Abstract
Objectives
The aim of this study was to test the ability of a commercially-available NLP tool to accurately extract exam quality-related and large polyp information from colonoscopy reports with varying report formats.
Background
Colonoscopy quality reporting often requires manual data abstraction. Natural language processing (NLP) is another option for extracting information; however, limited data exist on its ability to accurately extract exam quality and polyp findings from unstructured text in colonoscopy reports with different reporting formats.
Study Design
NLP strategies were developed using 500 colonoscopy reports from Kaiser Permanente Northern California and then tested using 300 separate colonoscopy reports that underwent manual chart review. Using findings from manual review as the reference standard, we evaluated the NLP tool’s sensitivity, specificity, positive predictive value (PPV), and accuracy for identifying colonoscopy exam indication, cecal intubation, bowel preparation adequacy, and polyps ≥10 mm.
Results
The NLP tool was highly accurate in identifying exam quality-related variables from colonoscopy reports. Compared to manual review, sensitivity for screening indication was 100% (95% confidence interval: 95.3%–100%), PPV was 90.6% (82.3%–95.8%), and accuracy was 98.2% (97.0%–99.4%). For cecal intubation, sensitivity was 99.6% (98.0%–100%), PPV was 100% (98.5%–100%), and accuracy was 99.8% (99.5%–100%). For bowel preparation adequacy, sensitivity was 100% (98.5%–100%), PPV was 100% (98.5%–100%), and accuracy was 100% (100%–100%). For polyp(s) ≥10 mm, sensitivity was 90.5% (69.6%–98.8%), PPV was 100% (82.4%–100%), and accuracy was 95.2% (88.8%–100%).
Conclusion
NLP yielded a high degree of accuracy for identifying exam quality-related and large polyp information from diverse types of colonoscopy reports.
Keywords: colonoscopy, quality, natural language processing
BACKGROUND
Colonoscopy is a common screening modality for colorectal cancer (CRC) in the United States [1]. Although observational studies have shown that colonoscopy screening is associated with lower CRC incidence and mortality [2–4], recent studies have reported that colonoscopy quality (e.g., physician adenoma detection rate) is associated with screening effectiveness [5–7]. Accordingly, multiple national gastroenterology societies have published quality indicators to improve colonoscopy performance [8–10], including the physician adenoma detection rate, cecal intubation rate, and bowel preparation quality [8–10]. However, the information needed to assess colonoscopy exam quality is often embedded in unstructured colonoscopy procedure reports of varying formats within health records, requiring time-consuming and costly manual data abstraction for accurate reporting.
Natural language processing (NLP) is a method for electronically analyzing and extracting information from unstructured free text (i.e., endoscopy procedure reports) that offers an efficient alternative to manual data abstraction [11–13]. However, most NLP tools to date evaluating colonoscopy reports have been locally developed, require extensive and costly programming efforts, and have limited applicability to diverse healthcare settings [14–18].
This study sought to test a commercially-available NLP tool that offers the potential for broader utilization and adoption. The primary aim was to test this NLP tool’s ability to accurately extract colonoscopy exam-related quality and polyp information from colonoscopy reports derived from multiple community-based medical centers with varying reporting formats, and to compare the findings to manual review of colonoscopy reports.
MATERIALS AND METHODS
Study setting
This cross-sectional study was conducted among members of Kaiser Permanente Northern California (KPNC), which is an integrated health care delivery organization that serves over 3.9 million members across 21 medical centers and hospitals in urban, suburban, and semirural regions within a large geographic area [19, 20]. Although part of a single healthcare delivery network, each medical center has substantial autonomy, including different endoscopic reporting methods between medical centers and over time, providing substantial variety in colonoscopy report formats for study. This research was approved by the KPNC Institutional Review Board.
Data sources
KPNC utilizes the EPIC platform (Epic Systems Corp, Verona, WI) for electronic health records; this platform is estimated to cover approximately half of the population in the United States [21]. All admission notes, history and physicals, progress notes, consult notes, endoscopy reports, and pathology reports are stored in the EPIC platform as unstructured text. For this study, we used colonoscopy reports within EPIC as source documents for colonoscopy quality information. Colonoscopy reports were in different formats reflecting the multiple medical centers within KPNC and formats changed over time (Supplemental Figure 1): some were dictated, others were typed in free text, and still others were created using standardized templates (e.g., via notewriter in EPIC) which were frequently supplemented by free text or phrases specific to each physician’s preference. No colonoscopy report type used fixed text fields amenable to direct data extraction from an associated relational database. Colonoscopic adenoma detection (from the pathology database) was ascertained using the Cerner CoPathPlus platform (Cerner Corp, Kansas City, KS) and Systematized Nomenclature of Medicine (SNOMED) coding. All pathology reports from each colonoscopy procedure were stored in the pathology database; colorectal “T” location codes were used to identify colon segment and “M” pathology codes to identify polyp/tissue histology (e.g., tubular adenoma, tubulovillous adenoma, and adenocarcinoma).
Variables from colonoscopy reports and derived variables
The colonoscopy exam quality-related measures of interest included variables present within colonoscopy reports and derived variables (Table 1). The following colonoscopy report variables were selected because they are recommended by national guidelines for assessing colonoscopy exam quality: exam indication (screening versus non-screening); extent of exam (cecal intubation); bowel preparation quality (adequate or inadequate); family history of CRC (first-degree relative with CRC and first-degree relative’s age at CRC diagnosis); presence of polyp(s) ≥10 mm (based on the clinician’s endoscopic assessment); and location of polyp(s) ≥10 mm [9,10]. These variables were used to calculate the following derived colonoscopy quality metrics: adenoma detection rate (the percentage of screening colonoscopies in which one more adenomas were detected); cecal intubation rate (the percentage of total colonoscopies in which the colonoscope reached either the cecum or terminal ileum); and adequate bowel preparation rate (the percentage of total colonoscopies in which the bowel preparation was excellent, good, satisfactory, or adequate).
Table 1.
List of variables of interest from colonoscopy reports and derived variables
| Variables present within colonoscopy reports |
|---|
| Screening indication |
| Family history of colorectal cancer |
| First-degree relative with colorectal cancer |
| Age of first-degree relative with colorectal cancer |
| Cecal intubation |
| Adequacy of bowel preparation |
| Presence of polyp(s) ≥10 mm |
| Location of polyp(s) ≥10 mm |
|
Derived variables
|
| Adenoma detection rate |
| Cecal intubation rate |
| Adequate bowel preparation rate |
Colonoscopy reports for development and validation of NLP query strategies
A total of 800 randomly selected colonoscopy reports from 2010 through 2015 were used for the development and validation of NLP query strategies. The sample size was selected out of convenience and is similar to prior studies evaluating NLP’s performance for colonoscopy-related variables [13, 15, 16, 18]. A total of 500 charts were used for the development of NLP query strategies and 300 separate charts were used for testing purposes.
Development of NLP query strategies
For NLP query strategy development, a board-certified gastroenterologist (JKL) manually reviewed the training set of 500 colonoscopy reports and assembled term and phrase variations for the variables of interest (Table 1). Query strategies were developed to flag the presence of these terms and phrases in the source colonoscopy reports using a commercially-available NLP software (Linguamatics I2E, www.linguamatics.com; United Kingdom). The NLP tool’s basic processing steps include parsing the report text into sentences, words, and sections, and then identifying key concepts by matching the text against clinical vocabularies (i.e., terms and phrases) [22]; the software provides a user-modifiable interface and the ability to use modules developed by other groups. Developing the NLP query strategy was an iterative process informed by theory, experimentation, logic, and domain knowledge. Query strategies were developed to incorporate focused negation so phrases such as ‘no family history of CRC’ and ‘no fhx crc’ would not be assigned as positive responses. Discrepancies in findings between use of the NLP query strategies and manual review of the 500 colonoscopy reports were investigated through error analysis and query strategies were then refined to reduce errors. Iterative development continued until the performance of the NLP tool reached a high level of accuracy.
Validation of NLP query strategies
The final query strategies were tested against a separate set of 300 colonoscopy reports that served as the reference standard for validation purposes. Two medical chart reviewers (one trained medical record abstractor, and one co-author (CDJ), without any knowledge of the NLP query strategies, independently manually reviewed each colonoscopy report and recorded the presence/absence of each variable of interest (Table 1). A total of 12 discrepancies between the two medical chart reviewers were resolved by an additional manual review with agreement by consensus (Supplemental Table 1).
Statistical analysis
Descriptive statistics were used to characterize the 300 colonoscopy patients in the validation sample. We evaluated the NLP tool’s sensitivity, specificity, positive predictive value (PPV), and accuracy for identifying the variables in Table 1 using the validated manual chart review as the gold standard. Sensitivity was defined as [true positives/(true positives + false negatives)]. Specificity was defined as [true negatives/(true negatives + false positives)]. PPV was defined as [true positives/(true positives + false positives)]. Accuracy was defined as [(true positives + true negatives)/(true positives + false positives + true negatives + false negatives)]. Lastly, we calculated the adenoma detection rate, cecal intubation rate, and adequate bowel preparation percentage by each data extraction method (i.e., NLP, validated manual review). STATA 13.0 (Stata Corporation, College Station, TX) was used for all statistical analyses.
RESULTS
Patient characteristics
The descriptive characteristics of the 300 colonoscopy patients in the validation sample are shown in Table 2. Mean age (±SD) was 62.8±8.6 years (range: 50–85), 51.0% of patients were female, 58.7% were white, and 54.7% had a Charlson-comorbidity score of 0. Of the 300 colonoscopy reports, 77 (25.7%) were screening exams; 282 (94.0%) reached the cecum; 237 (79.0%) had an adequate bowel preparation; 28 (9.3%) had a family history of CRC; 16 (5.3%) reported a first-degree relative with CRC and 14 (4.7%) reported the age of the first-degree relative with CRC; 21 (7.0%) had at least one polyp ≥10 mm; and 20 (6.7%) reported the location of polyp(s) ≥10 mm.
Table 2.
Patient characteristics of the validation sample
| Characteristics | n (%) |
|---|---|
| Total study sample | 300 (100.0) |
| Age, year | |
| 50–59 | 123 (41.0) |
| 60–69 | 112 (37.3) |
| 70–79 | 51 (17.0) |
| ≥80 | 14 (4.7) |
| Sex | |
| Female | 153 (51.0) |
| Male | 147 (49.0) |
| Race/Ethnicity | |
| White | 176 (58.7) |
| African American | 22 (7.3) |
| Hispanic | 38 (12.7) |
| Asian | 53 (17.7) |
| Other | 3 (1.0) |
| Missing | 8 (2.6) |
| Charlson co-morbidity score | |
| 0 | 164 (54.7) |
| 1 | 57 (19.0) |
| 2 | 47 (15.7) |
| 3+ | 12 (4.0) |
| Colonoscopy Indication | |
| Screening | 77 (25.7) |
| Non-screening | 223 (74.3) |
| Cecal intubation | 282 (94.0) |
| Bowel preparation adequacy | |
| Adequate | 237 (79.0) |
| Inadequate | 63 (21.0) |
| Family history of colorectal cancer | 28 (9.3) |
| First-degree relative with colorectal cancer | 16 (5.3) |
| Age of first-degree relative with colorectal cancer | 14 (4.7) |
| Presence of polyp(s) ≥10 mm | 21 (7.0) |
| Location of polyp(s) ≥10 mm | 20 (6.7) |
n, number
NLP performance
NLP performed well in comparison to the reference standard (Table 3). For example, PPV across all 8 variables ranged from 90.6% to 100%, sensitivity ranged from 78.6% to 100%, specificity ranged from 96.4% to 100%, and accuracy ranged from 89.3% to 100%. For screening indication, sensitivity was 100% (95% confidence interval (CI): 95.3%–100%), PPV was 90.6% (95% CI: 82.3%–95.8%), and accuracy was 98.2% (95% CI: 97.0%–99.4%). For cecal intubation, sensitivity was 99.6% (95% CI: 98.0%–100%), PPV was 100% (95% CI: 98.5%–100%), and accuracy was 99.8% (95% CI: 99.5%–100%). For adequate bowel preparation quality, sensitivity was 100% (95% CI: 98.5%–100%), PPV was 100% (95% CI: 98.5%–100%), and accuracy was 100% (95% CI: 100%–100%). For polyp(s) ≥10 mm, sensitivity was 90.5% (95% CI: 69.6%–98.8%), PPV was 100% (95% CI: 82.4%–100%), and accuracy was 95.2% (95% CI: 88.8%–100%) (Table 3).
Table 3.
Natural language processing tool’s performance as compared to a validated manual review of colonoscopy reports
| Variable | Sensitivity (95% CI) |
Specificity (95% CI) |
Positive Predictive Value (95% CI) |
Accuracy (95% CI) |
|---|---|---|---|---|
| Screening indication | 100 (95.3–100) | 96.4 (93.1–98.4) | 90.6 (82.3–95.8) | 98.2 (97.0–99.4) |
| Family history of CRC | 89.3 (71.8–97.7) | 100 (98.7–100) | 100 (86.3–100) | 94.6 (88.8–100) |
| First-degree relative with CRC | 100 (79.4–100) | 100 (98.7–100) | 100 (79.4–100) | 100 (100–100) |
| Age of first-degree relative with CRC | 78.6 (49.2–95.3) | 100 (98.7–100) | 100 (71.5–100) | 89.3 (78.1–100) |
| Cecal intubation (exam extent) | 99.6 (98–100) | 100 (81.5–100) | 100 (98.7–100) | 99.8 (99.5–100) |
| Adequate bowel preparation | 100 (98.5–100) | 100 (94.3–100) | 100 (98.5–100) | 100 (100–100) |
| Presence of polyp(s) ≥10 mm | 90.5 (69.6–98.8) | 100 (98.7–100) | 100 (82.4–100) | 95.2 (88.8–100) |
| Location of polyp(s) ≥10 mm | 90.0 (68.3–98.8) | 100 (98.7–100) | 100 (81.5–100) | 95.0 (88.3–100) |
CI, confidence interval; CRC, colorectal cancer
Colonoscopy quality reporting by method
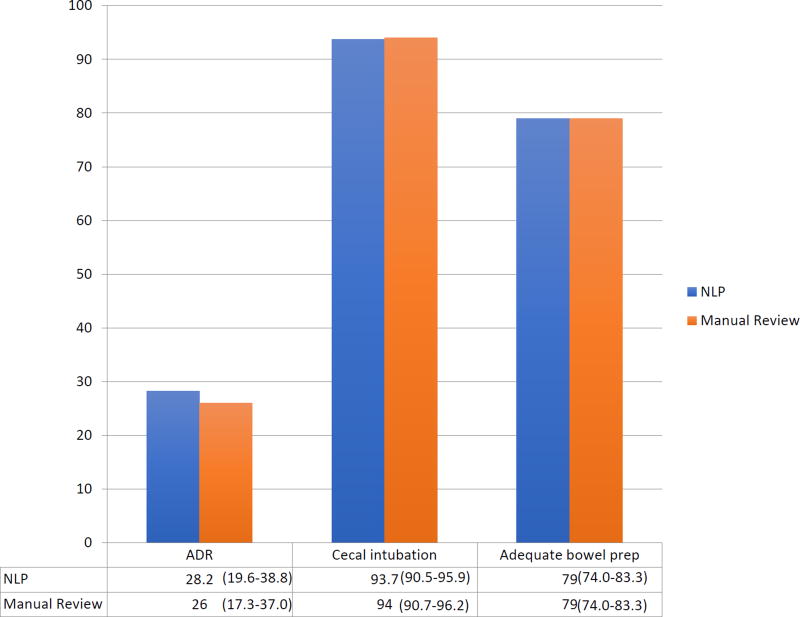
Manual review yielded an adenoma detection rate of 26.0% (95% CI: 17.3%–37.0%), cecal intubation rate of 94.0% (95% CI: 90.7%–96.2%), and an adequate bowel preparation rate of 79.0% (95% CI: 74.0%–83.3%) (Figure 1). By comparison, NLP data abstraction yielded an adenoma detection rate of 28.2% (95% CI: 19.6%–38.8%), cecal intubation rate of 93.7% (95% CI: 90.5%–95.9%), and an adequate bowel preparation rate of 79.0% (95% CI: 74.0%–83.3%).
Figure 1. Colonoscopy quality reporting by method.
ADR: adenoma detection rate
NLP: natural language processing
NLP error analysis
Although the NLP tool achieved a high-degree of accuracy for key findings within colonoscopy reports, examples where the NLP tool failed to correctly identify the target data element are shown in Supplemental Table 2. Most of these cases were related to the (a) disparate phrasing and abbreviations of the data elements, and (b) lack of refinement in word distances and negation terms around the data elements.
DISCUSSION
Colonoscopy is a common screening modality for CRC [1], and growing evidence suggests that exam quality may impact its effectiveness in reducing CRC incidence and mortality [2–4]. In 2015, the American Society of Gastrointestinal Endoscopy and the American College of Gastroenterology published a list of 15 quality indicators to improve colonoscopy safety and performance [9, 10]. However, key elements for assessing colonoscopy exam quality are difficult to extract because they are often embedded as unstructured text within health records, limiting the ability to report, monitor, and ultimately improve colonoscopy quality. In our study, we demonstrated that, compared to a validated manual review of colonoscopy reports, a commercially-available NLP tool yielded high levels of PPV, sensitivity, specificity, and accuracy for eight colonoscopy quality-related variables of interest. The NLP tool also yielded three derived measures of colonoscopy quality that were either identical or very close to those obtained by manual review, suggesting that NLP-based derived measures of colonoscopy quality may be sufficiently accurate for quality reporting. These findings have important implications for quality reporting given that millions of colonoscopies are performed annually (~14.2 million in 2002) [1].
To date, only a few studies have evaluated the role of NLP in extracting information from colonoscopy reports for assessing exam quality [14, 16–18]. Harkema et al. developed a Java-based NLP tool to extract 21 variables (e.g., exam indication, exam extent, bowel preparation quality) from colonoscopy and linked pathology reports, and found an average accuracy over all the variables of 89% (range: 62%–100%) compared to manual review [14]. Gawron et al. also used a Java-based NLP tool and reported a PPV of 96% for screening exams, 98% for completeness of colonoscopy, 98% for adequate bowel preparation quality, and 95% for histology of polyp as compared to manual review [18]. More recently, Raju et al. developed an NLP tool using C# programming language and reported an accuracy of 91.3% for identifying screening exams and 99.4% for adenoma diagnosis from colonoscopy reports and their associated pathology reports, as compared to manual record review; also overall adenoma detection rates by NLP and manual review methods were identical [17]. Although these locally-developed NLP tools were accurate for identifying key information from colonoscopy and/or pathology reports, several limitations minimized the applicability of these tools to other settings, including the need for expert programming support for installation and customization; their development and validation at a single center, thus limiting variation in colonoscopy report formats; and use of colonoscopy reports derived from highly structured template-driven software systems (e.g., Pentax EndoPro, ProVation) which limit linguistic variation. In contrast, the commercially-available NLP tool used in this study required no programming background and was adaptable to multiple colonoscopy report formats, including text-based reports. In addition, it has a unique plug-and-play feature where programmed query strategies can be distributed electronically (e.g., via email), resulting in faster deployment and easier adoption to other settings.
In our study, the calculation of adenoma detection rates, cecal intubation rates, and adequate bowel preparation percentages based on NLP findings were similar to calculations derived from manual review of colonoscopy reports. Thus, NLP may offer a degree of accuracy for colonoscopy quality reporting that is comparable to manual data abstraction, which itself is not without the possibility of error. In a multi-center Veteran Affairs study, board-certified gastroenterologists made incorrect assignments for colonoscopy findings during manual abstraction at a rate similar to NLP (21.1% versus 25.4%, respectively, P=0.07) [16]. More recently, Raju et al. reported that manual abstraction had an error rate of 12.2% and 1.7% for assigning screening status and adenoma diagnosis, respectively [17]. In our study, discordant findings were reported between the two independent manual record reviewers for 12 of 300 (4.0%) colonoscopy reports. Several factors may contribute to manual abstraction errors including lack of abstractor familiarity with medical/procedure terminology or the abstraction tool, fatigue, incomplete access to charts, and inadequate time for abstraction [23]. In our case, nearly all the errors by the two reviewers were either due to overlooking information during manual review or incorrectly entering the data elements of interest.
Information on family history of CRC is critical to initiating screening in high-risk populations, but such information is often embedded in unstructured text within progress notes and colonoscopy reports, making it difficult to ascertain. NLP in the current study had a high sensitivity (89.3%–100%) and PPV (100%) for identifying family history of CRC and first-degree relatives with CRC from colonoscopy reports; however, the tool was less sensitive (78.6%) in identifying the affected relative’s age of CRC diagnosis, due to lack of refinement in the query strategy to detect certain phrase variations of affected relative’s age of CRC diagnosis, a limitation that is relatively easy to remedy. Friedlin et al. reported their Java-based NLP tool identified family history information for 12 disease diagnoses (including CRC) from hospital admission notes with an overall sensitivity and PPV of 96% and 97%, respectively [24]. Recently, Harkema et al’s Java-based NLP tool identified family history of CRC from colonoscopy reports with a sensitivity and PPV of 96% and 86%, respectively [13]. Despite similar performance characteristics for identifying the presence of family history of CRC compared to our study, both NLP tools [13, 24] weren’t programmed to identify the affected family member with CRC or age of onset, critical elements needed for healthcare systems to optimize the screening and surveillance process for high-risk individuals.
Our study has several strengths. First, we utilized separate sets of randomly-selected colonoscopy reports for developing query strategies and subsequently testing the NLP strategies. We also utilized two independent medical record abstractors blinded to the NLP query strategies, and performed an additional manual review to resolve discrepancies in the creation of a validated reference standard. We were able to sample colonoscopy reports from a large number of medical centers within the healthcare system, which increased the variation in report formats, local terms, and phrases. The NLP software used in the study is commercially available and did not require prior programming experience for query development, which may ease dissemination and adoption of the tool in other settings. Also, although query strategies were built within the tool’s software package, potentially presenting barriers to dissemination, we designed the query framework to be easily exported to other users of this tool or adaptable to other NLP tools to foster future open-source availability.
There are also several limitations to the study that should be considered. Evaluation of the NLP tool was limited to a single healthcare system and the findings should be replicated in other healthcare settings (e.g., academic or safety-net healthcare systems). The tool requires machine-readable clinical text and does not work with print or scanned documents. The tool relied on physician reporting of cecal intubation and bowel preparation quality rather than photo-documentation. Also, in the calculation of adenoma detection rates, the tool only identified screening exams; the presence of an adenoma associated with an exam was obtained electronically from the pathology database and did not involve NLP abstraction.
In summary, we found that a commercially-available NLP tool was highly accurate in identifying key information from colonoscopy reports related to exam quality and polyp findings across multiple medical centers with different report formats. This automated method may allow for large numbers of colonoscopy reports to be quickly processed, facilitating the assessment of colonoscopy quality at the level of individual physicians, group providers, and hospitals. If validated in other healthcare settings, NLP has the potential to reduce the costs and burden associated with reporting quality metrics.
Supplementary Material
Acknowledgments
Primary Funding Source: National Cancer Institute (K07CA212057, U54 CA163262) and American Gastroenterological Association.
Funding: Funding for this study was provided by the American Gastroenterological Association Research Scholar Award (JKL), and the National Cancer Institute (K07CA212057 (JKL); U54 CA163262 (DAC, TRL, AGZ, CAD)).
Acknowledgements: not applicable
LIST OF ABBREVIATIONS
- CRC
colorectal cancer
- NLP
natural language processing
- KPNC
Kaiser Permanente Northern California
- PROSPR
Population-Based Research Optimizing Screening through Personalized Regimens
- SNOMED
Systematized Nomenclature of Medicine
- PPV
positive predictive value
- CI
confidence interval
Footnotes
Conflict of Interest Summary:
All authors have no conflict of interest in this manuscript. All authors have not published or submitted any related papers from the same study.
DECLARATIONS
Ethics approval and consent to participate: This study used de-identified colonoscopy reports obtained from Kaiser Permanente Northern California. The study was approved by the Kaiser Permanente Northern California Institutional Review Board.
Consent for publication: The authors have given consent for publication of this manuscript
Availability of data and materials: The natural language processing software is available from www.linguamatics.com. The validated queries used in this study is available via email request to the corresponding author.
Competing Interests: The authors declare that they have no competing interests
Authors’ contributions: JKL and DAC conceived of the study, developed the queries, performed the analysis and drafted the manuscript. AGZ, CAD, TRL, and WKZ participated in the analysis and helped draft the manuscript. AGZ, CAD, and TRL participated in the study design and helped draft the manuscript. CDJ helped manually review each colonoscopy report to create the gold standard report set and helped draft the manuscript. All authors have read and approved this manuscript.
References
- 1.Seeff LC, Richards TB, Shapiro JA, Nadel MR, Manninen DL, Given LS, Dong FB, Winges LD, McKenna MT. How many endoscopies are performed for colorectal cancer screening? Results from CDC’s survey of endoscopic capacity. Gastroenterology. 2004;127:1670–1677. doi: 10.1053/j.gastro.2004.09.051. [DOI] [PubMed] [Google Scholar]
- 2.Zauber AG, Winawer SJ, O’Brien, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, Shi W, Bond JH, Schapiro M, Panish JF, Stewart ET, Waye JD. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366:687–696. doi: 10.1056/NEJMoa1100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winawer SJ, Zauber AG, Ho MN, O'Brien MJ, Gottlieb LS, Sternberg SS, Waye JD, Schapiro M, Bond JH, Panish JF, et al. Prevention of colorectal cancer by colonoscopic polypectomy. The National Polyp Study Workgroup. N Engl J Med. 1993;329:1977–81. doi: 10.1056/NEJM199312303292701. [DOI] [PubMed] [Google Scholar]
- 4.Nishihara R, Wu K, Lochhead P, Morikawa T, Liao X, Qian ZR, Inamura K, Kim SA, Kuchiba A, Yamauchi M, Imamura Y, Willett WC, Rosner BA, Fuchs CS, Giovannucci E, Ogino S, Chan AT. Long-term colorectal cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369:1095–105. doi: 10.1056/NEJMoa1301969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corley DA, Jensen CD, Marks AR, Zhao WK, Lee JK, Doubeni CA, Zauber AG, de Boer J, Fireman BH, Schottinger JE, Quinn VP, Ghai NR, Levin TR, Quesenberry CP. Physician adenoma detection rate and interval colorectal cancer risk and mortality. N Engl J Med. 2014;370(14):1298–1306. doi: 10.1056/NEJMoa1309086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaminski MF, Regula J, Kraszewsk E, Polkowski M, Wojciechowska U, Didkowska J, Zwierko M, Rupinski M, Nowacki MP, Butruk E. Quality indicators for colonoscopy and the risk of interval cancer. N Engl J Med. 2010;362:1795–803. doi: 10.1056/NEJMoa0907667. [DOI] [PubMed] [Google Scholar]
- 7.Shaukat A, Rector TS, Church TR, Lederle FA, Kim AS, Rank JM, Allen JI. Longer withdrawal time is associated with a reduced incidence of interval cancer after screening colonoscopy. Gastroenterology. 2015;149:952–7. doi: 10.1053/j.gastro.2015.06.044. [DOI] [PubMed] [Google Scholar]
- 8.Rex DK, Petrini JL, Baron TH, Chak A, Cohen J, Deal SE, Hoffman B, Jacobson BC, Mergener K, Petersen BT, Safdi MA, Faigel DO, Pike IM. Quality indicators for colonoscopy. Gastrontest Endosc. 2006;63:S16–28. doi: 10.1016/j.gie.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 9.Rex DK, Schoenfeld PS, Cohen J, Pike IM, Adler DG, Fennerty MB, Lieb JG, Park WG, Rizk MK, Sawhney MS, Shaheen NJ, Wani S, Weinberg DS. Quality indicators for colonoscopy. Am J Gastroenterol. 2015;110(1):72–90. doi: 10.1038/ajg.2014.385. [DOI] [PubMed] [Google Scholar]
- 10.Rex DK, Schoenfeld PS, Cohen J, Pike IM, Adler DG, Fennerty MB, Lieb JG, Park WG, Rizk MK, Sawhney MS, Shaheen NJ, Wani S, Weinberg DS. Quality indicators for colonoscopy. Gastrointest Endosc. 2015;81:31–53. doi: 10.1016/j.gie.2014.07.058. [DOI] [PubMed] [Google Scholar]
- 11.Liddy ED. Encyclopedia of Library and Information Science. 2. NY: Marcel Decker, Inc; 2001. Natural Language Processing. [Google Scholar]
- 12.Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc. 2011;18:544–551. doi: 10.1136/amiajnl-2011-000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harkema H, Chapman WW, Saul M, Dellon ES, Schoen RE, Mehrotra A. Developing a natural language processing application for measuring the quality of colonoscopy procedures. J Am Med Inf Assoc. 2011;18:i150–6. doi: 10.1136/amiajnl-2011-000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehrotra A, Dellon ES, Schoen RE, Saul M, Bishehsari F, Farmer C, Harkema H. Applying a natural language processing tool to electronic health records to assess performance on colonoscopy quality measures. Gastrointest Endosc. 2012;75:1233–9. doi: 10.1016/j.gie.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imler TD, Morea J, Kahi C, Imperiale TF. Natural language processing accurately categorizes findings from colonscopy and pathology reports. Clin gastroenterol Hepatol. 2013;11:689–94. doi: 10.1016/j.cgh.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imler TD, Morea J, Kahi C, Sherer EA, Cardwell J, Johnson CS, Xu H, Ahnen D, Antaki F, Ashley C, Baffy G, Cho I, Dominitz J, Hou J, Korsten M, Nagar A, Promrat K, Robertson D, Saini S, Shergill A, Smalley W, Imperiale TF. Multi-center colonoscopy quality measurement utilizing natural language processing. Am J Gastroenterol. 2015;110:543–552. doi: 10.1038/ajg.2015.51. [DOI] [PubMed] [Google Scholar]
- 17.Raju GS, Lum PJ, Slack RS, Thirumurthi S, Lynch PM, Miller E, Weston BR, Davila ML, Bhutani MS, Shafi MA, Bresalier RS, Dekovich AA, Lee JH, Guha S, Pande M, Blechacz B, Rashid A, Routbort M, Shuttlesworth G, Mishra L, Stroehlein JR, Ross WA. Natural language processing as an alternative to manual reporting of colonoscopy quality metrics. Gastrointest Endosc. 2015;82:512–9. doi: 10.1016/j.gie.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gawron AJ, Thompson WK, Keswani RN, Rasmussen LV, Kho AN. Anatomic and advanced adenoma detection rates as quality metrics determined via natural language processing. Am J Gastroenterol. 2014;109(12):1844–9. doi: 10.1038/ajg.2014.147. [DOI] [PubMed] [Google Scholar]
- 19.Jensen CD, Doubeni CA, Quinn VP, Levin TR, Zauber AG, Schottinger JE, Marks AR, Zhao WK, Lee JK, Ghai NR, Schneider JL, Fireman BH, Quesenberry CP, Corley DA. Adjusting for patient demographics has minimal effects on rates of adenoma detection in a large community-based setting. Clin Gastroenterol Hepatol. 2015;13(4):739–46. doi: 10.1016/j.cgh.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JK, Jensen CD, Lee A, Doubeni CA, Zauber AG, Levin TR, Zhao WK, Corley DA. Development and validation of an algorithm for classifying colonoscopy indication. Gastrointest Endosc. 2015;81:575–582. doi: 10.1016/j.gie.2014.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moukheiber Z. [Accessed August 25, 2016];An interview with the most powerful woman in health care. Available at: http://www.forbes.com/sites/zinamoukheiber/2013/05/15/a-chat-with-epic-systems-ceo-judy-faulkner/
- 22.Liu V, Clark MP, Mendoza M, Saket R, Garner MN, Turk BJ, Escobar GJ. Automated identification of pneumonia in chest radiograph reports in critically ill patients. BMC Med Inform Decis Mak. 2013;13:90. doi: 10.1186/1472-6947-13-90. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zozus MN, Pieper C, Johnson CM, Johnson TR, Franklin A, Smith J, Zhang J. Factors affecting accuracy of data abstracted from medical records. PLoS One. 2015;10:e0138649. doi: 10.1371/journal.pone.0138649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedlin J, McDonald CJ. Using a natural language processing system to extract and code family history data from admission reports. AMIA Annu Symp Proc. 2006;925 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.