven S, Chen P, Zheng R, Demirci U. Advanced Materials. 2013;25:1137. doi: 10.1002/adma.201200285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Y, Duan X, Wei Q, Lieber CM. Science. 2001;291:630. doi: 10.1126/science.291.5504.630. [DOI] [PubMed] [Google Scholar]
- 7.Tanase M, Silevitch D, Hultgren A, Bauer L, Searson P, Meyer G, Reich D. Journal of Applied Physics. 2002;91:8549. [Google Scholar]
- 8.Hulteen JC, Van Duyne RP. Journal of Vacuum Science & Technology A: Vacuum Surfaces, and Films. 1995;13:1553. [Google Scholar]
- 9.Gracias DH, Tien J, Breen TL, Hsu C, Whitesides GM. Science. 2000;289:1170. doi: 10.1126/science.289.5482.1170. [DOI] [PubMed] [Google Scholar]; Jacobs HO, Tao AR, Schwartz A, Gracias DH, Whitesides GM. Science. 2002;296:323. doi: 10.1126/science.1069153. [DOI] [PubMed] [Google Scholar]
- 10.Lin RZ, Chang HY. Biotechnology journal. 2008;3:1172. doi: 10.1002/biot.200700228. [DOI] [PubMed] [Google Scholar]; Lu Y, Yin Y, Xia Y. Advanced materials. 2001;13:34. [Google Scholar]; Xia Y, Yin Y, Lu Y, McLellan J. Advanced Functional Materials. 2003;13:907. [Google Scholar]
- 11.Mesri EA, Cesarman E. Cell host & microbe. 2011;10:529. doi: 10.1016/j.chom.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fatehullah A, Tan SH, Barker N. Nature cell biology. 2016;18:246. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 13.Tasoglu S, Diller E, Guven S, Sitti M, Demirci U. Nature communications. 2014:5. doi: 10.1038/ncomms4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy LH, Arias JL, Nicolas J, Couvreur P. Chemical reviews. 2012;112:5818. doi: 10.1021/cr300068p. [DOI] [PubMed] [Google Scholar]
- 15.FDA Drug Safety Communication. FDA identifies no harmful effects to date with brain retention of gadolinium-based contrast agents for MRIs; review to continue. FDA; 2017. [Google Scholar]
- 16.Durmus NG, Tekin HC, Guven S, Sridhar K, Yildiz AA, Calibasi G, Ghiran I, Davis RW, Steinmetz LM, Demirci U. Proceedings of the National Academy of Sciences. 2015;112:E3661. doi: 10.1073/pnas.1509250112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wauthier E, Schmelzer E, Turner W, Zhang L, LeCluyse E, Ruiz J, Turner R, Furth ME, Kubota H, Lozoya O. Methods in cell biology. 2008;86:137. doi: 10.1016/S0091-679X(08)00008-3. [DOI] [PubMed] [Google Scholar]
- 18.Lecuit T, Lenne PF. Nature Reviews Molecular Cell Biology. 2007;8:633. doi: 10.1038/nrm2222. [DOI] [PubMed] [Google Scholar]
- 19.Foty RA, Steinberg MS. Developmental biology. 2005;278:255. doi: 10.1016/j.ydbio.2004.11.012. [DOI] [PubMed] [Google Scholar]; Foty RA, Pfleger CM, Forgacs G, Steinberg MS. Development. 1996;122:1611. doi: 10.1242/dev.122.5.1611. [DOI] [PubMed] [Google Scholar]; Jakab K, Norotte C, Marga F, Murphy K, Vunjak-Novakovic G, Forgacs G. Biofabrication. 2010;2:022001. doi: 10.1088/1758-5082/2/2/022001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Napolitano AP, Dean DM, Man AJ, Youssef J, Ho DN, Rago AP, Lech MP, Morgan JR. Biotechniques. 2007;43:494. doi: 10.2144/000112591. [DOI] [PubMed] [Google Scholar]
- 20.Weeber F, Ooft SN, Dijkstra KK, Voest EE. Cell Chemical Biology. 2017 doi: 10.1016/j.chembiol.2017.06.012. [DOI] [PubMed] [Google Scholar]; Astashkina A, Grainger DW. Advanced drug delivery reviews. 2014;69:1. doi: 10.1016/j.addr.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Zablotskii V, Polyakova T, Lunov O, Dejneka A. Scientific reports. 2016;6:37407. doi: 10.1038/srep37407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.