Abstract
Objective
We present a registration-based semi-automatic mandible segmentation (SAMS) pipeline designed to process a large number of computed tomography studies to segment three-dimensional (3D) mandibles.
Method
The pipeline consists of a manual preprocessing step, an automatic segmentation step, and a final manual post-processing step. The automatic portion uses a nonlinear diffeomorphic method to register each preprocessed input CT test scan on 54 reference templates aged birth to 19 years. This creates 54 segmentations, which are then combined into a single composite mandible.
Results
This pipeline was assessed using 20 mandibles from CT studies ages 1–19 years, segmented using both SAMS-processing and manual segmentation. Comparisons between the SAMS-processed and manually-segmented mandibles revealed 97% similarity agreement with comparable volumes. The resulting 3D mandibles were further enhanced with manual post-processing in specific regions.
Conclusions
Findings are indicative of a robust pipeline that reduces manual segmentation time by 75% and increases the feasibility of large-scale mandibular growth studies.
Keywords: Image Segmentation, Surface Reconstruction, Mandible, Automatic 3D Segmentation, Computed Tomography, Mandible Development
1. Introduction
With the advent of modern medical imaging technologies, automatic and semi-automatic methods have been developed to segment mandible volumes from medical imaging studies. Such computer-assisted segmentation methods help reduce the time needed to create precise and accurate 3D mandible models, and can be particularly beneficial for large-scale studies. These methods have been devised for application in dental surgical/treatment planning1,2, orthodontic diagnoses3, and dental forensics4 to reconstruct three dimensional (3D) mandible models from imaging studies such as panoramic x-rays, cone-beam computed tomography (CBCT), and multi-detector computed tomography (MDCT).
To study mandibular growth, a handful of studies have used 3D models by manually segmenting mandibles from MDCT.5–8 Most of these studies have used small age ranges and a restricted sample size. However, to enable a lifespan perspective on sex-specific mandibular growth including changes in surface area or regions of bone deposition or resorption9, a large number of 3D mandible models covering the entire age range is needed. Such a perspective can also help establish a developmental age and sex-specific normative reference, including a range of variability in typical growth, for use in diagnostics and/or treatment such as for application in orthodontic treatment planning10 or craniofacial surgical planning.11 To our knowledge, there has been only one mandibular development study utilizing reconstructed 3D mandibles.6
The heterogeneity of the mandibular morphology across individuals and age groups makes 3D mandible models difficult to extract from CT studies. The temporomandibular joint (TMJ) and dentition are often omitted from computer-assisted segmentation techniques due to difficulties in separating the condyle from the temporal bone and the dentition from its imaging artifacts. Since the growth and shifting of these structures affect mandibular growth and displacement, it is important to include them in mandibular growth studies and to establish a segmentation protocol that can segment the entire mandible.12,13
Given the morphological variations across individuals, manual segmentation of the entire mandible to generate 3D mandible models has been considered the ‘gold standard’ in both clinical and research settings.4 With this method, a human segmenter extracts the mandible from CT images through manual editing after 3D renderings or on a slice-by-slice basis. Manual segmentation requires 2–3 hours per mandible for a trained researcher to produce a complete 3D mandible model, and it is susceptible to inter- and intra-researcher variability. Thus, as noted earlier, manual segmentation is suboptimal for studying mandibular growth and development where large sample sizes are needed.
Both automatic and semi-automatic mandible segmentation techniques reduce potential human variability and error and speeds up the 3D segmentation process because human manual intervention or supervision is either not required (automatic segmentation) or limited (semi-automatic) to manual correction of segmentation errors.3,14–15 However, mandibular-focused automatic segmentation techniques, such as threshold-based, region growing, gray-level detection, active contour models, and statistical shape models (SSMs), often produce inaccurate segmentation of the teeth and the condyles, and some demonstrate only partial mandible delineation.2,15–17 Similarly, the available semi-automatic techniques are not particularly robust.3,14
Studies using automatic or semi-automatic mandible segmentation of images acquired using CBCT16–18 produce 3D models that are prone to low image quality and artifacts. In contrast, MDCT generates 3D models that show precise bony structures with high resolution and good signal-to-noise ratio, and this technology has been preferrentially used in developmental studies to assess morphological changes in the mandible.
This article presents a straightforward, easy-to-build semi-automatic mandible segmentation (SAMS) pipeline to generate accurate 3D models from MDCT. The aim of this pipeline is to generate a large number of 3D mandible models to study multidimensional mandibular development across the first two decades of life. We refer to this pipeline as semi-automatic because, although the majority of the processing is accomplished automatically, the initial (pre-processing) and final (post-processing) steps are manual. To demonstrate the utility of this pipeline, we present quantitative and qualitative assessments of the accuracy of the automatic portion of this pipeline in segmenting the mandible.
2. Methods
2.1 Image Acquisition / Dataset
An extant medical imaging database that covers the entire lifespan was used to establish the dataset that covers the first two decades of life to design and test this semi-automatic mandible segmentation method. The imaging database consisted of imaging studies collected retrospectively by our Vocal Tract Development Laboratory (VTLab), following University of Wisconsin–Madison Institutional Review Board (IRB) approval, with the aim of studying the growth of oral and pharyngeal structures in typically and atypically developing individuals. All imaging studies were stored in Digital Imaging and Communications in Medicine (DICOM) format, and all CT studies were acquired using General Electric helical CT scanners. Additional detail on the imaging database is provided in Vorperian et al.19 and Kelly et al.20 The dataset selected from the imaging database used in this study included 518 head and neck MDCT imaging studies from 266 typically developing patients, ranging in age from birth to 20 years, who were imaged for medical reasons not affecting the growth and development of the head and neck, and who were determined to have Class I bite (normognathic). All imaging studies in this dataset were visually inspected to ensure that the scan captured the whole mandible geometry while tolerating some dental artifacts. Additional inclusion criteria included: (1) slice thickness smaller or equal to 2.5 mm, (2) 14.0–22.0 cm FOV, and (3) 512 × 512 matrix size.
From this dataset, two groups of scans were formed. The first group (Group I) was used to create reference templates for the semi-automatic segmentation process. Group I consisted of 54 imaging studies (27 males / 27 females) that were carefully selected to represent an equal distribution of age and sex from age 13 months to 18 years, 8 months, and deemed to be an adequate number for reasonable pipeline processing speed without compromising segmentation accuracy. A second group of scans (Group II) was used to test the accuracy of the semi-automatic segmentation process. Group II consisted of 20 imaging studies (10 males / 10 females) that were randomly selected from the same dataset, after excluding cases selected for Group I, to equally represent both sexes in the age range of 0–20 years.
2.2 Manual Segmentation
Two-dimensional (2D) axial slices of all scans in Groups I and II were first reconstructed into 3D cubic volumes by a trained researcher using the Analyze 12.0 software package (AnalyzeDirect, Overland Park, KS).21 The reconstructed 3D cubic volume is referred to as the “reconstructed CT scan.” In this reconstruction, the anatomic geometry is preserved. The Volume Render and Volume Edit modules in Analyze 12.0 software were used to manually segment each 3D mandible model from the reconstructed CT scan. A global threshold that excludes all non-osseous tissues and provides the best visualization of mandible surface was determined and applied to each of the reconstructed CT scans as described in Whyms et al.22 and Kelly et al.20 The 3D mandible models were then extracted from the skeleton using Analyze 12.0. Cases with image artifacts were further edited slice-by-slice using multiplanar views (coronal, sagittal, and axial). These modules were also used to patch holes in the mandible models. Manually segmented mandibles and the reconstructed CT scans were then saved in Analyze75 [.hdr/.img] file format for the initial preprocessing step described in section 2.4.1. For additional details on segmentation modules and criteria, see Whyms et al.22 and Kelly et al.20
2.3 Reference Template Preparation
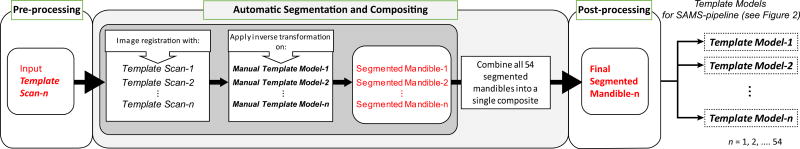
Each case in Group I – reference templates for this pipeline – consisted of a raw CT scan acquired as described in section 2.1 (template scan), and a manually segmented 3D mandible model as described in section 2.2 (template model). The 54 template scans and template models were preprocessed and cropped down to their minimum enclosing boxes using methods outlined in section 2.4.1. To reduce the influence of human variability in editing and artifacts during the manual segmentation of the template models, we ran each of the 54 template scans through the Semi-Automatic Mandible Segmentation (SAMS) pipeline described in section 2.4 (using the manually segmented template models from Group I). Thus, each initial, manually segmented template model was replaced with its post-processed, automatically segmented 3D mandible model. Once the post-processing steps were completed, generating a set of semi-automatically-segmented template models for each of the template scans, the reference templates were used for all subsequent SAMS processing. This template preparation process is represented in Figure 1.
Figure 1.
Flowchart describing template preparation to yield the 54 reference templates used in the Semi-Automatic Mandible Segmentation (SAMS) pipeline (see Figure 2). Each template is made up of a CT image (template scan) and a 3D mandible model (template model), both of which were processed through steps shown in this diagram/flowchart before being used as reference template for the pipeline. The template model was first segmented manually by a trained researcher, represented in the figure above as Manual Template Model. The template preparation process entailed the following three steps: Pre-processing, where each template scan and its Manual Template Model was cropped down into its minimal enclosing box. Automatic Segmentation and Compositing (gray-shaded box), where each pre-processed template scan was run through the SAMS pipeline with the pre-processed Manual Template Models as the deformable model to apply the deformation field of the registration between the input template scan and all of the template scans in the reference templates line-up. The resulting segmented mandibles were then combined into a single composite mandible. Post-processing: The post-processed segmented mandibles from this step were then used as the final template models in the SAMS pipeline.
2.4 Semi-Automatic Mandible Segmentation (SAMS)
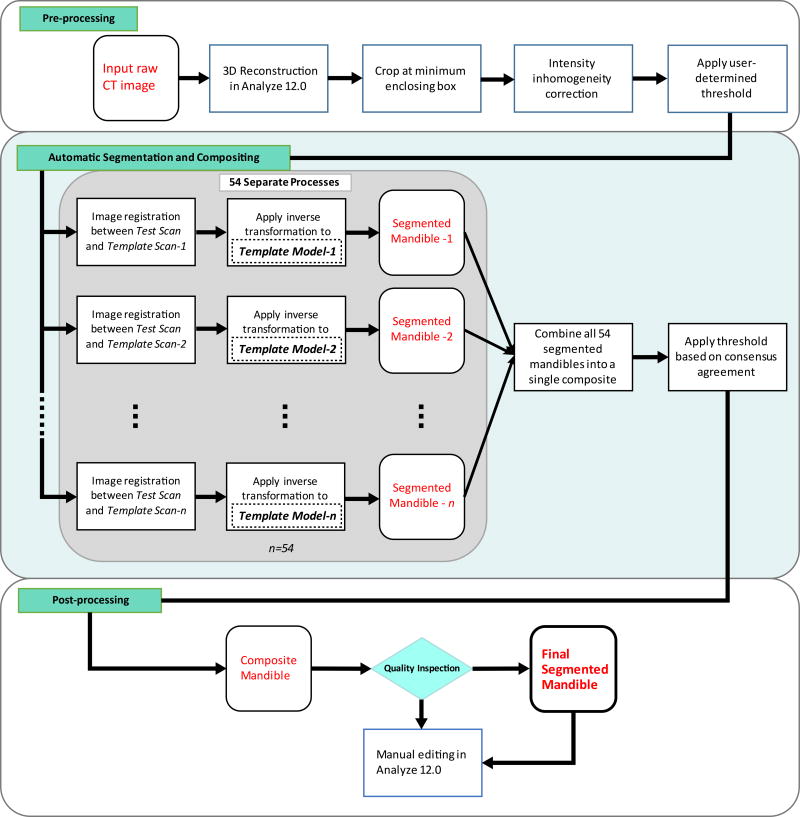
The semi-automatic mandible segmentation (SAMS) pipeline was designed to take in, as input, the preprocessed reconstructed CT scan, here referred to as “test scan,” and to output a 3D binary mask of the resulting segmented mandible. The pipeline, presented in Figure 2, first maps each test scan to each of the 54 reference templates (prepared as described in section 2.3), generating 54 independent registrations (these registrations can be done in parallel). Next, the output of the registrations are compiled into one single 3D mandible model, which is then manually post-processed by editing regions that need further refinement. The registration and processing tools are from the Advanced Normalization Tools (ANTs) package, fMRIB Software Library (FSL; the Oxford Centre for Functional MRI of the Brain)23 and ITK-SNAP’s Convert3D (C3D) module.24
Figure 2.
Flowchart of the SAMS pipeline. Pre-processing: An input raw CT image is reconstructed in 3D, cropped at its minimum enclosing box, and corrected for intensity inhomogeneity. A predetermined global threshold (Hounsfield Unit, HU) is applied. See text for details. Automatic Segmentation and Compositing: The preprocessed input CT image (test scan) is run through 54 separate ANTs-based nonlinear diffeomorphic registration processes. Each of the 54 processes results in its respective segmented mandible. The segmented mandibles are then composited into a single composite. Post-processing: The composite mandible is inspected to determine if manual editing in Analyze 12.0 is necessary. The gray-shaded box represents the automatic segmentation process with n=54.
2.4.1 Preprocessing
All reconstructed CT scans were examined by a rater using both Analyze 12.0 and FSL23 to collect: 1) A threshold in Hounsfield Units (HU) that removes non-osseous tissues (obtained using steps similar to threshold determination process described in section 2.2), and 2) the minimal enclosing box of the mandible, represented as minimum and maximum x-, y-, and z-coordinates that enclose the mandible. An automated preprocessing script was then used to take this information as input to apply a threshold to the reconstructed CT scan and crop the scan within the minimal enclosing box. Depending on the age and head orientation of the selected cases, the enclosing box dimensions ranged from a minimum of 80.63 × 43.60 × 61.65(mm) to a maximum of 133.60 × 90.35 × 107.93(mm), with cubic volumes of 216.72 cm3 to 1302.75 cm3. N4 bias correction was also applied to decrease inhomogeneity of the mandibular bone intensity.25 These scans were saved in zipped NIfTI format and ready for automatic segmentation. All manually segmented 3D mandible models created for Group I and Group II were preprocessed without the threshold application step.
2.4.2 Diffeomorphic Registration-based Automatic Segmentation
The diffeomorphic registration and segmentation portion of the pipeline is entirely automatic. Our scripts first register the test scan to each of the template scans of the 54 reference templates (prepared as described in 2.1), generating 54 separate, nonlinear diffeomorphic registration processes.26 Next, our scripts use the output from these registrations to transform the 3D mandible template model of the respective template scans into a mandible model that maps to the input test scan.
The initial step of using the test scan to affinely register to a template scan yields a deformation field output (deformation of warping the test scan to a template scan). Warping the test scan into the template scan space yields a transformation matrix (transformation of the test scan into the template scan). The inverse transformation of both of these outputs is applied to the mandible template model of the template scan, deforming the template model and propagating the deformed template model into the test scan’s native space. This deformed template model is similar to the mandibular structure in the test scan.
The ANTs parameters for the nonlinear diffeomorphic registrations were set using Mean-Squared Difference (MSQ) similarity metric with a window radius of 1, Gaussian regularization with sigma of 4, and a Greedy Symmetric Normalization (SyN) algorithm with a gradient step size value of 0.4. After applying the inverse transformation to each of the template models, the deformed template model was obtained. Next, ITK-SNAP’s convert3D (C3D) module24 was used to binarize these models, creating 54 independently segmented 3D binary mandibles for the input test scan.
Once all 54 registrations and segmentations were completed, the resulting 54 binary mandibles were merged into a single output in time using the fslmerge tool from the FSL toolkit23, to create a single composite mandible. The FSL toolkit then normalized the composite mandible based on its mean intensity and agreement, and removed composite voxels that were of less than 45% agreement, a percentage yielding the most accurate result during preliminary testing of the pipeline. This helped diminish the effect of aberrant variability in each individual model, giving to the final segmentation of a 3D mandible model of the test scan. A flowchart of our SAMS pipeline steps is shown in Figure 2.
2.4.3 Post-Pipeline Inspection and Touch up
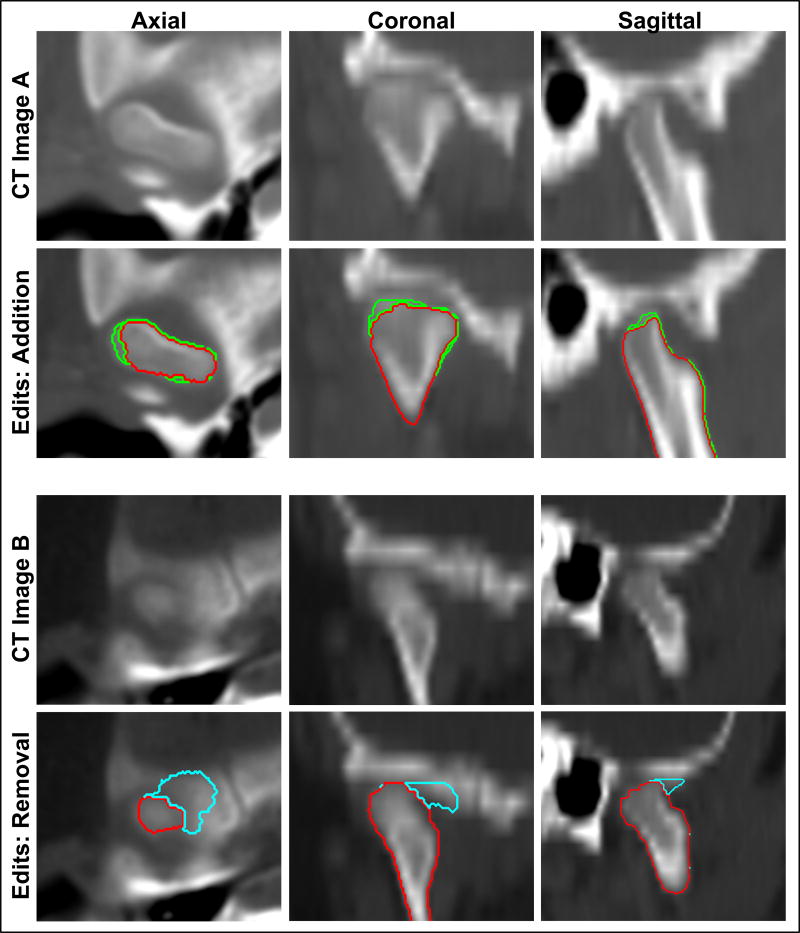
The mandibles processed with the automatic portion of our SAMS pipeline, henceforth referred to as SAMS-processed mandibles, were converted to triangle meshes using the marching cubes algorithm implemented in MATLAB (The MathWorks Inc., Natick, MA, USA)27 to allow for visualization of the 3D mandible.28 The 3D meshes were visually inspected to carefully evaluate for regions that needed manual editing. Mandibles with artifacts were converted into Analyze 12.0 software-compatible Object Map file format for researchers to manually edit anatomic inaccuracies such as regions that were over-segmented and needed removal, or regions that were under-segmented and needed “patch-up” or addition of voxels. An example of the post-processing editing is shown in Figure 3.
Figure 3.
Examples of post-processing addition and removal of voxels for under-segmented and over-segmented regions displayed in all three orientations. Top panel: CT image A displays the temporomandibular joint region of a male case (13 years 10 months old). Red represents the automatically segmented condyle region while green represents the voxel additions edited manually by a trained researcher. Lower panel: CT image B displays the temporomandibular joint region of another male case (2 years and 5 months old). Cyan represents the over-segmented region, originally segmented automatically, but edited for removal manually by a trained researcher.
2.5 Cluster Computing
This pipeline is designed to work with cluster computing resources due to the need to segment a large amount of input data, where a parallel computing environment allows more resources to run multiple template-based CT image registration jobs at the same time. It is, however, not limited to the environment described in this study if appropriate configurations were made to ensure sufficient computing power. Using a parallel strategy, the registration and compositing steps of the SAMS pipeline were entirely automatic, and completed using the high-throughput computing (HTC) resources –HTCondor – offered by the Center of High Throughput Computing (CHTC) at the University of Wisconsin-Madison. Each of the 54 nonlinear diffeomorphic registrations was submitted to HTCondor as an independent job and assigned to Linux machines with requirements set at a minimum of 1 CPU, 6GB disk space and 4GB memory. These jobs were coordinated using the DAGMan (Directed Acrylic Graph Manager) meta-scheduler for HTCondor to ensure that the compositing step would only be initiated when all 54 registration jobs were successful.29
2.6 Segmentation Accuracy Evaluation
To evaluate the accuracy of the automatic portion of our SAMS pipeline, defined as the process of section 2.4.2 that was executed by nonlinear diffeomorphic registrations and its subsequent compositing step, we carried out quantitative and qualitative assessments using the cases in Group II (described in section 2.1), where each of the 20 cases had both a SAMS-processed mandible and a manually segmented mandible. All statistical analyses below were carried out with MATLAB27 and R packages (R Foundation for Statistical Computing, Vienna, Austria).30
2.6.1 Quantitative Assessment
We compared the accuracy of SAMS-processed mandibles against the manually segmented mandibles by computing: 1) the overlap probability using a modified version of the Dice coefficient31 which is a similarity measure, 2) the intraclass correlation coefficients (ICC)32 of the segmented mandibles’ volumetric measurements as a measure of similarity, and 3) the Bland-Altman analysis to assess agreement of the volumetric measurements from the two methods.33
To estimate the overlap probability between mandibles segmented using automatic and manual segmentation techniques, we computed the Dice coefficient defined as:
where |A| and |B| are size of the individual objects and A B is the size of the intersection or the overlap of the object.31 The definition of the Dice coefficient has been extended for multiple objects as follows. Given n objects A1, A2 … An, the Dice coefficient is defined as:
The Dice coefficient gives number between 0 and 1 and provides the measure of overlap between the objects. If we further normalize the object size such that |A1| = |A2| = … |An| = 1, then the Dice coefficients can be written as:
The Dice coefficient can be viewed as the overlap probability of binary segmentations, and a measure of the overall accuracy of segmentation. Comparing only between the manually segmented and automatically segmented mandible, as |A1| and |A2| respectively, the closer the resulting values was to 1, the better agreement existed between the two mandibles.
In addition to the above described measure of overlap, the similarity between the two sets of 3D mandible volumes was assessed by computing the ICC using a two-way mixed-model with single measure of consistency.32 ICC also used a range of 0–1, where the closer the value to 1, the higher the similarity/reproducibility. For this study, we considered an ICC value greater than 0.75 as excellent, values between 0.5–0.75 as acceptable, and values less than 0.49 as not acceptable.
Since we did not set a priori ‘ground truth’ for this study but assumed the manual segmentation as a “reference standard,” the Bland-Altman method was also used to provide a downstream assessment of the difference and agreement of volumes produced using the two segmentation techniques.33 Bland-Altman is able to infer correlational information of the two sets of measurements without requiring a reference standard to be established a priori, which is ideal for our dataset, where manual segmentations have known limitations.34
2.6.2 Qualitative Assessment
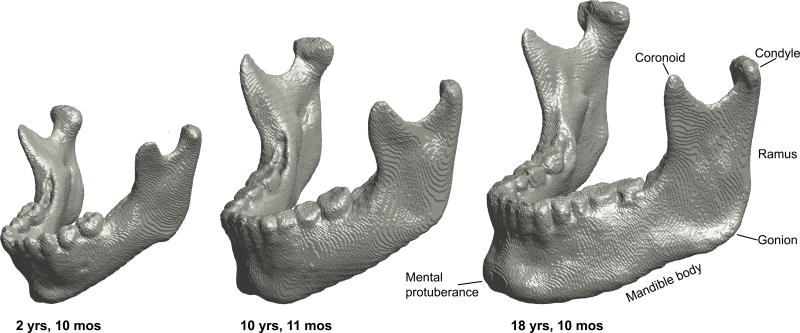
For the qualitative assessment, we developed a rating scale to rate the general or overall accuracy of the mandible models, as well as specific mandibular regions that can be problematic for modeling. Figure 4 shows the major mandibular regions using three SAMS-processed male mandible models ages 2 years 10 months, 10 years 11 months and 18 years 10 months, highlighting major developmental changes in size and shape. The rating scale ranged from 1 to 5, with higher numbers representing more accuracy with respect to the original scan. Appendix A shows the rating scale used to rate two general categories for overall mandible accuracy and usability for anatomical landmark-placement, and to determine the models’ general utility. Appendix A also shows the scale used to rate three region-specific categories, namely the teeth, the condyles, and the coronoid processes with detailed descriptions of the 1–5 ratings for each region.
Figure 4.
3D models of three male SAMS post-processed mandibles at age 2 years 10 months (left), 10 years 11 months (middle) and 18 years 10 months (right) showcasing the developmental changes in size and shape. The characteristic downward and forward growth of the mandible is well-depicted with the increased length of the ramus, length of the mandible body, and protrusion of the mental protuberance. Note also the changes in the location of the gonion relative to the condyle.
Two raters who were experienced in segmenting mandibles from CT scans independently rated a total of 40 models from the 20 subjects in Group II. Raters were blinded to whether the mandible models were automatically-segmented (SAMS-processed mandibles that were segmented through the automatic portion of the pipeline and have yet to be processed through the final post-processing step) or manually segmented. ICC, used in the previous section to compare volumetric measurements, is a typical measure to assess reliability of multiple raters examining the same set of data.35 It was therefore used again to conduct a reliability analysis between the two raters based on a two-way mixed model with average measure of consistency. ICC between the two raters was calculated for each of the general and specific regions of interest to determine if the ratings of the two segmentation methods were reliably similar among raters.
3. Results
3.1.1 Accuracy Evaluation: Quantitative results
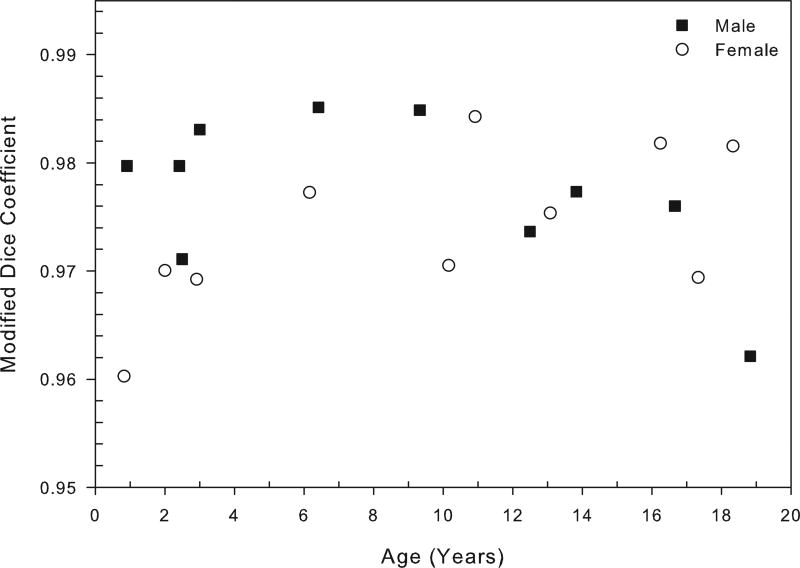
Using the 20 CT studies in Group II, the average similarity overlap between the manually segmented mandibles and automatically segmented mandibles, as measured by the modified Dice coefficient, was 0.976 (SD=0.106) indicating very high segmentation accuracy. Average similarity overlap for male cases was 0.977 (SD=0.103), and for female cases was 0.974 (SD=0.110). Figure 3 displays the modified Dice coefficient values as a function of age for each of the 20 mandibles. All male and female cases show a high Dice coefficient reflective of over 95% segmentation accuracy (Figure 5).
Figure 5.
Overlap probability between manually and automatically segmented mandibles for each of the twenty mandibles in Group II using the measure of modified Dice coefficient as a function of age. Mean value of similarity measure is 0.976 (SD=0.106), range=0.960 to 0.985. See text for details.
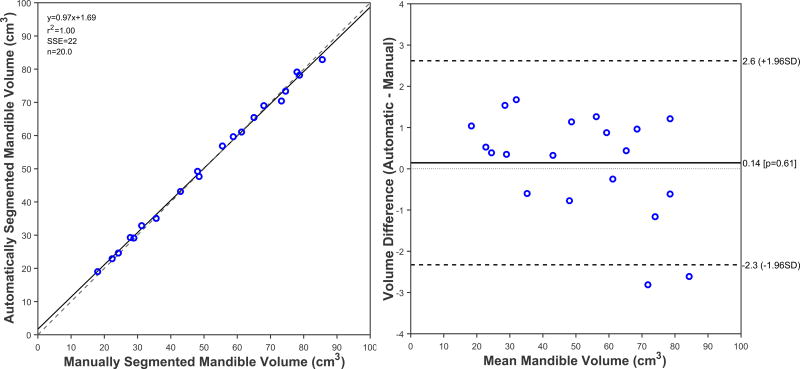
The ICC for volume between manual and automatic segmentation methods was 0.998 (P<0.001; 95% CI, 0.996 to 0.999), showing high level of agreement (99%) for volume measurements between mandibles segmented using two different methods. The regression line between the two methods also showed good agreement between volumes obtained from manual and automatic segmentation as seen in Figure 6a. The Bland-Altman plot showed a slight over-segmentation effect of 0.14 cm3 between volumes of automatically segmented mandibles and manually segmented mandibles, but otherwise showed a small range of differences with most data points placed within the limits of agreement (Figure 6b).
Figure 6.
Results of the regression (6a, left panel) and Bland-Altman analysis (6b, right panel). Figure 6a shows the linear regression plot between volumetric measurements of the 20 mandibles segmented automatically using SAMS pipeline, and the manually segmented mandibles. Solid line is the line of best fit, the dashed line is the identity line. Figure 6b displays the Bland-Altman scatter plot representing the difference between mandible volumes obtained from manual and automatic segmentation methods. Solid line is the mean difference (0.14cm3, P=0.61) the dashed lines are two standard deviations away from the mean difference (±1.96 SD).
3.1.2 Accuracy Evaluation: Qualitative results
Tables 1 and 2 provide a summary of the ratings and ICC for the general and region-specific ratings. Qualitative analysis findings show a generally favorable view of both manually and automatically segmented mandibles. For interrater reliability between the two raters, the ICC value was 0.73 (P< 0.001) which is considered to be acceptable/good reliability.
Table 1.
Ratings for automatic and manual segmentation: Mean of ratings (1–5; low-high) placed by two raters across 20 mandibles of each methods. See Appendix A for definition of ratings.
| Measure | Rater 1 Mean | Rater 2 Mean | Average Mean | |||
|---|---|---|---|---|---|---|
| Auto | Manual | Auto | Manual | Auto | Manual | |
| All | 3.69 (0.10) | 4.50 (0.06) | 3.73 (0.14) | 4.30 (0.08) | 3.71 (0.11) | 4.40 (0.06) |
| Teeth | 4.05 (0.20) | 4.30 (0.16) | 4.25 (0.29) | 4.45 (0.22) | 4.15 (0.20) | 4.38 (0.13) |
| Condyle | 3.20 (0.20) | 4.20 (0.12) | 2.85 (0.36) | 3.90 (0.18) | 3.03 (0.25) | 4.05 (0.12) |
| Coronoid | 4.20 (0.26) | 4.95 (0.05) | 3.90 (0.34) | 4.80 (0.12) | 4.05 (0.29) | 4.88 (0.06) |
| General | 3.40 (0.18) | 4.30 (0.13) | 3.85 (0.23) | 4.10 (0.18) | 3.63 (0.19) | 4.20 (0.12) |
| Usability | 3.60 (0.23) | 4.75 (0.10) | 3.80 (0.25) | 4.25 (0.16) | 3.70 (0.23) | 4.50 (0.12) |
All values are in form of Mean (Standard Error)
Table 2.
Intraclass Correlation Coefficient (ICC) of rating for automatic and manual segmentation
| Measure | Rater 1 | Rater 2 | Average | |||
|---|---|---|---|---|---|---|
| ICC | P | ICC | P | ICC | P | |
| All | 0.491 | <.001 | 0.638 | <.001 | 0.583 | <.001 |
| Teeth | 0.760 | .002 | 0.876 | <.001 | 0.814 | <.001 |
| Condyle | 0.038 | .466 | 0.524 | .057 | 0.469 | .088 |
| Coronoid | 0.168 | .346 | 0.048 | .458 | 0.128 | .384 |
| General | 0.652 | .013 | 0.864 | <.001 | 0.771 | .001 |
| Usability | 0.280 | .240 | 0.596 | .027 | 0.489 | .076 |
ICC = Intraclass Correlation
P = p-value of F-test
Mean ratings across all rating categories, mandibles, and between raters revealed an average score of 3.71 (SE=0.11) for the automatically segmented mandibles, and a score of 4.40 (SE=0.06) for the manually segmented models, where both values were rated on a scale of 1–5. Of the three mandible regions set for comparison, for manually segmented mandibles, the coronoid processes showed the highest ratings, with a mean score of 4.88 (SE=0.06), while for automatically segmented mandibles, the teeth region had the highest ratings, with a mean score of 4.15 (SE=0.20). The condyles had the lowest ratings for both segmentation methods, with a mean score of 3.03 (SE=1.25) for automatically segmented mandibles and a mean score of 4.05 (SE=0.66) for manually segmented mandibles.
Ratings on the general shape of the mandible had an ICC of 0.771 (P=0.001), indicative of high agreement between raters for the overall shape of the mandibles generated between the two segmentation methods. However, the low agreement for coronoid processes and condyles implies that these regions are different between the two segmentation methods. Similarly, ratings on the usability for landmark placement also showed that the automatically segmented mandibles had lower usability compare to manually segmented mandibles (mean score of 3.70, SE=0.23; vs. mean score of 4.50, SE=0.12).
3.2 Computational Cost
The average disk usage for each registration and compositing job ranged from 4–20 GB, while memory usage ranged from 1–10 GB. The average computational time for each registration job was around 3 hours. The time needed to segment a mandible for one input test scan (54 registration jobs plus one compositing job) ranged from 3–8 hours, depending on the pooling and coordination schedule as described in section 2.4, because registration jobs were submitted and run in parallel. The automatic portion of the pipeline was completed in less than 12 hours for all 20 of the preprocessed test scans. The total duration of segmenting whole mandibles from 20 subjects, including the manual pre-processing and post-processing steps, was between 16 and 18 hours.
4. Discussion
This study documents the steps used for an easy-to-build semi-automatic mandible segmentation pipeline and to document its performance in maintaining high precision of anatomical accuracy with minimal experimental bias. Quantitative assessments indicate that the SAMS pipeline presented here is able to consistently produce 3D mandible models of the same quality as mandible models generated manually by trained researchers, with less variability. The difference in overlap and volume measurements between manually and automatically segmented mandibles indicated high agreement between mandibles from these two segmentation methods. As for qualitative assessment, the two raters generally favored the manually segmented mandibles over the automatically segmented models. Ratings evaluating the overall shape and usability of the models also favored the manually segmented mandibles, but indicated fair ratings for automatically segmented mandibles.
Differences between the two segmentation methods can be clearly visualized in Figures 7 and 8, with superimposition of an automatically segmented mandible on the manually segmented mandible from the same case. The overall body is similar except in the condyles, coronoid processes, and teeth, further confirming our expectation that automatic segmentations are lacking in these three regions, most evidently in condyles due to bone density ambiguity in the TMJ. Manual editing of 3D models presented the full anatomy, but showed condyles with irregular surfaces and superior borders. Automatic segmentation methods could render condyles with even surfaces, but often overestimated or underestimated the surfaces, requiring manual editing to fix the absent or additional voxels, as seen in Figure 3. Similarly, automatic segmentation typically underestimated the coronoid processes, likely due to lower bone density, and needed post processing for enhancement. The manually segmented mandible captured a better-defined anatomy of the condyles and coronoid processes, but showed rather irregular teeth edges and borders (Figure 7). In the dental region, automatic segmentation seemed to under-segment but more closely outlined the teeth borders than the manually segmented teeth region (Figure 7). The lower ratings of the automatically segmented dental region were likely due to dental artifacts that had to be manually edited during post-processing.
Figure 7.
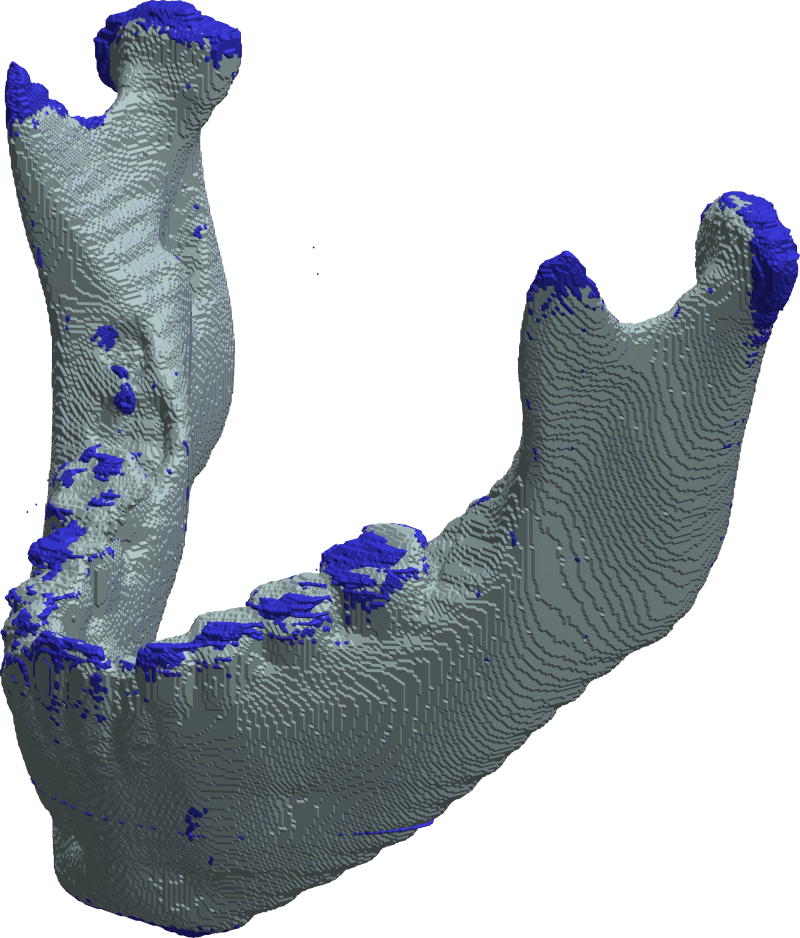
3D superimposition of surface models of an automatically generated mandible segmentation (gray) over a manually created mandible segmentation (blue) of a female case at the age of 6 years 2 months old. The automatic segmentation had no manual adjustment applied. This example shows that manual segmentation captures the anatomy in coronoid processes and condyles better, but produces an uneven surface in the teeth region. The modified Dice coefficient for this case was 0.977 (SD=0.104).
Figure 8.
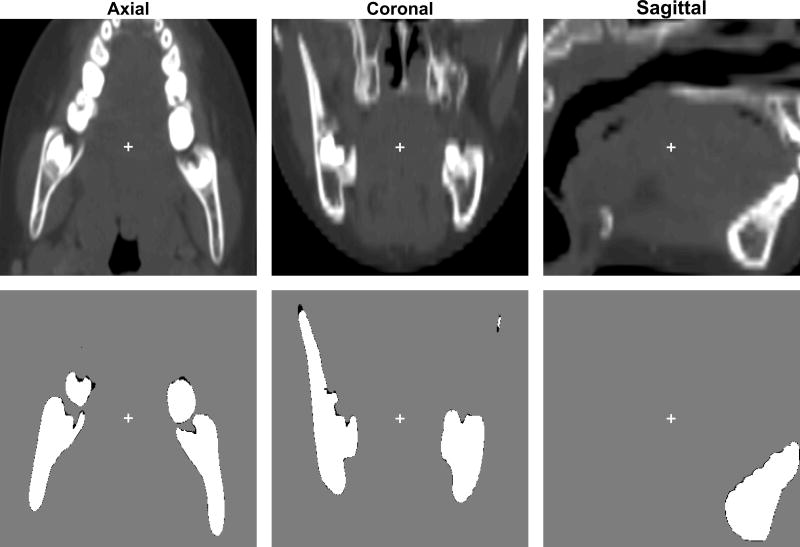
2D superimposition of the mandible described in Figure 2 lining up against its raw CT images in all three anatomical views. Second rows represented overlaps of automatically segmented mandible (white) and manually-segmented mandible (black) of a female case at the age of 6 years 2 months old. Modified Dice coefficient in this case is 0.977 (SD=0.104). The crosses in each image represent the center of the minimal enclosing box that contains the mandible.
The small difference in volume between the mandibles segmented using the two different methods suggested that the SAMS-processed mandibles, even before post processing, are comparable to manually segmented mandibles. Thus, the automatic portion of the SAMS pipeline presented in this study successfully produce mandible models that closely approximate manually-segmented mandible models, and only require minimal post-processing in limited regions. Such a pipeline provides more consistency in the segmentation of the mandibular structure from CT imaging studies, and reduces the effort needed to produce highly accurate mandible models.
The processing of all 20 cases in Group II through the SAMS pipeline, from preprocessing, automatic segmentation, and post-processing took only around 16–18 hours, in comparison to manual segmentation of all 20 cases, which could take over 80 hours for a trained researcher to complete. Since the registration jobs are parallelizable, increasing the number of imaging studies to be processed for each job submission does not necessarily increase the processing time, making such automatic registration appealing for studies that require large sample sizes. Also, there may be potential for further automation to replace the manual preprocessing steps, further increasing the efficiency of the method.
The high success rate of the SAMS pipeline in this study is in part due to the reference template parameters optimized for the age range of our data set, but this pipeline can be easily modified to better meet user needs and preferences. For example, the reference templates can be modified to become age-specific or population-based by switching out the 54 reference templates and replaced with cases adjusted to different age range or cases of a specific disordered group.
The ultimate purpose in developing the SAMS pipeline was to make it feasible to conduct a large-scale, across-the-lifespan analysis of human mandibular growth using a multidimensional perspective. Such knowledge will not only help advance the understanding on typical mandibular growth where developmental sexual dimorphism of the mandible emerges, but also make it feasible to establish a normative reference for clinical application. Examples of the latter include assessment of typical growth or deviation from typical growth36, planning for orthodontic treatment10 or surgical reconstruction37, as well as predictions necessary in the forensic sciences.38 Similar segmentation pipelines applied to other structures of interest could make large-scale studies more feasible by reducing the time and effort required to produce accurate 3D anatomical models.
Conclusion
The SAMS pipeline we describe in this article performed well, with a significant reduction in the number of man-hours spent segmenting cases, although some cases required manual inspection and touching up. The pipeline is easy to configure for different age and disease groups, as well as different anatomical structures. Users can adjust the parameters and reference templates as needed according to their study focus. The assessment in this study showed that the pipeline is efficient and able to produce accurate 3D mandible models, permitting large-scale segmentation of mandibles for development-focused surface morphological study in the future. Further investigation is warranted into the performance of the semi-automatic mandible segmentation pipeline to accurately segment mandibles from imaging studies of older adults or of atypically developing mandibles, such as from individuals with Down syndrome.
Acknowledgments
This work was supported by NIH research grant R01 DC006282 (MRI and CT Studies of the Developing Vocal Tract, Houri K. Vorperian, Principal Investigator) from the National Institute on Deafness and other Communication Disorders (NIDCD), and by a core grant P30 HD03352 and U54 HD090256 to the Waisman Center (Albee Messing, PI) for research support from the National Institute of Child Health and Human Development (NICHD).
The authors thank Katelyn Kassulke Tillman and Abigail Lamers for serving as raters; Simon Lank for assistance in the development and testing of the pipeline parameters; Ellie Fisher, Chantal Van Ginkel and Courtney Miller for assistance in the pre-processing steps and quality assurance checks at different stages of the pipeline. We also thank Lauren Michael of the Center of High-Throughput Computing (CHTC) at the University of Wisconsin-Madison for the support and guidance she provided for the cluster computing portion of this pipeline. Finally, we are grateful to Jacqueline Houtman for comments on an earlier version of this manuscript.
Appendix A
Mandible Segmentation Rating Scale: Qualitative Assessment
This form contains the rating descriptions for the different mandibular regions of interest.
General ratings
Overall
General rating based on entire model, taking into account ratings for teeth, condyle, and coronoid, as well as any discontinuity in the rest of the model.
Landmarking
General rating based on ability to accurately place landmarks, using model as the main reference.
Regions-specific ratings
Teeth
individual teeth are not discernable or missing, unable to place any landmarks
front teeth discernable, still unable to identify individual molars, able to place some landmarks
front teeth and posterior point of last erupted molar visible, superior and lateral borders of teeth still messy AND/OR small portions missing, able to place all landmarks (endomolare, sublingual fossa, posterior dental border)
all individual teeth discernable, superior border still messy, able to easily place all landmarks
all individual teeth discernable, clean superior borders, able to easily place all landmarks
Condyles
large portions of superior and/or lateral borders of condyle obviously missing, unable to place any landmarks
model contains full condyle but includes large amount of surrounding tissue that should easily have been removed, able to place some landmarks
superior and lateral borders rough, but generally visible AND/OR missing small portions of condyle, such that the borders do not match the true anatomy/DICOM, able to place all landmarks
clean lateral condyle borders, superior borders of condyle may be slightly rough, able to easily place all landmarks
superior and lateral borders of both condyle and coronoid clean and clearly visible, able to easily place all landmarks
Coronoid Processes
coronoid process missing, unable to place any landmarks
parts of coronoid process obviously cut-off, able to place some landmarks
model contains full coronoid but includes some surrounding tissue that should easily have been removed AND/OR missing small sections of the coronoid, such that the borders do not match the true anatomy/DICOM, able to place all landmarks
superior and lateral borders slightly rough, but generally visible, able to easily place all landmarks
full coronoid process clean and fully visible, able to easily place all landmarks
Footnotes
Disclosure:
All authors declare no conflict of interest.
Contributor Information
Ying Ji Chuang, Vocal Tract Development Lab, Waisman Center, University of Wisconsin-Madison.
Benjamin M. Doherty, Vocal Tract Development Lab, Waisman Center, University of Wisconsin-Madison.
Nagesh Adluru, Vocal Tract Development Lab, Waisman Center, University of Wisconsin-Madison.
Moo K. Chung, Vocal Tract Development Lab, Waisman Center, University of Wisconsin-Madison; Dept of Biostatistics and Medical Informatics, University of Wisconsin-Madison.
Houri K. Vorperian, Vocal Tract Development Lab, Waisman Center, University of Wisconsin-Madison.
References
- 1.Ezzat KA, Kandil AH, Fawzi SA. A novel computerized system to simulate orthodontic treatment plan. International Journal of Applied Engineering Research. 2016;11(8):5673–5681. [Google Scholar]
- 2.Moreno S, Caicedo S, Strulovic T, et al. Paper presented at: 2010 International Conference on Computer Vision and Graphics. Berlin, Heidelberg: 2010. Inferior maxillary bone tissue classification in 3D CT images. [Google Scholar]
- 3.Rueda S, Gil JA, Pichery R, et al. Automatic segmentation of jaw tissues in CT using active appearance models and semi-automatic landmarking. Med Image Comput Comput Assist Interv. 2006;9(Pt 1):167–174. doi: 10.1007/11866565_21. [DOI] [PubMed] [Google Scholar]
- 4.Abdi AH, Kasaei S, Mehdizadeh M. Automatic segmentation of mandible in panoramic x-ray. J Med Imaging (Bellingham) 2015;2(4):044003. doi: 10.1117/1.JMI.2.4.044003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andresen PR, Bookstein FL, Conradsen K, et al. Surface-bounded growth modeling applied to human mandibles. IEEE Trans Med Imaging. 2000;19(11):1053–1063. doi: 10.1109/42.896780. [DOI] [PubMed] [Google Scholar]
- 6.Coquerelle M, Bookstein FL, Braga J, et al. Sexual dimorphism of the human mandible and its association with dental development. Am J Phys Anthropol. 2011;145(2):192–202. doi: 10.1002/ajpa.21485. [DOI] [PubMed] [Google Scholar]
- 7.Hilger KB, Larsen R, Wrobel MC. Growth modeling of human mandibles using non-euclidean metrics. Medical image analysis. 2003;7(4):425–433. doi: 10.1016/s1361-8415(03)00034-3. [DOI] [PubMed] [Google Scholar]
- 8.Reynolds M, Reynolds M, Adeeb S, et al. 3-D volumetric evaluation of human mandibular growth. Open Biomed Eng J. 2011;5:83–89. doi: 10.2174/1874120701105010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung MK, Qiu A, Seo S, et al. Unified heat kernel regression for diffusion, kernel smoothing and wavelets on manifolds and its application to mandible growth modeling in CT images. Medical image analysis. 2015;22(1):63–76. doi: 10.1016/j.media.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacob HB, Buschang PH. Mandibular growth comparisons of class I and class II divisions 1 skeletofacial patterns. The Angle Orthodontist. 2014;84(5) doi: 10.2319/100113-719.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smartt JM, Jr, Low DW, Bartlett SP. The pediatric mandible: II. Management of traumatic injury or fracture. Plast Reconstr Surg. 2005;116(2):28e–41e. doi: 10.1097/01.prs.0000173445.10908.f8. [DOI] [PubMed] [Google Scholar]
- 12.Krarup S, Darvann TA, Larsen P, et al. Three-dimensional analysis of mandibular growth and tooth eruption. J Anat. 2005;207(5):669–682. doi: 10.1111/j.1469-7580.2005.00479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bjork A, Skieller V. Normal and abnormal growth of the mandible. A synthesis of longitudinal cephalometric implant studies over a period of 25 years. Eur J Orthod. 1983;5(1):1–46. doi: 10.1093/ejo/5.1.1. [DOI] [PubMed] [Google Scholar]
- 14.Gamboa A, Cosa A, Benet F, et al. 2012 9th IEEE International Symposium on Biomedical Imaging (ISBI) Barcelona: May, 2012. A semiautomatic segmentation method, solid tissue classification and 3d reconstruction of mandible from computed tomography imaging for biomechanical analysis. [Google Scholar]
- 15.Barandiaran I, Macia I, Berckmann E, et al. An automatic segmentation and reconstruction of mandibular structures from CT-data. In: Corchado E, Yin H, editors. Intelligent Data Engineering and Automated Learning - Ideal. Vol. 5788. Heidelberg: Springer; 2009. pp. 649–655. [Google Scholar]
- 16.Brandariz M, Barreira N, Penedo MG, et al. Automatic segmentation of the mandible in cone-beam computer tomography images; Paper presented at: 2014 IEEE 27th International Symposium on Computer-Based Medical System; 2014. [Google Scholar]
- 17.Gollmer S, Buzug M. Paper presented at: 9th IEEE International Symposium on Biomedical Imaging (ISBI) Barcelona: 2012. Fully automatic shape constrained mandible segmentation from cone-beam CT data. [Google Scholar]
- 18.Wang L, Chen KC, Gao Y, et al. Automated bone segmentation from dental CBCT images using patch-based sparse representation and convex optimization. Med Phys. 2014;41(4):043503. doi: 10.1118/1.4868455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vorperian HK, Wang S, Chung MK, et al. Anatomic development of the oral and pharyngeal portions of the vocal tract: An imaging study. J Acoust Soc Am. 2009;125(3):1666–1678. doi: 10.1121/1.3075589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly MP, Vorperian HK, Wang Y, et al. Characterizing mandibular growth using three-dimensional imaging techniques and anatomic landmarks. Arch Oral Biol. 2017;77:27–38. doi: 10.1016/j.archoralbio.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Analyze 12.0 [computer program] Overland Park, Kansas: AnalyzeDirect; [Google Scholar]
- 22.Whyms BJ, Vorperian HK, Gentry LR, et al. The effect of computed tomographic scanner parameters and 3-dimensional volume rendering techniques on the accuracy of linear, angular, and volumetric measurements of the mandible. Oral Surg Oral Med Oral Pathol Oral Radiol. 2013;115(5):682–691. doi: 10.1016/j.oooo.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenkinson M, Beckmann CF, Behrens TE, et al. Fsl. Neuroimage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 24.Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Tustison NJ, Avants BB, Cook PA, et al. N4itk: Improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Avants BB, Tustison NJ, Song G, et al. A reproducible evaluation of ants similarity metric performance in brain image registration. Neuroimage. 2011;54(3):2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MATLAB [computer program]. Version 9.0.0. Natick, Massachusetts, USA: The MathWorks Inc.; [Google Scholar]
- 28.Lorensen WE, Cline HE. Marching cubes: A high resolution 3d surface construction algorithm. Proceedings of the 14th Annual Conference on Computer Graphics and Interactive Techniques. 1987 [Google Scholar]
- 29.Couvares P, Kosar T, Roy A, et al. Workflow in condor. In: Taylor IJ, Deelman E, Gannon DB, et al., editors. Workflows for e-science. Springer Press; 2007. [Google Scholar]
- 30.R: A language and environment for statistical computing [computer program] Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 31.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]
- 32.Shrout PE, Fleiss JL. Intraclass correlations: Uses in assessing rater reliability. Psychol Bull. 1979;86(2):420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- 33.Chung D, Chung MK, Durtschi RB, et al. Measurement consistency from magnetic resonance images. Acad Radiol. 2008;15(10):1322–1330. doi: 10.1016/j.acra.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bland JM, Altman DG. Applying the right statistics: Analyses of measurement studies. Ultrasound Obstet Gynecol. 2003;22(1):85–93. doi: 10.1002/uog.122. [DOI] [PubMed] [Google Scholar]
- 35.Koch GG. Intraclass correlation coefficient. 4. New York, NY: John Wiley & Sons; 1982. [Google Scholar]
- 36.Bjork A. Prediction of mandibular growth rotation. Am J Orthod. 1969;55(6):585–599. doi: 10.1016/0002-9416(69)90036-0. [DOI] [PubMed] [Google Scholar]
- 37.Xia JJ, Shevchenko L, Gateno J, et al. Outcome study of computer-aided surgical simulation in the treatment of patients with craniomaxillofacial deformities. Journal of Oral Maxillofacial Surgery. 2011;69(7):2014–2024. doi: 10.1016/j.joms.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abduo J, Bennamoun M. Three-dimensional image registration as a tool for forensic odontology a preliminary investigation. Am J Foren Med Path. 2013;34(3):260–266. doi: 10.1097/PAF.0b013e31829f6a29. [DOI] [PubMed] [Google Scholar]