Abstract
Background & Aims
The incidence of colorectal cancer (CRC) in individuals younger than 50 years old is increasing. We sought to ascertain the proportion of young CRC cases associated with genetic predisposition.
Methods
We performed a retrospective study of individuals diagnosed with CRC at an age younger than 50 years, evaluated by the clinical genetics service at a single tertiary care cancer center from 1998 through 2015. We collected data on patient histories, tumor phenotypes, and results of germline DNA sequencing. For subjects with uninformative clinical evaluations, germline DNA samples were (re)sequenced using a research-based next-generation sequencing multigene panel. The primary outcome was identification of a pathogenic germline mutation associated with cancer predisposition.
Results
Of 430 young CRC cases, 111 (26%) had a first-degree relative with CRC. Forty-one of the subjects with CRC (10%) had tumors with histologic evidence for mismatch repair deficiency. Of 315 subjects who underwent clinical germline sequencing, 79 had mutations associated with a hereditary cancer syndrome and 21 had variants of uncertain significance. Fifty-six subjects had pathogenic variants associated with Lynch syndrome (25 with mutations in MSH2, 24 with mutations in MLH1, 5 with mutations in MSH6, and 2 with mutations in PMS2) and 10 subjects had pathogenic variants associated with familial adenomatous polyposis. Thirteen subjects had mutations in other cancer-associated genes (8 in MUTYH, 2 in SMAD4, 1 in BRCA1, 1 in TP53, and 1 in CHEK2), all identified through multigene panel tests. Among 117 patients with uninformative clinical evaluations, next-generation sequence analysis using a multigene panel detected actionable germline variants in 6 patients (5%). Only 43 of the 85 subjects with germline mutations associated with a hereditary cancer syndrome (51%) reported a CRC diagnosis in a first-degree relative.
Conclusions
Approximately 1 in 5 individuals diagnosed with CRC at age younger than 50 years carries a germline mutation associated with cancer; nearly half of these do not have clinical histories typically associated with the identified syndrome. Germline testing with multigene cancer panels should be considered for all young patients with CRC.
Keywords: NGS, FDR, APC, risk
Introduction
Although implementation of routine screening has resulted in overall reductions in both colorectal cancer (CRC) incidence and mortality among individuals age>50(1), CRC incidence among individuals age<50 years is rising and is projected to double by 2030 (2–5). Since nearly 1 in 10 new CRC diagnoses involves individuals younger than age 50(6), there is a need to identify young individuals at increased risk who would benefit from early screening.
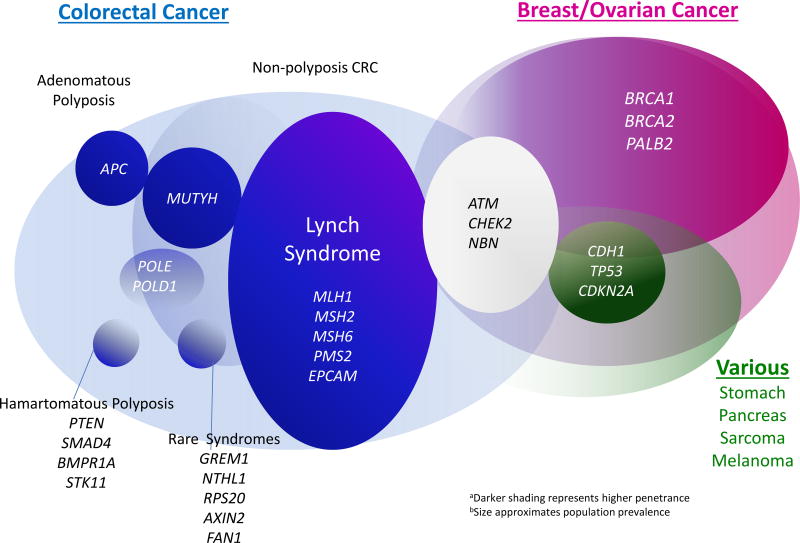
Clinical guidelines for CRC risk stratification rely largely on patients’ family history of CRC and number and histology of colorectal polyps (7). Historically, hereditary cancer syndromes have been implicated in 3–5% of CRCs overall (8), with individuals who harbor germline mutations in specific genes at high risk for developing CRC at young ages. Lynch Syndrome, the most common hereditary cancer syndrome associated with predisposition to CRC, is associated with germline mutations in DNA mismatch repair (MMR) genes MLH1, MSH2, MSH6, PMS2, and EPCAM. Lynch Syndrome has been consistently implicated in 2–3% of unselected CRC cases(9–12) justifying the recommendation that all CRC tumors be screened for mismatch repair deficiency with microsatellite instability (MSI) or immunohistochemistry for DNA MMR proteins(13). Familial Adenomatous Polyposis (FAP) associated with germline mutations in the APC tumor suppressor gene has been implicated in 1% of CRCs. Although APC mutation carriers typically develop classic colonic polyposis (100–1000s adenomas), attenuated phenotypes (20–100 polyps) are increasingly recognized, prompting the recommendation that individuals with >20 colorectal polyps be offered genetic testing(14–17). Germline mutations in additional high and moderate penetrance cancer genes have also been associated with increased risk for colorectal neoplasia (Figure 1).(12, 15, 18)
Figure 1.
Genes implicated in High and Moderate Penetrance Hereditary Predisposition to Cancer
To date, the clinical practice standard has been to offer germline genetic testing selectively to patients who meet clinical criteria for known hereditary cancer syndromes, with sequencing of germline DNA limited to highly penetrant genes associated with specific clinical phenotypes. However, recent studies which have extended germline genetic testing using multigene panels to patients with breast cancer, ovarian cancer, and/or CRC have discovered germline mutations that would not have been predicted based on the patients’ clinical histories (12, 19–22). Although diagnosis of CRC at age<50 is an indication for referral for genetic evaluation(17), prevalence estimates for germline mutations in young CRC patients range from 5–35% (23–30) depending on how subjects were ascertained and evaluated. Current algorithms used in clinical genetic risk assessment rely on family history and tumor phenotype to identify individuals who warrant germline DNA sequencing of genes selected based on clinical history. Given the variability in disease spectrum and potential overlap in clinical manifestations of germline mutations associated with moderate or high cancer risk, we hypothesized that current clinical approaches fail to detect a significant proportion of young CRC patients with actionable germline mutations. We conducted a retrospective review of outcomes of clinical genetic evaluations of patients diagnosed with CRC age<50 and examined the incremental yield of employing next generation sequencing (NGS) multigene panels for retesting patients whose initial evaluations were clinically uninformative.
Patients and Methods
We conducted retrospective chart reviews of individuals diagnosed with CRC age<50 evaluated by the clinical genetics service at a single tertiary care cancer center (University of Michigan Comprehensive Cancer Center) between 1998–2015. All subjects had undergone formal genetic risk assessment performed by a genetic counselor and physician in the cancer genetics specialty clinic as part of clinical care. Subjects whose personal and/or family history met criteria for clinical genetic testing for one or more hereditary cancer syndromes underwent genetic counseling about the risks and benefits of clinical genetic testing and submitted a peripheral blood sample, from which germline DNA was isolated for clinical sequencing. The decisions regarding which genes to sequence for each subject were made by clinicians according to clinical best practices at the time of the patient’s evaluation. All patients evaluated were invited to participate in the IRB-approved cancer genetics registry and donate a peripheral blood sample for research. Subject demographics, clinical history, family cancer history (3 generation pedigrees), colonoscopy findings, CRC stage, tumor location, tumor histology and phenotype, and outcomes of clinical genetic testing were obtained through review of medical records.
Clinical Genetic Testing
Clinical sequencing of germline DNA for mutations in one or more genes was performed at one of seven CLIA-certified commercial genetic testing laboratories using either Sanger sequencing or next generation sequencing (NGS) in accordance with current practice standards. The specific genes sequenced for each subject were selected by the clinicians at the time of the clinical visit, based on review of each subject’s personal and family history. Sequence variants were reported by the clinical testing laboratory as pathogenic, likely pathogenic, variant of uncertain significance (VUS), likely benign, or benign according to consensus guidelines.(31)
Research-based Sequencing
Archived samples of germline DNA from subjects whose clinical genetic evaluation was uninformative either because 1) they did not undergo clinical germline DNA sequencing or 2) clinical sequencing failed to identify a pathogenic germline mutation, were submitted for research-based NGS sequencing using one of two multigene panels. Initially, 16 germline samples were sequenced using a commercially available cancer gene panel which included 124 genes implicated in various cancer types (Human Comprehensive Cancer Panel V1, Qiagen, Germany). Since several clinically relevant colorectal cancer genes (e.g. MUTYH, PMS2, POLE, POLD1) were not included on this particular multigene panel, sequencing of the remaining germline samples (N=101) was performed using a custom design NGS panel of 67 genes (Agilent SureSelect XT2 Custom Panel) selected by the investigators based on their associations with highly penetrant hereditary cancer syndromes, moderate increases in cancer risk, or suspected roles as somatic drivers. Genes included on each of the research-based multigene panels are listed in Appendix A.
DNA library preparation and NGS sequencing were carried out by the University of Michigan Sequencing core according to manufacturer’s recommended protocols on Illumina HiSeq instruments with target read depths of >50×. Bioinformatics analysis of NGS sequence data was performed by the UM Bioinformatics Core, with read mapping, variant calling and annotation performed using the Genome Analysis Toolkit (GATK) v3.3-2 using Broad Institute Best Practice guidelines. Reads were aligned to the hg19 human reference genome with BWA v0.7.8 and variants identified using the Broad UnifiedGenotyper with standard parameters and hard filters. Variants were annotated using GoldenHelix VarSeq v1.1.4 (Golden Helix, Bozeman, MT, USA) to draw attention to truncating variants (nonsense, frameshift deletions/insertions, highly conserved splice site mutations); RefSeq v105v2 gene models were used for annotation.
Risk Assessment
For each subject in whom a germline mutation was identified, family history of cancer (3 generation pedigree), personal history, and tumor/polyp histopathologic features were retrospectively re-examined by two clinicians and correlated with PREMM1,2,6 model scores(32) and NCCN criteria for genetic testing for CRC predisposition (17). Subjects were categorized according to level of suspicion for possible genetic predisposition (high, moderate, low) and each reviewer noted whether or not the subject met clinical diagnostic criteria for the genetic syndrome corresponding to the germline mutation identified.
Results
Four hundred and thirty patients with CRC diagnosed age<50 presented for clinical genetic evaluation during the 17 year study period. The average age of diagnosis of CRC was 40 years (range 16–49), with 38 individuals (8.8%) diagnosed at age<30. The majority (59%) of tumors were located in the distal colon or rectum and 10% had somatic testing demonstrating DNA mismatch repair deficient phenotypes with high levels of microsatellite instability (MSI-H). Twenty four (5.6%) subjects had polyposis phenotypes (defined as >20 polyps) at the time of their cancer diagnosis. Only 111 (26%) young CRC patients reported a family history of CRC in a first degree relative. (Table 1)
Table 1.
Characteristics of Subjects with CRC age<50
| Characteristics of Patients with Young Onset Colorectal Cancer (N=430) |
|||||
|---|---|---|---|---|---|
|
| |||||
| All Cases N=430 (100%) |
Positive for Mutation N=79 (18.4%) |
No mutation N=215 (50.0%) |
VUS N=21 (4.9%) |
Not tested N=115 (26.7%) |
|
|
| |||||
| Sex | |||||
| Female | 213 (49.5%) | 41 (51.9%) | 103 (47.9%) | 13 (61.9%) | 56 (48.7%) |
| Male | 217 (50.5%) | 38 (48.1%) | 112 (52.1%) | 8 (38.1%) | 59 (51.3%) |
|
| |||||
| Mean age dx (range) | 40.0 | 37.2 | 41.1 | 37.1 | 40.5 |
|
| |||||
| Race | |||||
| White | 388 (90.2%) | 75 (94.9%) | 198 (92.1%) | 15 (71.4%) | 100 (87.0%) |
| African American | 17 (4.0 %) | 0 (0.0%) | 6 (2.8%) | 4 (19.0%) | 7 (6.1%) |
| Asian | 17 (4.0%) | 4 (5.1%) | 6 (2.8%) | 2 (9.5%) | 5 (4.3%) |
| Other/unknown | 8 (1.9%) | 0 (0.0%) | 5 (2.3%) | 0 (0.0%) | 3 (2.6%) |
|
| |||||
| MSI-Status* | |||||
| MSI-H | 41 (9.5%) | 17 (21.5%) | 20 (9.3%) | 1 (4.8%) | 3 (2.6%) |
| MSS | 163 (37.9%) | 6 (7.6%) | 111 (51.6%) | 12 (57.1%) | 34 (29.6%) |
| Unknown | 226 (52.6%) | 56 (70.9%) | 84 (39.1%) | 8 (38.1%) | 78 (67.8%) |
|
| |||||
| Family history* | |||||
| FDR with CRC | 111 (25.8%) | 42 (53.2%) | 35 (16.3%) | 7 (33.3%) | 27 (23.5%) |
| Any relative with CRC | 246 (57.2%) | 66 (83.5%) | 109 (50.7%) | 13 (61.9%) | 58 (50.4%) |
| No history of CRC | 181 (42.1%) | 13 (16.5%) | 106 (49.3%) | 8 (38.1%) | 54 (47.0%) |
| Unknown | 3 (0.7%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 3 (2.6%) |
|
| |||||
| Tumor stage* | |||||
| 0 | 2 (0.5%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 2 (1.7%) |
| I | 52 (12.1%) | 20 (25.3%) | 23 (10.7%) | 3 (14.3%) | 6 (5.2%) |
| II | 43 (10.0%) | 11 (13.9%) | 27 (12.6%) | 0 (0.0%) | 5 (4.3%) |
| III | 124 (28.8%) | 13 (16.5%) | 76 (35.3%) | 8 (38.1%) | 27 (23.5%) |
| IV | 59 (13.7%) | 4 (5.1%) | 22 (10.2%) | 5 (23.8%) | 28 (24.3%) |
| Unknown | 149 (34.7%) | 30 (38.0%) | 67 (31.2%) | 5 (23.8%) | 47 (40.9%) |
|
| |||||
| CRC Site* | |||||
| Right colon | 115 (26.7%) | 28 (35.4%) | 50 (23.3%) | 5 (23.8%) | 32 (27.8%) |
| Left colon | 182 (42.3%) | 24 (30.4%) | 109 (50.7%) | 10 (47.6%) | 39 (33.9%) |
| Rectum | 72 (16.7%) | 6 (7.6%) | 47 (21.9%) | 5 (23.8%) | 14 (12.2%) |
| Not specified | 61 (14.2%) | 21 (26.6%) | 9 (4.2%) | 1 (4.8%) | 30 (26.1%) |
p<0.05 difference between mutation positive and mutation negative group
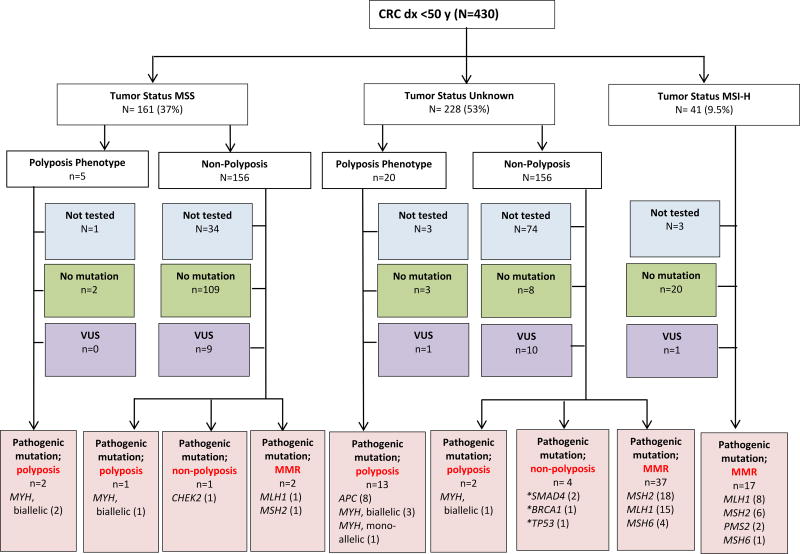
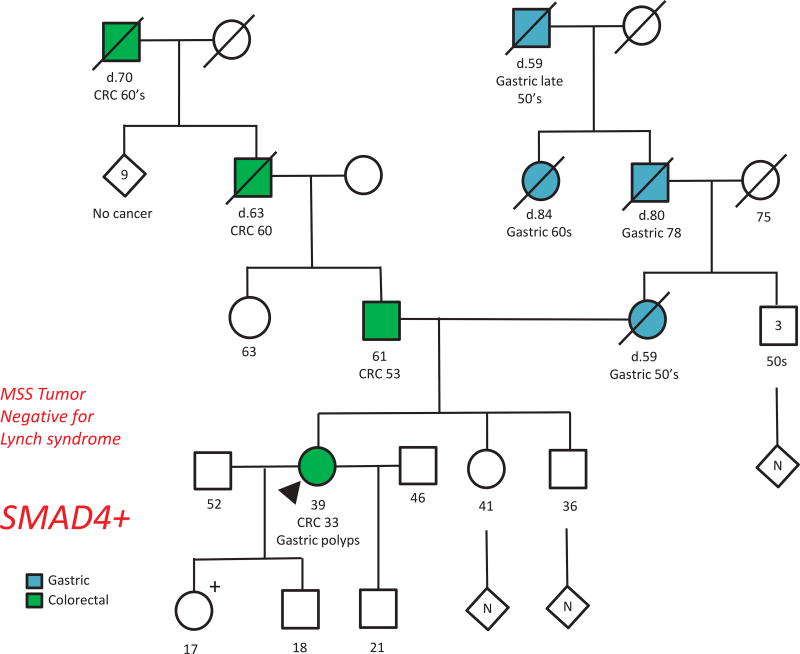
Clinical genetic testing with germline DNA sequencing by a CLIA-certified laboratory had been performed in 315 (73%) subjects, with multigene panel tests employed in 22 (7%). Pathogenic germline mutations in genes known to be associated with cancer predisposition were identified in 79 (18% of entire cohort, 25% of individuals who underwent clinical sequencing), with variants of uncertain significance (VUS) identified in an additional 21 (6.7%) individuals (9 (41%) VUS were identified on multigene panels) (Figure 2). The pathogenic germline mutations most frequently identified involved DNA MMR genes associated with Lynch Syndrome (N=56, 13% of entire cohort, 71% of all germline mutations), followed by APC (N=10, 2.3% of entire cohort, 13% of germline mutations) and MUTYH (N=8, 1.9% of entire cohort, 10% of germline mutations). Five individuals had pathogenic mutations in other genes known to be associated with genetic predisposition to cancer (SMAD4=2, BRCA1=1,TP53=1, CHEK2=1), all but one of these were identified on multigene panel tests. One patient diagnosed with a SMAD4 mutation on a multigene panel test had previously undergone a clinically uninformative genetic evaluation for a presumed diagnosis of attenuated adenomatous polyposis, with tumor testing demonstrating mismatch repair proficiency and germline sequencing revealing no pathogenic mutations in APC or MUTYH (Figure 3).
Figure 2.
Outcomes of genetic evaluation in subjects with CRC age<50
Figure 3.
Pedigree of Patient with a germline SMAD4 mutation identified on multigene panel
39F patient diagnosed with CRC at age 36 and report of “multiple” colorectal adenomas. At her initial presentation clinical genetic evaluation demonstrated that the colorectal tumor was microsatellite stable (MSS) and germline DNA sequencing identified no pathogenic mutations in APC or MUTYH. Patient returned 3 years later with anemia and underwent upper endoscopy which revealed numerous large gastric polyps which were described by outside pathology as hyperplastic. Clinical genetic testing with a multigene panel identified a pathogenic germline mutation in SMAD4, confirming diagnosis of Juvenile Polyposis Syndrome (JPS).
Comparison of individuals with and without germline mutations demonstrated that mutation carriers were more likely to have tumors located proximal to the splenic flexure (35% vs 23%), more likely to report a family history of CRC in a FDR (53% vs 16%), and less likely to have advanced disease stage (III–IV) at diagnosis (22% vs 46%). Phenotype of colorectal neoplasia was correlated with mutation status, with germline mutations more common among individuals whose tumors were MMR deficient (17/41 (40%), p=0.07) or who presented with multiple (>20) colorectal polyps clinical polyposis (15/25 (60%), p=0.1).
Research based NGS sequencing
Clinical germline sequencing was not performed in 115 (27%) subjects with young onset CRC, in most cases because patients failed to meet insurance criteria for genetic testing. Germline DNA from 117 subjects with CRC age<50 with prior “negative” clinical genetic evaluations (26 had previously undergone limited germline sequencing, while 91 had no prior germline testing) underwent targeted sequencing using the research multigene panels. An average of 5 germline variants were identified per subject (124 or 67 genes sequenced). Likely pathogenic germline mutations associated with known hereditary cancer syndromes were identified in 6 (5%) subjects (1 each in PMS2, MSH6, MUTYH, TP53, POLE, APC). Two subjects with TP53 and PMS2 mutations had mismatch repair proficient CRC tumors and had not previously had germline sequencing because authorization for clinical testing had been denied by their insurance company. The subject with MSH6 germline mutation also had a mismatch repair proficient tumor and had previously undergone clinical sequencing of MUTYH only. The subject with POLE germline mutation had <5 colorectal polyps and had previously undergone clinical sequencing for Lynch Syndrome with no mutations identified in the associated MMR genes. We made attempts to recontact the subjects with pathogenic variants identified on research-based testing to notify them that there was now “additional clinical genetic testing available that may be important for their care.” We were able to speak with only 2/6 subjects, both of whom declined to return for clinical confirmatory testing.
Effectiveness of Current Risk Assessment Algorithms for identifying mutation carriers
In all, clinical and research-based sequencing identified germline mutations in 85/430 (20%) individuals with CRC diagnosed at age<50. Among individuals with germline MMR mutations associated with Lynch Syndrome, only 29/58 (50%) met Amsterdam or Modified Amsterdam criteria and 4/58 (7%) had MMR proficient tumors. Among mutation carriers of any gene, 42/85 (49%) had no FDR affected with CRC. PREMM1,2,6 scores calculated including the proband’s CRC diagnosis were above the 5% threshold for referral for genetic testing in all but 2 mutation carriers (monoallelic MUTYH and CHEK2, both had mismatch repair proficient tumors and no family history of CRC). Among individuals with germline mutations in genes typically associated with polyposis phenotypes (APC, MUTYH, SMAD4, BMPR1A, POLE) 4/24 (17%) did not meet the threshold of >20 polyps required by most insurance carriers for coverage of germline sequencing. The individual with the BRCA1 mutation had no family history of CRC, but reported his mother and maternal aunt had been diagnosed with breast cancer at ages 36 and 50, respectively. After review of family history, polyp number and histology, and tumor MMR phenotype, 69/85 (81%) mutation carriers met NCCN diagnostic criteria for the syndrome corresponding to the germline mutation.
Discussion
In this retrospective cohort of patients referred for genetic evaluation based on personal history of CRC diagnosed age<50, 79/430 (18%) had pathogenic germline mutations associated with cancer predisposition identified through clinical sequencing. Further research-based multigene panel sequencing in a subset of subjects with previously uninformative genetic evaluations identified germline mutations in an additional 6 individuals, suggesting that as many as 1 in 5 young onset CRC cases can be attributed to hereditary cancer syndromes. While germline mutations in genes associated with Lynch Syndrome and FAP were the most common findings, approximately 1 in 8 mutation carriers had an alternate genetic diagnosis. Nearly 1 in 4 mutation carriers reported no family history of CRC in FDR and/or failed to meet diagnostic criteria associated with their germline mutation, suggesting the need to expand clinical genetic testing in young CRC patients beyond the current practice of selectively offering germline sequencing to only those who present with “classic” phenotypes typically associated with highly penetrant CRC syndromes.
Our findings complement those of other recent cohort studies seeking to define the contribution of genetics in the changing epidemiology of CRC. Historically, it had been estimated that hereditary cancer syndromes were implicated in up to 5% of all CRC cases.(5, 12) Based on previous published studies which noted that most young CRC patients had no family history of cancer with germline mutations identified in only 5–10% (23, 25), it had been assumed that the vast majority of cases were not “genetic.” However, it is important to note that these studies relied on family history and tumor phenotypes to select individuals for germline testing, with sequencing focusing largely on genes associated with Lynch Syndrome and FAP. More recent studies have demonstrated that germline sequencing using NGS multigene panels increases the yield of genetic testing, identifying pathogenic variants in as many as 10% of unselected CRC patients of all ages,(12) with half of the mutations involving high or moderate penetrance genes not usually considered in CRC (e.g. TP53, BRCA1, BRCA2, CHEK2, ATM)(12, 19, 22, 33). The Ohio Colorectal Cancer Prevention Initiative employed population-based ascertainment of individuals with CRC age<50 with universal germline genetic testing using a NGS panel of 25 cancer genes (APC, ATM, BARD1, BMPR1A, BRCA1, BRCA2, BRIP1, CDH1, CDK4, CDKN2A, CHEK2, EPCAM, MLH1, MSH2, MSH6, MUTYH, NBN, PALB2, PMS2, PTEN, RAD51C, RAD51D, SMAD4, STK11,TP53) identifying pathogenic germline variants in 72/450 (16%) (30). Among very young CRC patients (age<35) undergoing clinical genetics evaluations at a Texas academic cancer center, highly-penetrant hereditary cancer syndromes were identified in 67/193 (35%)(29). In our cohort of CRC patients age<50 referred for genetic evaluation, a combined approach of clinically-directed testing guided by the patients’ personal and family history, followed by expanded NGS (re)sequencing, identified pathogenic germline variants in cancer genes in 85/430 (20%). Together, these studies confirm that 1) the prevalence of genetic predisposition in young CRC patients is significantly higher than had been previously assumed and 2) current clinical strategies which rely on family history and tumor phenotypes to select individuals for germline testing often miss mutation carriers.
While our finding that 1 in 5 young CRC patients carries a germline mutation in a cancer predisposition gene falls in between the estimates from recent population-based (1 in 6)(30) and clinic based (1 in 3)(29) cohorts, it is worth noting the differences among these studies. All of the Ohio CRC patients age<50 underwent comprehensive germline testing with a 25 gene NGS multigene panel irrespective of family history or clinical phenotype. In contrast, in the Texas genetics clinic CRC patients age<35 were evaluated according to standard clinical practice, with syndrome specific genetic testing based on phenotype. In our study, CRC patients age<50 all had clinical evaluations as per “best practices” at the time, but only 2/3 actually went on to have clinical germline testing, most of which was performed with Sanger sequencing for ≤ 5 genes selected based on clinical phenotype. Only 1 in 3 subjects in our cohort had clinical sequencing using NGS multigene panels, therefore mutations in genes not traditionally associated with CRC (BRCA1, BRCA2, TP53, PALB2, CDKN2A) and or moderate penetrance cancer risks (CHEK2, ATM) accounted for a smaller proportion of the genetic diagnoses in our cohort compared to the Ohio cohort (4% vs 30%)(30). While our research-based NGS panels of 67 and 124 genes contained several cancer-relevant genes not evaluated in the Ohio study (e.g. POLE, POLD1, AXIN2), our large multigene panels did yield an average of 5 VUS per subject, many of these in genes whose roles in pathogenesis of CRC have not been well-characterized.
Our study adds to the literature in several important ways. First, it illustrates the shortcomings of clinical algorithms used to identify patients likely to benefit from genetic testing. Although the population prevalence of Lynch Syndrome is similar to that of Hereditary Breast Ovarian Cancer, young patients with CRC are significantly less likely than young women with breast cancer to be referred for genetic evaluation (34, 35). Even when young CRC patients present to genetics clinics, approximately one third do not undergo germline sequencing because they fail to meet clinical criteria required by their insurance for coverage of testing. Our finding of germline mutations in 20% of in young CRC patients is similar to the prevalence in women with ovarian cancer(20) for whom universal genetic testing is now recommended regardless of age at diagnosis or family history(36). Thus, a compelling case can be made for offering germline testing to all young CRC patients. Our finding that 1 in 20 CRC patients with “negative” clinical genetic evaluations were subsequently found to have germline mutations during (re)sequencing with an expanded multigene panel highlights the variability in disease penetrance, potential for overlap in phenotypes associated with known syndromes, and attests to the difficulty in predicting genotype based on history alone. Most of the patients in our study underwent their clinical evaluations before multigene panels were available. Although VUS results are more common with multigene panel testing,(37) the higher diagnostic yield and cost savings compared with stepwise disease specific testing are powerful arguments in favor of multigene panel tests, especially for patients in which the differential diagnosis includes more than one syndrome.(38)
We recognize our study has limitations. As 90% of study subjects were Caucasian, we are unable to determine whether there are racial differences in prevalence of germline mutations which might contribute to observed disparities in CRC incidence and survival(39, 40). As this is a retrospective analysis of patients referred for genetic evaluation at a tertiary care academic cancer center, we acknowledge the potential for ascertainment bias. Earlier studies have reported higher prevalence of Lynch Syndrome among young CRC patients evaluated in genetics clinics as compared to other healthcare settings, presumably because patients with a family history of CRC are more likely to be referred for genetic evaluation(26). Indeed, we made diagnoses of Lynch Syndrome in 13% of our clinic-based CRC patients age<50, compared with 8% of those in the Ohio population-based study(29, 30). Yet given that 75% of our subjects reported no FDR with CRC we feel the role of ascertainment bias was limited, and unlikely to lead to a significant overestimate of germline mutations. In fact, since all high and moderate/low penetrance cancer genes were not systematically evaluated and fewer than 1/3 subjects underwent sequencing using NGS multigene panels, the true prevalence of germline mutations in our cohort may be even higher. Since our research based sequencing was not comprehensive and analysis pipelines were not optimized for identifying insertions/deletions, it is possible that additional actionable mutations in known or novel cancer predisposition genes could have been missed.
Our findings illustrate the complexities of clinical care in the rapidly-changing landscape of genomic medicine and highlight some of the barriers to effective implementation of genetic risk assessment in “real world” settings. Although screening of CRC tumors for DNA mismatch repair deficiency is standard of care; tumor phenotype data were not available for half of the subjects in our cohort. And even when universal tumor testing was properly implemented, use of MSI-H tumor phenotype as a pre-requisite for germline testing resulted in clinically missed highly actionable genetic diagnoses of TP53, PMS2 and MSH6 mutations. Although genetic risk models (such as PREMM1,2,6) have proven useful for identifying individuals at risk for Lynch Syndrome in ambulatory settings,(41) these rely heavily on family history and have lower sensitivity for MSH6 and PMS2 carriers (42). As recent population-based studies suggest PMS2 and MSH6 are the most prevalent pathogenic germline DNA MMR variants, (43) lower disease penetrance and variability in MMR phenotype permit many of these families to escape clinical diagnosis (44). Since approximately 1 in 279 individuals carry germline MMR mutations(45), making the diagnosis has immediate implications not only for surgical decision-making (e.g. hemicolectomy vs subtotal colectomy) and choice of oncologic therapy (e.g. immune checkpoint inhibitor for hypermutated tumors) for CRC patients, but also for cancer surveillance for members of their immediate and extended families.
Although CRC incidence and mortality have declined overall since the implementation of CRC screening, they continue to rise among individuals age<50 (5). Early and more frequent colonoscopy is effective in reducing CRC incidence and mortality in individuals with Lynch Syndrome, and specialized surveillance has been recommended for individuals with germline mutations in other genes associated with high and moderate cancer risk, although for many of these (e.g. CHEK2 and ATM) the magnitude of risk increase for CRC and optimal surveillance interval remain to be defined.(17) While the time trends in incidence suggest dietary, environmental, and behavioral exposures (rather than “genetics”) are potential contributors to recent rise of CRC in young patients (46), additional study of germline and somatic characteristics associated with these colorectal neoplasms may provide important clues to pathogenesis. Interim advances in sequencing technologies, discovery of novel genes (e.g. NTHL1, BUB1, FAN1, FANCM, RSP20) (47) not included on currently available multigene panels, and growing appreciation for variability in disease penetrance and expressivity mean that for some patients today’s “negative” test may not be the final answer. Patients with suspicious histories and uninformative genetic test results should be counseled to check back periodically as additional information will likely become available that may inform approaches to cancer risk stratification, prevention and treatment.
In summary, we found that approximately 1 in 5 CRC patients diagnosed at age<50 carries a germline mutation in a gene associated with cancer predisposition. Nearly half of individuals with a genetic diagnosis did not exhibit the family history typically associated with the corresponding hereditary cancer syndrome. Making the diagnosis of a hereditary cancer syndrome has significant clinical implications, not only for care of the patient with CRC but also for management of their at-risk family members. As CRC is one of the most preventable cancers, it is important to identify individuals at high risk for colorectal neoplasia who would benefit from specialized surveillance for CRC (and potentially other extracolonic cancers) beginning long before age 50. Given rising incidence of young onset CRC and the availability of effective interventions, individuals with CRC age<50 should be referred for genetic evaluation, with strong consideration for use of multigene panels for germline sequencing.
Acknowledgments
The authors would like to thank Eva Yokosawa for her assistance with data compilation
Funding:
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers P30CA046592 and P50CA130810. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Appendix A
Classification of Genes Sequenced on Multigene Research Panels
Description
Bolded genes were included on both the 124 gene Qiagen Comprehensive Cancer panel and 67 gene Agilent custom panel. Underlined genes were included on the Agilent custom panel only. All others were included on the Qiagen Comprehensive Cancer panel only.
Classification of N=154 Genes
| Grouping | Genes |
|---|---|
| High Penetrance Syndromes, known to be associated with CRC risk | APC, BMPR1A, BRCA1, BRCA2, CDH1, GREM1, PTEN, SMAD4, STK11, TP53, EPCAM, MLH1, MSH2, MSH6, MUTYH, PMS2, POLD1, POLE, AXIN2 |
| High Penetrance Syndromes, associated primarily with risk of other cancers (non CRC) | BRCA1, BRCA2, CDKN2a, CDK4, MEN1, PALB2, RB1, SDHB, VHL, WT1, MET, PTCH1, RET, SMARCA4, SMARCB1, |
| Genes known to be associated with Moderate Penetrance Cancer Risk | ATM, BAP1, BARD1, BRIP1, CHEK2, FAT1, SMAD3, SOX9 IGF2R, HOXB12, NF1, NF2, NBN, RAD51, |
| Other genes potentially associated with cancer risk | |
| Oncogenes | AURKA, CASP8 (FLICE), ERBB3, JAK2, KIT (CD117), MYC, NRAS, PIK3CA ROS1, RUNX1 (AML1), SRC |
| Signal Transduction | |
| TGFβ | SMAD2, SMAD7, TGFBR2 |
| WNT | CYLD, CTNNB1, FAM123B, HNF1A, PPP2R1A |
| Notch | NOTCH1 FBXW7, GATA2, NOTCH2 |
| PI3K/AKT/PTEN | AKT1, AKT3, CBL, ERBB2, FLT1 (VEGFR1), FLT3, KDR (VEGFR3), PDGFRA, PDGFRB, PIK3C2A, PIK3R1, PIK3R5 |
| G-Protein Coupled Receptor | GNA11, GNAQ, GNAS, SMO, TSHR |
| Fibroblast Growth Factor | FGFR1, FGFR2, FGFR3, PTPN11 |
| Other | ERBB4, FKBP9, IGF1, IGF2, KRAS, MPL, MTOR, PRKAR1A, |
| Apoptosis | CTNNA1, TERT BRAF, CTSL1, FOXL2, GATA1, HRAS, KLLN, MAP2K4 (JNKK1), NPM1, TNFAIP3 |
| Angiogenesis | FIGF (VEGFD), FLT4 (VEGFR2), PTGS2 (COX2), |
| Adhesion | GALNT12, MAP2K1 (MEK1), PRKCE |
| Cell Cycle | ABL1, CDC73, PTPRC, TET2, TNKS, XPO1 |
| DNA Damage Response | ATR, EGFR, MSH3, PARP1 |
| Epigenetics | SYNE1 ARID1A, ASXL1, DNMT3A, EZH2, HDAC4, KDM6A, SETD2, TRRAP |
| Inflammatory Response | CEBPA, CSF1R, MYD88, PARP4 |
| Immune Response | ALK, CARD11, CRLF2, IL7R, JAK3, POLR3A, SOCS1 |
| Hypoxia | CREBBP, EP300 HSP90B1 (TRA1), NOS1 (nNOS) |
| Other | ABCC1, GRIN2A, IDH1, IDH2, NTN3, TOP1 |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: Dr. Stoffel and Ms. Koeppe had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study Concept and Design: Stoffel, Rozek, Everett, Koeppe,
Acquisition, Analysis or Interpretation of Data: Stoffel, Koeppe, Williams, Osborne, Everett, Hanson, Rozek, Ulintz, Kiel
Drafting of the Manuscript: Stoffel, Koeppe, Rozek
Critical Revision of the Manuscript for Important Intellectual Content: Stoffel, Rozek, Koeppe, Gruber, Everett, Osborne, Hanson, Williams, Ulintz
Statistical Analysis: Stoffel, Koeppe, Ulintz, Kiel
Disclosures:
The authors report no conflicts of interest related to this work.
References
- 1.Edwards BK, Ward E, Kohler BA, Eheman C, Zauber AG, Anderson RN, Jemal A, Schymura MJ, Lansdorp-Vogelaar I, Seeff LC, van Ballegooijen M, Goede SL, Ries LA. Annual report to the nation on the status of cancer, 1975–2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer. 2010;116(3):544–73. doi: 10.1002/cncr.24760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer JE, Narang T, Schnoll-Sussman FH, Pochapin MB, Christos PJ, Sherr DL. Increasing incidence of rectal cancer in patients aged younger than 40 years: an analysis of the surveillance, epidemiology, and end results database. Cancer. 2010;116(18):4354–9. doi: 10.1002/cncr.25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Jemal A, Ward EM. Increase in incidence of colorectal cancer among young men and women in the United States. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18(6):1695–8. doi: 10.1158/1055-9965.EPI-09-0186. [DOI] [PubMed] [Google Scholar]
- 4.Bailey CE, Hu CY, You YN, Bednarski BK, Rodriguez-Bigas MA, Skibber JM, Cantor SB, Chang GJ. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975–2010. JAMA surgery. 2015;150(1):17–22. doi: 10.1001/jamasurg.2014.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Fedewa SA, Anderson WF, Miller KD, Ma J, Rosenberg PS, Jemal A. Colorectal Cancer Incidence Patterns in the United States, 1974–2013. Journal of the National Cancer Institute. 2017;109(8) doi: 10.1093/jnci/djw322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdelsattar ZM, Wong SL, Regenbogen SE, Jomaa DM, Hardiman KM, Hendren S. Colorectal cancer outcomes and treatment patterns in patients too young for average-risk screening. Cancer. 2016;122(6):929–34. doi: 10.1002/cncr.29716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levin B, Lieberman DA, McFarland B, Andrews KS, Brooks D, Bond J, Dash C, Giardiello FM, Glick S, Johnson D, Johnson CD, Levin TR, Pickhardt PJ, Rex DK, Smith RA, Thorson A, Winawer SJ American Cancer Society Colorectal Cancer Advisory G; Force USM-ST, American College of Radiology Colon Cancer C. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology. 2008;134(5):1570–95. doi: 10.1053/j.gastro.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138(6):2044–58. doi: 10.1053/j.gastro.2010.01.054. Epub 2010/04/28. S0016-5085(10)00168-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Clendenning M, Sotamaa K, Prior T, Westman JA, Panescu J, Fix D, Lockman J, LaJeunesse J, Comeras I, de la Chapelle A. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol. 2008;26(35):5783–8. doi: 10.1200/JCO.2008.17.5950. Epub 2008/09/24. JCO.2008.17.5950 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moreira L, Balaguer F, Lindor N, de la Chapelle A, Hampel H, Aaltonen LA, Hopper JL, Le Marchand L, Gallinger S, Newcomb PA, Haile R, Thibodeau SN, Gunawardena S, Jenkins MA, Buchanan DD, Potter JD, Baron JA, Ahnen DJ, Moreno V, Andreu M, Ponz de Leon M, Rustgi AK, Castells A Consortium E. Identification of Lynch syndrome among patients with colorectal cancer. Jama. 2012;308(15):1555–65. doi: 10.1001/jama.2012.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, Chadwick RB, Kaariainen H, Eskelinen M, Jarvinen H, Mecklin JP, de la Chapelle A. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. The New England journal of medicine. 1998;338(21):1481–7. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- 12.Yurgelun MB, Kulke MH, Fuchs CS, Allen BA, Uno H, Hornick JL, Ukaegbu CI, Brais LK, McNamara PG, Mayer RJ, Schrag D, Meyerhardt JA, Ng K, Kidd J, Singh N, Hartman AR, Wenstrup RJ, Syngal S. Cancer Susceptibility Gene Mutations in Individuals With Colorectal Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2017;35(10):1086–95. doi: 10.1200/JCO.2016.71.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evaluation of Genomic Applications in P, Prevention Working G. Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genetics in medicine : official journal of the American College of Medical Genetics. 2009;11(1):35–41. doi: 10.1097/GIM.0b013e31818fa2ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grover S, Kastrinos F, Steyerberg EW, Cook EF, Dewanwala A, Burbidge LA, Wenstrup RJ, Syngal S. Prevalence and phenotypes of APC and MUTYH mutations in patients with multiple colorectal adenomas. Jama. 2012;308(5):485–92. doi: 10.1001/jama.2012.8780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Syngal S, Brand RE, Church JM, Giardiello FM, Hampel HL, Burt RW American College of G. ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. The American journal of gastroenterology. 2015;110(2):223–62. doi: 10.1038/ajg.2014.435. quiz 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giardiello FM, Allen JI, Axilbund JE, Boland CR, Burke CA, Burt RW, Church JM, Dominitz JA, Johnson DA, Kaltenbach T, Levin TR, Lieberman DA, Robertson DJ, Syngal S, Rex DK Cancer USM-STFoC. Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US Multi-Society Task Force on colorectal cancer. Gastroenterology. 2014;147(2):502–26. doi: 10.1053/j.gastro.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 17.NCCN. Genetic/Familial High Risk Assessment: Breast and/or Ovarian and Colorectal. NCCN Clinical Practice Guidelines in Oncology. 2016 v. 2.2017 nccn.org.
- 18.Broderick P, Dobbins SE, Chubb D, Kinnersley B, Dunlop MG, Tomlinson I, Houlston RS. Validation of Recently Proposed Colorectal Cancer Susceptibility Gene Variants in an Analysis of Families and Patients-a Systematic Review. Gastroenterology. 2017;152(1):75–7. e4. doi: 10.1053/j.gastro.2016.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yurgelun MB, Allen B, Kaldate RR, Bowles KR, Judkins T, Kaushik P, Roa BB, Wenstrup RJ, Hartman AR, Syngal S. Identification of a Variety of Mutations in Cancer Predisposition Genes in Patients with Suspected Lynch Syndrome. Gastroenterology. 2015 doi: 10.1053/j.gastro.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, Roeb W, Agnew KJ, Stray SM, Wickramanayake A, Norquist B, Pennington KP, Garcia RL, King MC, Swisher EM. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(44):18032–7. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurian AW, Hare EE, Mills MA, Kingham KE, McPherson L, Whittemore AS, McGuire V, Ladabaum U, Kobayashi Y, Lincoln SE, Cargill M, Ford JM. Clinical Evaluation of a Multiple-Gene Sequencing Panel for Hereditary Cancer Risk Assessment. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014 doi: 10.1200/JCO.2013.53.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cragun D, Radford C, Dolinsky JS, Caldwell M, Chao E, Pal T. Panel-based testing for inherited colorectal cancer: a descriptive study of clinical testing performed by a US laboratory. Clin Genet. 2014;86(6):510–20. doi: 10.1111/cge.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Limburg PJ, Harmsen WS, Chen HH, Gallinger S, Haile RW, Baron JA, Casey G, Woods MO, Thibodeau SN, Lindor NM. Prevalence of alterations in DNA mismatch repair genes in patients with young-onset colorectal cancer. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2011;9(6):497–502. doi: 10.1016/j.cgh.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giraldez MD, Balaguer F, Bujanda L, Cuatrecasas M, Munoz J, Alonso-Espinaco V, Larzabal M, Petit A, Gonzalo V, Ocana T, Moreira L, Enriquez-Navascues JM, Boland CR, Goel A, Castells A, Castellvi-Bel S. MSH6 and MUTYH deficiency is a frequent event in early-onset colorectal cancer. Clin Cancer Res. 2010;16(22):5402–13. doi: 10.1158/1078-0432.CCR-10-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang DT, Pai RK, Rybicki LA, Dimaio MA, Limaye M, Jayachandran P, Koong AC, Kunz PA, Fisher GA, Ford JM, Welton M, Shelton A, Ma L, Arber DA, Pai RK. Clinicopathologic and molecular features of sporadic early-onset colorectal adenocarcinoma: an adenocarcinoma with frequent signet ring cell differentiation, rectal and sigmoid involvement, and adverse morphologic features. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2012;25(8):1128–39. doi: 10.1038/modpathol.2012.61. [DOI] [PubMed] [Google Scholar]
- 26.Terdiman JP, Levin TR, Allen BA, Gum JR, Jr, Fishbach A, Conrad PG, Miller GA, Weinberg V, Bachman R, Bergoffen J, Stembridge A, Toribara NW, Sleisenger MH, Kim YS. Hereditary nonpolyposis colorectal cancer in young colorectal cancer patients: high-risk clinic versus population-based registry. Gastroenterology. 2002;122(4):940–7. doi: 10.1053/gast.2002.32537. [DOI] [PubMed] [Google Scholar]
- 27.Goel A, Nagasaka T, Spiegel J, Meyer R, Lichliter WE, Boland CR. Low frequency of Lynch syndrome among young patients with non-familial colorectal cancer. Clin Gastroenterol Hepatol. 2010;8(11):966–71. doi: 10.1016/j.cgh.2010.06.030. Epub 2010/07/27. S1542-3565(10)00676-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen S, Wang W, Lee S, Nafa K, Lee J, Romans K, Watson P, Gruber SB, Euhus D, Kinzler KW, Jass J, Gallinger S, Lindor NM, Casey G, Ellis N, Giardiello FM, Offit K, Parmigiani G. Prediction of germline mutations and cancer risk in the Lynch syndrome. Jama. 2006;296(12):1479–87. doi: 10.1001/jama.296.12.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mork ME, You YN, Ying J, Bannon SA, Lynch PM, Rodriguez-Bigas MA, Vilar E. High Prevalence of Hereditary Cancer Syndromes in Adolescents and Young Adults With Colorectal Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33(31):3544–9. doi: 10.1200/JCO.2015.61.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearlman R, Frankel WL, Swanson B, Zhao W, Yilmaz A, Miller K, Bacher J, Bigley C, Nelsen L, Goodfellow PJ, Goldberg RM, Paskett E, Shields PG, Freudenheim JL, Stanich PP, Lattimer I, Arnold M, Liyanarachchi S, Kalady M, Heald B, Greenwood C, Paquette I, Prues M, Draper DJ, Lindeman C, Kuebler JP, Reynolds K, Brell JM, Shaper AA, Mahesh S, Buie N, Weeman K, Shine K, Haut M, Edwards J, Bastola S, Wickham K, Khanduja KS, Zacks R, Pritchard CC, Shirts BH, Jacobson A, Allen B, de la Chapelle A, Hampel H Ohio Colorectal Cancer Prevention Initiative Study G. Prevalence and Spectrum of Germline Cancer Susceptibility Gene Mutations Among Patients With Early-Onset Colorectal Cancer. JAMA oncology. 2016 doi: 10.1001/jamaoncol.2016.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL Committee ALQA. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine : official journal of the American College of Medical Genetics. 2015;17(5):405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kastrinos F, Steyerberg EW, Mercado R, Balmana J, Holter S, Gallinger S, Siegmund KD, Church JM, Jenkins MA, Lindor NM, Thibodeau SN, Burbidge LA, Wenstrup RJ, Syngal S. The PREMM(1,2,6) model predicts risk of MLH1, MSH2, and MSH6 germline mutations based on cancer history. Gastroenterology. 2011;140(1):73–81. doi: 10.1053/j.gastro.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yurgelun MB, Masciari S, Joshi VA, Mercado RC, Lindor NM, Gallinger S, Hopper JL, Jenkins MA, Buchanan DD, Newcomb PA, Potter JD, Haile RW, Kucherlapati R, Syngal S Colon Cancer Family R. Germline TP53 Mutations in Patients With Early-Onset Colorectal Cancer in the Colon Cancer Family Registry. JAMA oncology. 2015;1(2):214–21. doi: 10.1001/jamaoncol.2015.0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pujol P, Lyonnet DS, Frebourg T, Blin J, Picot MC, Lasset C, Dugast C, Berthet P, de Paillerets BB, Sobol H, Grandjouan S, Soubrier F, Buecher B, Guimbaud R, Lidereau R, Jonveaux P, Houdayer C, Giraud S, Olschwang S, Nogue E, Galibert V, Bara C, Nowak F, Khayat D, Nogues C. Lack of referral for genetic counseling and testing in BRCA1/2 and Lynch syndromes: a nationwide study based on 240,134 consultations and 134,652 genetic tests. Breast cancer research and treatment. 2013;141(1):135–44. doi: 10.1007/s10549-013-2669-9. [DOI] [PubMed] [Google Scholar]
- 35.Idos GKA, McDonnel K, et al. The GI Gap in Genetic Testing for Inherited Susceptibility to Cancer. American College of Gastroenterology Annual Meeting. 2015 Abstract P159. [Abstract]. In press. [Google Scholar]
- 36.NCCN. Genetic/Familial High Risk Assessment: Breast and ovarian. NCCN Clinical Practice Guidelines in Oncology. 2016 v. 2.2016 nccn.org.
- 37.Robson ME, Bradbury AR, Arun B, Domchek SM, Ford JM, Hampel HL, Lipkin SM, Syngal S, Wollins DS, Lindor NM. American Society of Clinical Oncology Policy Statement Update: Genetic and Genomic Testing for Cancer Susceptibility. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33(31):3660–7. doi: 10.1200/JCO.2015.63.0996. [DOI] [PubMed] [Google Scholar]
- 38.Gallego CJ, Shirts BH, Bennette CS, Guzauskas G, Amendola LM, Horike-Pyne M, Hisama FM, Pritchard CC, Grady WM, Burke W, Jarvik GP, Veenstra DL. Next-Generation Sequencing Panels for the Diagnosis of Colorectal Cancer and Polyposis Syndromes: A Cost-Effectiveness Analysis. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33(18):2084–91. doi: 10.1200/JCO.2014.59.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guindalini RS, Win AK, Gulden C, Lindor NM, Newcomb PA, Haile RW, Raymond V, Stoffel E, Hall M, Llor X, Ukaegbu CI, Solomon I, Weitzel J, Kalady M, Blanco A, Terdiman J, Shuttlesworth GA, Lynch PM, Hampel H, Lynch HT, Jenkins MA, Olopade OI, Kupfer SS. Mutation spectrum and risk of colorectal cancer in African American families with Lynch syndrome. Gastroenterology. 2015;149(6):1446–53. doi: 10.1053/j.gastro.2015.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siegel RL, Miller KD, Jemal A. Colorectal Cancer Mortality Rates in Adults Aged 20 to 54 Years in the United States, 1970–2014. Jama. 2017;318(6):572–4. doi: 10.1001/jama.2017.7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luba DG, DiSario JA, Rock C, Saraiya D, Moyes K, Brown K, Rushton K, Ogara MM, Raphael M, Zimmerman D, Garrido K, Silguero E, Nelson J, Yurgelun MB, Kastrinos F, Wenstrup RJ, Syngal S. Community Practice Implementation of a Self-administered Version of PREMM1,2,6 to Assess Risk for Lynch Syndrome. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2017 doi: 10.1016/j.cgh.2017.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kastrinos F, Uno H, Ukaegbu C, Alvero C, McFarland A, Yurgelun MB, Kulke MH, Schrag D, Meyerhardt JA, Fuchs CS, Mayer RJ, Ng K, Steyerberg EW, Syngal S. Development and Validation of the PREMM5 Model for Comprehensive Risk Assessment of Lynch Syndrome. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2017;35(19):2165–72. doi: 10.1200/JCO.2016.69.6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haraldsdottir S, Rafnar T, Frankel WL, Einarsdottir S, Sigurdsson A, Hampel H, Snaebjornsson P, Masson G, Weng D, Arngrimsson R, Kehr B, Yilmaz A, Haraldsson S, Sulem P, Stefansson T, Shields PG, Sigurdsson F, Bekaii-Saab T, Moller PH, Steinarsdottir M, Alexiusdottir K, Hitchins M, Pritchard CC, de la Chapelle A, Jonasson JG, Goldberg RM, Stefansson K. Comprehensive population-wide analysis of Lynch syndrome in Iceland reveals founder mutations in MSH6 and PMS2. Nat Commun. 2017;8:14755. doi: 10.1038/ncomms14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodenberger ML, Thomas BC, Riegert-Johnson D, Boland CR, Plon SE, Clendenning M, Win AK, Senter L, Lipkin SM, Stadler ZK, Macrae FA, Lynch HT, Weitzel JN, de la Chapelle A, Syngal S, Lynch P, Parry S, Jenkins MA, Gallinger S, Holter S, Aronson M, Newcomb PA, Burnett T, Le Marchand L, Pichurin P, Hampel H, Terdiman JP, Lu KH, Thibodeau S, Lindor NM. PMS2 monoallelic mutation carriers: the known unknown. Genetics in medicine : official journal of the American College of Medical Genetics. 2016;18(1):13–9. doi: 10.1038/gim.2015.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Win AK, Jenkins MA, Dowty JG, Antoniou AC, Lee A, Giles GG, Buchanan DD, Clendenning M, Rosty C, Ahnen DJ, Thibodeau SN, Casey G, Gallinger S, Le Marchand L, Haile RW, Potter JD, Zheng Y, Lindor NM, Newcomb PA, Hopper JL, MacInnis RJ. Prevalence and Penetrance of Major Genes and Polygenes for Colorectal Cancer. Cancer Epidemiol Biomarkers Prev. 2017;26(3):404–12. doi: 10.1158/1055-9965.EPI-16-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahnen DJ, Wade SW, Jones WF, Sifri R, Mendoza Silveiras J, Greenamyer J, Guiffre S, Axilbund J, Spiegel A, You YN. The increasing incidence of young-onset colorectal cancer: a call to action. Mayo Clin Proc. 2014;89(2):216–24. doi: 10.1016/j.mayocp.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 47.Yurgelun MB, Boland CR. "New" Cancer Genes and Inherited Colorectal Cancer Risk: Caveat Emptor. Gastroenterology. 2017;152(1):12–3. doi: 10.1053/j.gastro.2016.11.027. [DOI] [PubMed] [Google Scholar]