Abstract
Background
The integration of traditional contact tracing with HIV sequence analyses offers opportunities to mitigate some of barriers to effective network construction. We utilized combined analyses during an outbreak investigation of spatiotemporally clustered acute HIV infections to evaluate if the observed clustering was the product of a single outbreak.
Methods
We investigated acute and recent HIV index cases reported in North Carolina from 2013–2014 and their reported contacts. Contact tracing networks were constructed with surveillance data and compared with phylogenetic transmission clusters involving an index case using available HIV-1 pol sequences including 1672 references. Clusters were defined as clades ≥2 sequences with <1.5% genetic distance and bootstrap ≥98% on maximum-likelihood phylogenies.
Results
In total, 68 index cases and 210 contacts (71 HIV-infected) were reported. The contact tracing network involved 58 components with low overall density (1.2% statewide); 33% of first-degree contacts could not be located. Among 38/68 (56%) index cases and 34/71 (48%) contacts with sequences, 13 phylogenetic clusters were identified (size 2–4 members). Four clusters connected network components that were not linked in contact tracing. The largest component (n=28 cases) included two distinct phylogenetic clusters and spanned two regions.
Conclusions
We identified the concurrent expansion of multiple small transmission clusters rather than a single outbreak among the largely disconnected contact tracing network. Integration of phylogenetic analyses provided timely information on transmission networks during the investigation. Our findings highlight the potential of combined methods to better identify high risk networks for intervention.
Keywords: HIV-1, contact tracing, molecular epidemiology, phylogenetics, outbreak
INTRODUCTION
Identifying and responding to newly acquired HIV infections remains a critical component of HIV control in North Carolina (NC), where over 1,300 new HIV diagnoses are reported annually.1 Although only a fraction of these new diagnoses are detected during acute HIV infection (AHI) or recent HIV infection (RHI),1 incident infections represent the leading edge of the HIV epidemic. Due in part to elevated viremia during acute infection, an estimated 30–50% of transmissions occur in the six months following HIV acquisition.2,3 Thus, detection of AHI and RHI present an important opportunity for secondary prevention via rapid initiation of antiretroviral therapy (ART) and provision of transmission prevention counseling. In pursuit of this goal, NC has actively screened routine HIV tests at public testing sites for AHI since 2002 through the NC Screening and Tracing for Active HIV-1 Transmission (NC-STAT) program.4 Individuals identified with AHI and RHI are prioritized for expedited counseling, contact tracing, and referral services. From 2003–2012, 3.4% of all HIV diagnoses in NC were detected as AHI by NC-STAT and 96.6% of these cases successfully reached for interview.4
Although contact tracing remains central to HIV outbreak control, detecting HIV transmission sources and networks is often limited; contact tracing is resource-intensive and subject to participant willingness and recall bias.5 Additionally, networks involving anonymous or unreachable partners may go unrecognized despite contact tracing efforts. The integration of traditional contact tracing with HIV sequence analyses may offer opportunities to mitigate some of these barriers. HIV pol sequences, derived from routinely collected drug resistance genotypes, are increasingly used to reconstruct putative HIV transmission clusters through phylogenetic inference in the population of interest.6 The combination of these phylogenetic analyses with epidemiologic data provides a unique view of population-level transmission patterns,7 and can be used to evaluate potential and identified HIV outbreaks.8–10
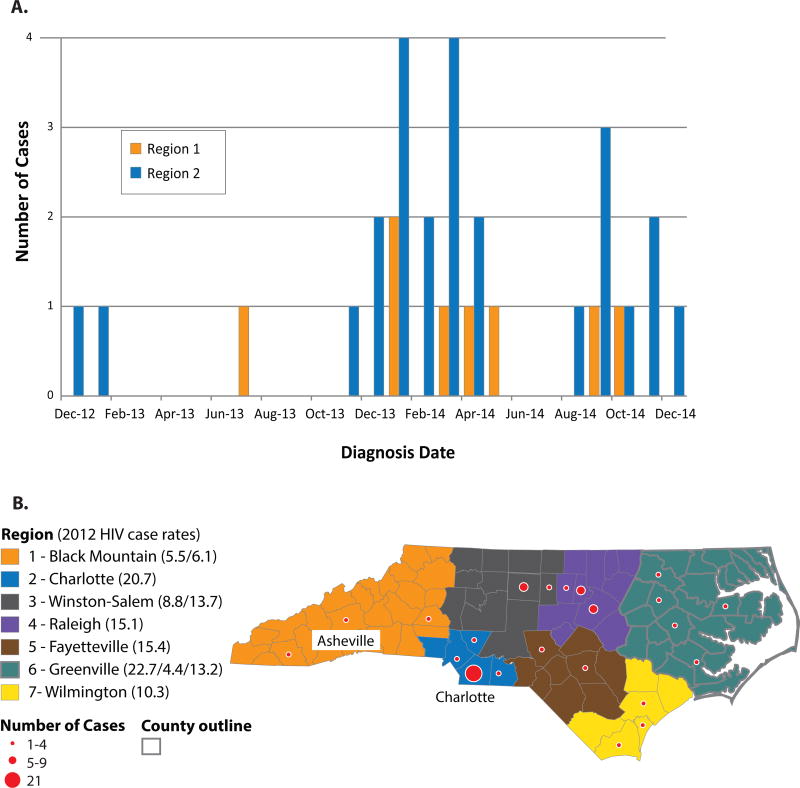
We investigated contact tracing and HIV phylogenetic networks that involved AHI cases after spatiotemporal clustering of AHI cases was detected in NC in early 2014. Between December 2013 and February 2014, more than three times as many AHI among men who have sex with men (MSM) were reported in two regions in western NC (Regions 1 and 2; Figure 1) compared to the same time frame during the previous year. HIV transmission in NC is predominately sexual, with most new cases reported among MSM (61%), and is concentrated in urban and suburban areas.1 Metropolitan Region 2 (Charlotte) has the highest HIV cases rate, although Region 1 is among the lowest in the state.1 The apparent spatiotemporal clustering in these regions triggered an investigation by the NC Department of Health and Human Services (NC-DHHS) in February 2014 out of concern for a potentially undetected outbreak. As an exploratory component of the multidisciplinary investigation, we characterized the contact tracing networks involving index cases diagnosed during AHI and RHI in Regions 1 and 2, as well as throughout the state, to determine whether the new infections were phylogenetically linked.
Figure 1.
A. Trends in acute HIV diagnoses in North Carolina reported in Regions 1 (Black Mountain) and 2 (Charlotte) from 2012–2014. An outbreak investigation was triggered after the observed spatiotemporal spike in acute HIV diagnoses from December 2013 – February 2014. B. Map of North Carolina regions for HIV field services. Locations of acute HIV infections by county are indicated by red circles. As reference, the estimated 2012 newly diagnosed HIV infection rates per 100,000 population for North Carolina Regional Networks of Care and Prevention1 are provided in parentheses after each HIV Field Service Region. Regions with multiple values include several Regional Care Networks
MATERIALS AND METHODS
Study Population
Following an increase in AHI cases reported in Regions 1 and 2 in early 2014, we investigated all index cases diagnosed with AHI or RHI in NC from December 2013 – December 2014 and their reported contacts (Figure 1). Additionally, index cases reported in Region 1 and 2 from December 2012 - November 2013 were included to assess potential genetic relatedness to these earlier cases in the putative outbreak foci. All index cases were reported to NC-DHHS through the NC-STAT program11 or by acute care settings (i.e. emergency departments, urgent or primary care clinics). AHI is defined as either: 1) positive antibody test with seronegative documentation within the previous 30 days, or 2) reactive fourth generation antigen/antibody combination test with a negative/ indeterminate multispot assay and a detectable HIV RNA (since November 2013) or 3) negative/indeterminate antibody test and detectable HIV RNA (NC-STAT through RNA pooling of negative tests11 2002–2013). Cases with RHI are similarly investigated by NC-DHHS and defined as a positive antibody test with seronegative documentation and/or symptoms compatible with AHI within three months of diagnosis. Medical and surveillance records are extensively reviewed by STAT program medical staff for suspected RHI cases without confirmed seronegative testing to determine if STAT investigation warranted. As this study was initially conducted in partnership with a NC-DHHS public health outbreak investigation, no additional informed consent was needed. The Institutional Review Board at the University of North Carolina approved the study.
Contact Tracing Networks
In NC, disease intervention specialists (DIS) perform routine contact tracing for all HIV-infected cases and their named contacts. DIS expedite routine follow-up for AHI and RHI cases through NC-STAT, contacting individuals within 72 hours of HIV-positive testing results. Named sexual and injection drug use (IDU) contacts within the 8 (AHI) or 12 weeks (RHI) prior to diagnosis are notified of HIV exposure, counseled, and tested for HIV infection within 72 hours of the case interview when possible. High-risk social contacts reported by the index case are also investigated. For cases with newly diagnosed chronic HIV infection, all named contacts reported within 12 months prior to diagnosis are investigated.
Relevant contact tracing, demographic, and clinical data for index and contact cases were abstracted from the NC Electronic Disease Surveillance system. We classified first-degree contacts of index cases as HIV-infected (previous or new chronic diagnosis), HIV-uninfected, or HIV-status unknown. We defined a new, chronic HIV diagnosis as a case diagnosed ≤30 days prior to or any time after the date of the index’s HIV diagnosis. Contacts that were also diagnosed during AHI or RHI were classified as index cases. Further contact tracing data (second-degree contacts to the index) were included for first-degree contacts who had either a: 1) new HIV diagnosis, or 2) HIV diagnosis anytime in 2013 or 2014. Demographic data included age at the time of the investigation, race/ethnicity, sex, and county of residence. Clinical data included HIV testing and diagnosis dates, testing locations, initial HIV RNA levels for index cases, and the HIV RNA level reported to NC-DHHS for HIV-infected contacts most proximal to the time of DIS investigation.
We constructed networks using contact tracing data for index cases, their reported contacts, and applicable second-degree contacts to the index cases. While DIS perform name-based contact tracing, all analyses were conducted on de-identified datasets. Data were symmetrized and an undirected network was constructed using UCINET 6.486 and NetDraw 2.134 (Analytic Technologies, Lexington, KY). Mean index node degrees and overall graph density were calculated for the statewide network and the subset network including contacts epidemiologically linked to index cases from Regions 1 and 2. Isolated nodes were removed prior to density calculations. We evaluated the demographic and clinical features of independent network components, defined as all index cases and contacts linked by any epidemiological relationship.
HIV Sequences and Phylogenetic Inference
During the investigation, NC-DHHS requested available HIV-1 pol sequences for all index cases and HIV-infected contacts from laboratories serving clinical providers in NC. These sequences were generated by genotype assays for drug resistance including HIV GenoSure MG® and GenoSure PRIme® (Laboratory Corporation of America Holdings, Research Triangle Park, NC and Monogram Biosciences, South San Francisco, CA), or TRUGENE® HIV-1 assay (Siemens Healthcare Diagnostics, Tarrytown, NY). All spanned full-length protease and partial reverse transcriptase; a minority included integrase. Additional partial pol sequences (n=1672) from participants in the UNC Center for AIDS Research HIV Clinical Cohort were included as background references. These sequences were sampled from 1997–2011 and were the largest sequence dataset available in NC during the investigation (GenBank JX160108-JX161480). Sequences were aligned using MUSCLE12 and edited manually, stripping gapped positions; final sequence length was 1497 nucleotides. Drug resistance mutations (DRM) and subtype assignments were identified using The Stanford HIV Web Service (Sierra v.1.1) to query the Stanford HIVdb Program.13 Major DRMs were selected using the 2009 World Health Organization standardized surveillance list.14
We constructed maximum-likelihood phylogenies in RAxML v.7.0.415 with the general time reversible model of nucleotide substitution and assessed statistical support of clades with 1000 bootstrap replicates. We defined transmission clusters as clades with bootstrap support ≥98% that included at least one index case sequence with 0.015 substitutions/site pairwise genetic distance from another sequence from another index or any contact case. Additionally, we identified transmission clusters that were part of larger, genetically divergent clades, and those that involved index cases in clades with a reference sequence. All analyses and results were shared with NC-DHHS on a regular basis during the approximately 12-month investigation period to inform the ongoing outbreak investigation in a timely manner.
RESULTS
Study Population
In total, 68 index cases were identified (Table 1); 65 were index cases reported statewide from December 2013 – December 2014 (55 AHI and 13 RHI) and three were AHI cases reported in Region 2 in from 2012–2013. Nearly half of the index cases (n=32/68) were diagnosed in Regions 1 or 2. In comparison, from 2003–2012, an average of 23 AHI cases were diagnosed annually statewide (range 15–32).4 Almost all index cases (n=66/68) were reached by DIS. Index cases were predominately male (n=58/68, 85%) and median age was 25 (IQR 22–35) years (Table 1). Most index cases were black (n=45/68, 66%), although racial composition varied by region; 86% (6/7) of index cases in Region 1 were white, compared to only 24% (6/25) in Region 2. Among the RHI cases, most (10/13; 77%) were categorized based on documented negative HIV-testing within three months of diagnosis.
Table 1.
Demographic and clinical characteristics of index cases with acute HIV infection (n=68) reported in North Carolina and their contacts identified through contact tracing (n=210).
| Contact | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Characteristic | Index | HIV-infecteda n (%) |
HIV-negative n (%) |
HIV-status unknownb n (%) |
||
|
| ||||||
| Total | 68 | 71 | 66 | 73 | ||
| Reached for interview | 66 (97) | 68 (96) | 66 (100) | 14 (19) | ||
| Sex | ||||||
| Male | 58 (85) | 68 (96) | 58 (88) | 62 (85) | ||
| Female | 10 (15) | 1 (1) | 7 (11) | 4 (5) | ||
| Unknown/missing | 0 (0) | 2 (3) | 1 (2) | 7 (10) | ||
| Age at investigation | ||||||
| <30 years | 42 (62) | 44 (62) | 43 (65) | 28 (38) | ||
| ≥30 years | 26 (38) | 23 (32) | 22 (33) | 26 (36) | ||
| Unknown/missing | 0 (0) | 4 (6) | 1 (2) | 19 (26) | ||
| Race/ethnicity | ||||||
| Black, non-Hispanic | 45 (66) | 51 (72) | 37 (56) | 37 (51) | ||
| White, non-Hispanic | 21 (31) | 12 (17) | 24 (36) | 18 (25) | ||
| Other/missing | 2 (3) | 8 (11) | 5 (8) | 18 (25) | ||
| Region of residence / % with HIV sequences | ||||||
| 1 – Black Mountain | 7 (10) | / 71% | 5 (7) | / 0% | 10 (15) | 2 (3) |
| 2 – Charlotte | 25 (37) | / 68% | 29 (41) | / 52% | 21 (32) | 18 (25) |
| 3 – Winston-Salem | 9 (13) | / 56% | 5 (7) | / 80% | 6 (9) | 7 (10) |
| 4 - Raleigh | 14 (21) | / 50% | 15 (21) | / 60% | 3 (5) | 12 (16) |
| 5 - Fayetteville | 4 (6) | / 0% | 6 (8) | / 50% | 7 (11) | 1 (1) |
| 6 – Greenville | 6 (9) | / 50% | 2 (3) | / 100% | 11 (17) | 7 (10) |
| 7 - Wilmington | 3 (4) | / 33% | 3 (4) | / 33% | 1 (2) | 0 (0) |
| Out of State/unknown | 0 | 6 (8) | / 0% | 7 (11) | 26 (36) | |
| HIV previously diagnosed | --- | 54 (76) | --- | --- | ||
| Diagnosed ≥5 years agoc | --- | 21 (39) | --- | --- | ||
| HIV RNA>1000 copies/mL or missingc,d | --- | 38 (70) | --- | --- | ||
Includes 20 second-degree HIV-infected contacts
Includes one second-degree contact with unknown HIV-status
Proportion of those with previously diagnosed HIV
HIV RNA viral load reported to the NC DHHS most closely to the time of index case investigation among the 54 contacts with previous HIV diagnoses
Some columns may not sum to 100% due to rounding
From the 66 index cases interviewed, 210 unique contacts were reported (Table 1). Of these, 189 were first-degree contacts (yielding 197 partnerships: three contacts were named more than once and four contacts were index cases) and 21 were second-degree contacts. All partnerships were sexual (93%) or social (7%), and none were identified as IDU. Index cases reported a mean of 3.0 first-degree contacts (SD 2.0). Of the 189 first-degree contacts, 51 (27%) were HIV-infected, 66 (35%) were HIV-uninfected, and 72 (38%) had unknown HIV status. Nearly one-third of the 189 first-degree contacts (33%, n=62/189) could not be located for DIS interviews; 38/189 (61%) were anonymous partnerships and 24/189 (39%) had incomplete information or could not be found. Fifteen contacts resided outside of NC.
Most HIV-infected first-degree contacts (71%; 36/51) were previously diagnosed with a median of 4.2 (IQR 1.4–6.8) years elapsed between contact and case diagnoses. Two contacts were named by more than one index case and were assessed at the time of the first index case’s diagnosis. Most previously diagnosed contacts (75%) had HIV viremia (RNA>1000 copies/mL) at their most recent care visit or had no HIV RNA reported, suggesting a lack of care engagement. The 15 first-degree contacts with new HIV diagnoses were diagnosed an average of 10 days after the corresponding index case (range 25 days prior to 39 days after index case). Of the 21 second-degree contacts, 20 (95%) were HIV-infected and one (5%) had an unknown HIV status.
Contact Tracing Networks
In the statewide network, we identified 58 independent components (median 4 [range 1–28] individuals per component) [Figure, Supplemental Digital Content 1]. The mean degree in the statewide network was 1.7 (SD = 1.5) and the overall network density was low at 1.2% of actual contact ties among all possible connections in the network. Within the investigation focal area (Regions 1 and 2), the overall density was slightly higher at 2.5%. Statewide, 52% components (30/58) included at least one individual from these regions.
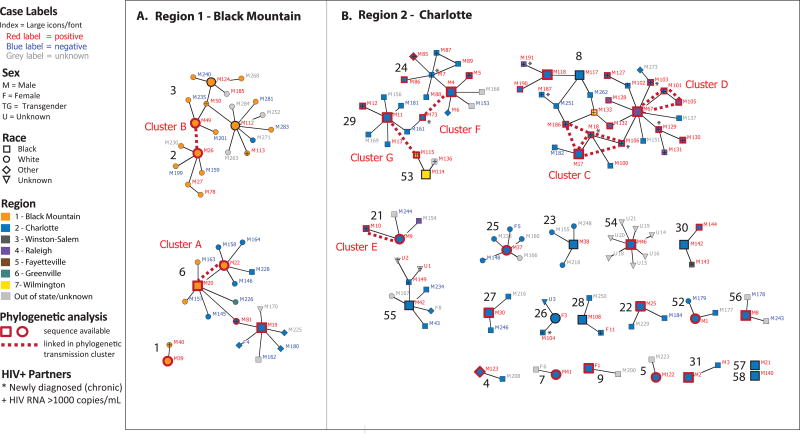
The 32 index cases reported in Regions 1 and 2 were distributed in 26 components, including two index cases unreachable by DIS (Figure 2). The network included 118 discrete contacts (100 first-degree and 18 second-degree) with median component size of three (range 2–28). Most components (85%: 22/26) contained index cases exclusively from Region 2. However, one (component #6) contained index cases from both Regions 1 and 2, and three components (components #1, 2, and 3) contained index cases exclusively from Region 1. The mean index node degrees by contacts’ HIV statuses were 0.8 (SD=0.9) for HIV-infected contacts, 1.0 (SD=1.2) for HIV-negative contacts, and 1.3 (SD=1.5) for contacts with unknown HIV status.
Figure 2.
Contact tracing network components stemming from index cases diagnosed during acute HIV infection (n=32) and their reported partners (n=118 non-index cases) investigated in two regions in North Carolina, 2012–2014. Components are labeled with black numbers and index cases nodes are large while contacts nodes are small. HIV-infected cases and contacts have red labels and those with available sequences are indicated with red outlines. Dashed lines depict cases linked in phylogenetic clusters and are labeled with cluster names (A–G). Panel A. Region 1 (Black Mountain) involves 4 components, including components 3 and 2 which are linked by phylogenetic cluster (B) but not by contact tracing. Panel B. Region 2 (Charlotte), where most index cases were found in small components not linked in clusters. One large component (8) involves two distinct phylogenetic clusters (C & D) and multiple index cases not in clusters. Clusters F and G phylogenetically link cases that are not connected in contact trancing components.
HIV-1 Sequence Analyses and Comparison to Contact Network
HIV-1 pol sequences were available for 56% (38/68) of the index cases and 48% (34/71) HIV-infected contacts (Table 1). Sequence availability did vary by region with a higher proportion in Region 1 (71%) and Region 2 (68%). Time from diagnosis to drug resistance sequencing was shorter for index cases compared to contacts (median 19 [IQR 12–34] versus 43 [IQR 17–529] days for contacts). All sequences from index cases were sampled prior to ART; antiretroviral exposure data were not available for contacts. All sequences were subtype B.
We identified 13 transmission clusters involving >one index or an index and any contact (Table 2; Figure, Supplemental Digital Content 2). These clusters included 30 individuals (range 2–4 members), involving 42% (16/38) of the index cases and 41% (14/34) of the contacts with sequences. Six clusters (46%) involved an index from Region 1 or 2; only three clusters spanned more than one region. While most clusters contained only MSM (n=11) there two clusters involved women (Clusters L and M). Additionally, nearly one-third of clusters (31%; 4/13) included a previously diagnosed contact. Many clusters were sub-clusters on highly supported clades (with larger genetic distance spans) with reference sequences (Figure, Supplemental Digital Content 2). Additionally, two index cases that did not cluster with a case or contact sequence clustered closely (<1.5% pairwise genetic distance difference) with a reference sequence.
Table 2.
Characteristics of phylogenetic transmission clusters involving sequences from index cases diagnosed during acute HIV infection in North Carolina.
| Transmission Cluster |
Network Component |
ID | Region | Case | Race | Sex | Diagnosis Year |
DRM |
|---|---|---|---|---|---|---|---|---|
| A | 6 | M20 | 1 – Black Mountain | Index | Black | M | 2014 | --- |
| 6 | M22 | 1 – Black Mountain | Index | White | M | 2014 | --- | |
|
| ||||||||
| B | 2 | M26 | 1 – Black Mountain | Index | White | M | 2014 | --- |
| 3 | M49 | 1 – Black Mountain | Index | White | M | 2014 | --- | |
|
| ||||||||
| C | 8 | M17 | 2 – Charlotte | Index | Black | M | 2013 | K103N, P225H |
| 8 | M106 | 2 – Charlotte | First-degree | Black | M | 2014 | K103N | |
| 8 | M18 | 2 – Charlotte | First-degree | Black | M | 2013 | K103N | |
| 8 | M186 | 2 – Charlotte | Second-degree | Black | M | 2014 | K103N | |
|
| ||||||||
| D | 8 | M67 | 4 – Raleigh | Index | Black | M | 2014 | --- |
| 8 | M101 | 2 – Charlotte | First-degree | Black | M | 2002a | --- | |
| 8 | M105 | 4 – Raleigh | Second-degree | Black | M | 2013a | --- | |
|
| ||||||||
| E | 21 | M9 | 2 – Charlotte | Index | White | M | 2014 | L100I, K103N |
| 21 | M10 | 2 – Charlotte | First-degree | Black | M | 2012a | L100I, K103N | |
|
| ||||||||
| F | 24 | M4 | 2 – Charlotte | Index | Black/M | M | 2014 | --- |
| 29 | M73 | 2 – Charlotte | Second-degree | Black/M | M | 2014 | --- | |
|
| ||||||||
| G | 29 | M11 | 2 – Charlotte | Index | Black / M | M | 2013 | --- |
| 53 | M115 | 7 - Wilmington | First-degree | Black / M | M | 2006a | --- | |
|
| ||||||||
| H | 51 | M14 | 3 – Winston-Salem | Index | Black / M | M | 2014 | --- |
| 51 | M16 | 3 – Winston-Salem | First-degree | Black / M | M | 2014 | --- | |
|
| ||||||||
| I | 47 | M35 | 3 – Winston-Salem | Index | Black / M | M | 2014 | --- |
| 47 | M36 | 3 – Winston-Salem | First-degree | Black / M | M | 2014a | --- | |
|
| ||||||||
| J | 43 | M116 | 4 – Raleigh | Index | Black/M | M | 2014 | K101E, K103N |
| 43 | M141 | 5 - Fayetteville | First-degree | Black/M | M | 2014 | K101E, K103N | |
|
| ||||||||
| K | 36 | M47 | 4 – Raleigh | Index | Black / M | M | 2014 | --- |
| 36 | M48 | 4 – Raleigh | First-degree | Black / M | M | 2014 | --- | |
|
| ||||||||
| L | 19 | F13 | 4 – Raleigh | Index | White/F | F | 2014 | --- |
| 19 | M135 | 4 – Raleigh | First-degree | White/M | M | 2014 | --- | |
|
| ||||||||
| M | 39 | F10 | 6 – Greenville | Index | Black | F | 2014 | K103N |
| 40 | F15 | 6 – Greenville | Index | Black | F | 2014 | K103N | |
| 40 | M138 | 6 – Greenville | First-degree | Black | M | 2014 | K103N | |
M, Male; F, Female; DRM, Drug Resistance Mutation
Contact previously diagnosed (>30 days prior to the index case)
Major DRMs to non-nucleoside reverse transcriptase inhibitors (NNRTIs) were common. Six (16%) index cases had transmitted drug resistance (TDR); five of these were in four clusters where all members shared the same mutations, including all with K103N (Table 2). Among contacts, 38% (13/34) had a major DRM (nearly all to NNRTIs). No major DRMs in protease or integrase (among 20 integrase sequences) were identified among index or contact cases.
While a single clustered outbreak was not identified, there are notable findings in comparing the contact and phylogenetic networks. Four clusters connected more than one network component revealing links not apparent from contact tracing (including MSM Clusters B, F, and G in in Regions 1 and 2). Additionally, components #40 and #39 in Region 6, involving two women and one man, were phylogenetically linked (Cluster M). The largest component (#8, n=28 cases) included members identified in distinct phylogenetic clusters (Clusters C and D; Figure 2). This component included 18 HIV-infected contacts and four index cases from Regions 2 and 4, demonstrating geographic bridging of clusters and components.
DISCUSSION
We conducted an integrated phylogenetic and contact tracing investigation to assess for a connected HIV outbreak after the spatiotemporal clustering of AHI was reported in NC. The analyses demonstrate that the observed spatiotemporal clustering was not a product of the expansion of a single linked cluster, but rather of the concurrent expansion of several smaller clusters. These ongoing HIV transmission chains were embedded in sexual network components that appeared largely disconnected both within and between regions across the state. The rise in AHI observed may be a random data artifact or the result of improved screening or reporting of AHI. Nonetheless, our study demonstrates the feasibility and applicability of incorporating HIV sequence analyses into an AHI outbreak investigation.
This investigation represents a novel integration of routinely generated contact tracing and sequence data to assess a potential HIV outbreak. Genetic networks have been widely used retrospectively to characterize local,16 national,17 and international HIV clusters18 and contact tracing has long served as a backbone of infection control. However, only a few studies integrated both methods during an HIV investigation8,19 or retrospectively to confirm outbreaks attributed to IDU.9 Among a cluster of HIV cases among rural MSM, phylogenetics showed HIV transmission arose from separate sources.8 In another study of MSM, sexual network components that were not connected through contact tracing were genetically linked.19 These findings are congruent with our results showing multiple transmission chains and links between network components. Additionally, our analyses show significant clustering of TDR, which highlights an additional advantage of tracking sequence data. Clustering analyses may inform public health efforts in the event of a future TDR outbreak. All TDR in our study included K103N, a mutation that is increasingly linked to ART-naïve transmission sources.20,21
Our primary objective was to test for a genetically clustered outbreak, rather than identify likely transmitters to AHI index cases. However, social and genetic networks combined can inform the use of genetic linkage to identify potential transmission partners.22 Recently proposed methods to integrate both networks are all reliant on relative completeness of both networks and minimal time delays between infection, diagnosis, contact tracing, and sequencing.23–26 Our study is limited in that not all diagnosed cases had a sequence available for analysis, thus providing a restricted view of the transmission network. Further, nearly one-third of contacts were not located and may be important connections in the transmission network. These analyses also cannot confirm direction of transmission, and unsampled third parties, including undiagnosed persons, may be involved in the transmission chain. Thus, observed phylogenetic linkages may be indirect in reality. Despite these limitations, over half of the index cases in the investigation foci had sequences, and phylogenetic analyses showed multiple clusters rather than one cluster; adding more sequences may increase the size and number of these clusters. With availability of more complete data, future investigation into source attribution can further inform local HIV transmission dynamics and outbreak investigations. Future analyses would also benefit from the inclusion of sequencing data from neighboring states, particularly given high number of out of state partners reported in contact tracing.
The network analyses revealed a low yield from HIV case finding, whether previously or newly diagnosed. Both network densities statewide and in the investigation foci were low, indicating that cases (diagnosed or undiagnosed) in the community are not being identified and that chronically-infected individuals may be important contributors to ongoing transmission.27 Prior studies have shown variable success of HIV case finding from contact tracing.28,29 Contact tracing of AHI may theoretically yield higher case finding compared to chronic cases because DIS expedites the investigation and direct attention to higher risk networks. Additionally, AHI cases may have less recall bias due to the condensed infectious period for potential contacts. However, the index node degree was low at 0.8 for HIV-infected contacts and higher at 1.3 for HIV-unknown status contacts. The mean index node degree for HIV-infected contacts indicates the mean number of contacts named by index cases who were diagnosed with HIV, excluding any potential HIV-infected contacts who could not be reached by DIS and any anonymous contacts. These findings largely stem from the substantial proportion of cases that could not be located and altogether indicate the need for improved case detection.
Our exploratory investigation revealed multiple HIV transmissions clusters, involving both new and previously diagnosed cases, which reinforces the need for a comprehensive HIV prevention strategy. Enhanced prevention measures towards transmission clusters and highly connected contact networks could be considered, including intensified, expedited partner and linkage to care services for HIV-infected cases and pre-exposure prophylaxis for HIV-negative cases. Additionally, innovative methods are needed to better identify high-risk networks and to improve case finding. In this study, no cases reported IDU contacts which is consistent with our earlier phylogenetic studies in NC showing predominately sexual transmission.16 However, continued surveillance is important given increased rates of opioid drug and the recent HIV outbreak associated with IDU in Indiana.9 In support of these efforts, NC-DHHS is currently establishing a formal system for routine analysis of HIV sequences as is done through sequences reported through the National HIV Surveillance System.30 Such an expanded surveillance system will allow for more timely and complete identification of HIV transmission clusters, a limitation of this study. Combined with ongoing analysis of surveillance data, such a system could allow for improved epidemic monitoring including for TDR, outbreak detection, and DIS response.
Conclusions
The observed spatiotemporal clustering in this putative outbreak was a product of the concurrent expansion of several smaller clusters, rather than a single outbreak cluster. Integration of contact tracing and phylogenetic networks may provide insights into HIV transmission dynamics and may be conducted in near real-time to support outbreak investigations. Further development of methods to enhance the detection of and response to HIV transmission and contact networks is needed.
Supplementary Material
Acknowledgments
We thank the disease intervention specialists of North Carolina and the people they worked with and served during this investigation.
Funding Sources: Research reported in this publication was supported by the National Insitute of Allergy and Infectious Diseases of the National Institutes of Health under Award Numbers K08AI112432-0 and 5T32AI070114-10. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
References
- 1.North Carolina HIV/STD Surveillance Unit. North Carolina HIV/STD Epidemiologic Profile 2013. Issued March 2015. North Carolina Department of Health and Human Services; Raleigh, North Carolina: [Accessed July 12, 2016]. 2015. Available from: http://epi.publichealth.nc.gov/cd/stds/figures/Epi_Profile_2013.pdf. [Google Scholar]
- 2.Brenner BG, Roger M, Routy JP, et al. High rates of forward transmission events after acute/early HIV-1 infection. The Journal of Infectious Diseases. 2007;195(7):951–959. doi: 10.1086/512088. [DOI] [PubMed] [Google Scholar]
- 3.Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS medicine. 2008;5(3):e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuruc JD, Cope AB, Sampson LA, et al. Ten Years of Screening and Testing for Acute HIV Infection in North Carolina. Journal of acquired immune deficiency syndromes (1999) 2016;71(1):111–119. doi: 10.1097/QAI.0000000000000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Resik S, Lemey P, Ping L, et al. Limitations to contact tracing and phylogenetic analysis in establishing HIV type 1 transmission networks in Cuba. AIDS Res Hum Retroviruses. 2007;23(3):347–356. doi: 10.1089/aid.2006.0158. [DOI] [PubMed] [Google Scholar]
- 6.Hassan AS, Pybus OG, Sanders EJ, Albert J, Esbjornsson J. Defining HIV-1 transmission clusters based on sequence data. AIDS. 2017;31(9):1211–1222. doi: 10.1097/QAD.0000000000001470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenner B, Wainberg MA, Roger M. Phylogenetic inferences on HIV-1 transmission: implications for the design of prevention and treatment interventions. AIDS. 2013;27(7):1045–1057. doi: 10.1097/QAD.0b013e32835cffd9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nett RJ, Bartschi JL, Ellis GM, et al. Two clusters of HIV-1 infection, rural Idaho, USA, 2008. Emerg Infect Dis. 2010;16(11):1807–1809. doi: 10.3201/eid1611.100857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters PJ, Pontones P, Hoover KW, et al. HIV Infection Linked to Injection Use of Oxymorphone in Indiana, 2014–2015. New England Journal of Medicine. 2016;375(3):229–239. doi: 10.1056/NEJMoa1515195. [DOI] [PubMed] [Google Scholar]
- 10.Poon AF, Gustafson R, Daly P, et al. Near real-time monitoring of HIV transmission hotspots from routine HIV genotyping: an implementation case study. Lancet HIV. 2016;3(5):e231–238. doi: 10.1016/S2352-3018(16)00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pilcher CD, Fiscus SA, Nguyen TQ, et al. Detection of acute infections during HIV testing in North Carolina. New England Journal of Medicine. 2005;352(18):1873–1883. doi: 10.1056/NEJMoa042291. [DOI] [PubMed] [Google Scholar]
- 12.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis. 2006;42(11):1608–1618. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett DE, Camacho RJ, Otelea D, et al. Drug Resistance Mutations for Surveillance of Transmitted HIV-1 Drug-Resistance: 2009 Update. PLoS One. 2009;4(3):e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 16.Dennis AM, Hue S, Hurt CB, et al. Phylogenetic insights into regional HIV transmission. AIDS. 2012;26(14):1813–1822. doi: 10.1097/QAD.0b013e3283573244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aldous JL, Pond SK, Poon A, et al. Characterizing HIV Transmission Networks Across the United States. Clin Infect Dis. 2012;55(8):1135–1143. doi: 10.1093/cid/cis612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wertheim JO, Leigh Brown AJ, Hepler NL, et al. The global transmission network of HIV-1. The Journal of infectious diseases. 2013;209(2):304–313. doi: 10.1093/infdis/jit524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peters PJ, Gay C, Beagle S, Shankar A, Switzer WM, Hightow-Weidman LB. HIV Infection Among Partners of HIV-Infected Black Men Who Have Sex with Men - North Carolina, 2011–2013. MMWR Morb Mortal Wkly Rep. 2014;63(5):90–94. [PMC free article] [PubMed] [Google Scholar]
- 20.Mbisa JL, Fearnhill E, Dunn DT, et al. Evidence of Self-Sustaining Drug Resistant HIV-1 Lineages Among Untreated Patients in the United Kingdom. Clin Infect Dis. 2015;61(5):829–836. doi: 10.1093/cid/civ393. [DOI] [PubMed] [Google Scholar]
- 21.Drescher SM, von Wyl V, Yang WL, et al. Treatment-naive individuals are the major source of transmitted HIV-1 drug resistance in men who have sex with men in the Swiss HIV Cohort Study. Clin Infect Dis. 2014;58(2):285–294. doi: 10.1093/cid/cit694. [DOI] [PubMed] [Google Scholar]
- 22.Wertheim JO, Kosakovsky Pond SL, Forgione LA, et al. Social and Genetic Networks of HIV-1 Transmission in New York City. PLoS Pathogens. 2017;13(1):e1006000. doi: 10.1371/journal.ppat.1006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCloskey RM, Liang RH, Poon AFY. Reconstructing contact network parameters from viral phylogenies. Virus Evolution. 2016;2(2):vew029. doi: 10.1093/ve/vew029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villandre L, Stephens DA, Labbe A, et al. Assessment of Overlap of Phylogenetic Transmission Clusters and Communities in Simple Sexual Contact Networks: Applications to HIV-1. PLoS One. 2016;11(2):e0148459. doi: 10.1371/journal.pone.0148459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jombart T, Cori A, Didelot X, Cauchemez S, Fraser C, Ferguson N. Bayesian Reconstruction of Disease Outbreaks by Combining Epidemiologic and Genomic Data. PLoS Computational Biology. 2014;10(1):e1003457. doi: 10.1371/journal.pcbi.1003457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zarrabi N, Prosperi MCF, Belleman RG, et al. Combining social and genetic networks to study HIV transmission in mixing risk groups. The European Physical Journal Special Topics. 2013;222(6):1377–1387. [Google Scholar]
- 27.Cope AB, Powers KA, Kuruc JD, et al. Ongoing HIV Transmission and the HIV Care Continuum in North Carolina. PLoS ONE. 2015;10(6):e0127950. doi: 10.1371/journal.pone.0127950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golden MR, Hogben M, Potterat JJ, Handsfield HH. HIV Partner Notification in the United States: A National Survey of Program Coverage and Outcomes. Sexually Transmitted Diseases. 2004;31(12):709–712. doi: 10.1097/01.olq.0000145847.65523.43. [DOI] [PubMed] [Google Scholar]
- 29.Brewer DD. Case-Finding Effectiveness of Partner Notification and Cluster Investigation for Sexually Transmitted Diseases/HIV. Sexually Transmitted Diseases. 2005;32(2):78–83. doi: 10.1097/01.olq.0000153574.38764.0e. [DOI] [PubMed] [Google Scholar]
- 30.Oster AM, Wertheim JO, Hernandez AL, Ocfemia M, Saduvala N, Hall HI. Using Molecular HIV Surveillance Data to Understand Transmission Between Subpopulations in the United States. Journal of acquired immune deficiency syndromes (1999) 2015;70(4):444–451. doi: 10.1097/QAI.0000000000000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.