Abstract
The development of metal complexes that promote degradation of nucleic acids has garnered significant interest as a result of their broad range of potential application. This review focuses on recent progress in the design and synthesis of metal complexes as artificial nucleases that promote either hydrolytic or oxidative cleavage of nucleic acids. Illustrative examples demonstrate the versatility of artificial nucleases for in vitro applications as molecular tools to address biochemical problems, as well as their potential use as therapeutic agents. We also address future challenges for improvement and avenues for further investigation.
Introduction
Nucleic acids are a major class of biopolymers that play an important role in the central dogma; namely, DNA contains genetic information that can be transcribed into mRNA and subsequently translated to form proteins [1]. Both DNA and RNA are composed of polymeric backbones composed of phosphodiesters linked to deoxyribose and ribose, respectively [1]. The half-life for hydrolysis of DNA under physiological conditions is estimated to be over 521 year [2]; however, more rapid reaction is promoted by enzyme catalysts (nucleases), termed DNase and RNase, respectively [3]. The nuclease function is ubiquitous and involved in a variety of biological activities, including nucleic acid synthesis, recombination, regulation, processing and degradation [3]. Restriction endonucleases have been widely used in molecular cloning [3]. In addition to the natural nucleases, engineered DNase and RNase enzymes also have broad application in vitro and in vivo. For example, sequence-selective DNA strand-breaks promoted by zinc finger nucleases, transcription activator-like effector nucleases, and CRISPR/Cas systems have been employed as powerful tools for gene editing [4].
Despite the outstanding properties of conventional protein nucleases, these are not without their challenges [5]. Problems associated with protein-based nucleases include a limited pool of natural nucleases and substrate selectivity, solution instability, and a lack of membrane permeability. To address these problems and potentially overcome the aforementioned problems, extensive effort has been made over several decades toward developing low-molecular-weight metal complexes as “artificial nucleases” and mimics of nuclease function [6]. In this review, we briefly summarize recent progress in the design of metal complexes that mediate nuclease activity against DNA or RNA, either through hydrolytic or oxidative cleavage mechanisms. Also considered are the application of artificial nucleases, and their potential chemical and biological advantages in comparison to conventional protein-based nucleases. We also discuss the problems and challenges associated with artificial nucleases, especially for in vivo applications, and summarize design considerations for artificial nucleases that can potentially resolve those problems in vivo.
Hydrolytic and oxidative cleavage of nucleic acids
For most natural metallonucleases, divalent magnesium is the catalytic metal of choice, in part as a results of its high natural availability and high Lewis acidity, while in a few other cases divalent Zn or Ca are used [7]. Recent designs of novel catalytic DNA’s (DNAzymes) that promote DNA and RNA cleavage also require Zn, Mg, Cr, or Mn as cofactors [8]. Some recent reviews have summarized the role of metal cofactors in metallonucleases and the design of DNAzymes, and so these will not be further discussed here [8]. Inspired by natural metallonucleases, a large number of studies have reported the design of strongly Lewis acidic metal complexes as artificial nucleases that promote hydrolysis of the nucleic acid phosphate backbone. These include novel Cu(II) [9,10], Cr(III) [11], Zn(II) [12], Ce(IV) [13] Zr(IV) [14], La(III) [15], Fe(III) [16], and Co(III) [17] complexes. Mechanistically, these involve the formation of a metal-phosphate intermediate that facilitates hydrolysis of the metal-bound ester (Figure 1). DNA cleavage rate is dependent on the Lewis acidity of the metal and the ease of formation of the metal-bound hydroxide nucleophile. In addition, RNA is more susceptible to hydrolysis, relative to DNA, due to the nucleophilic attack of RNA backbone by the nearby 2′ OH group that can be potentially deprotonated by metal complexes (Figure 1).
Figure 1.
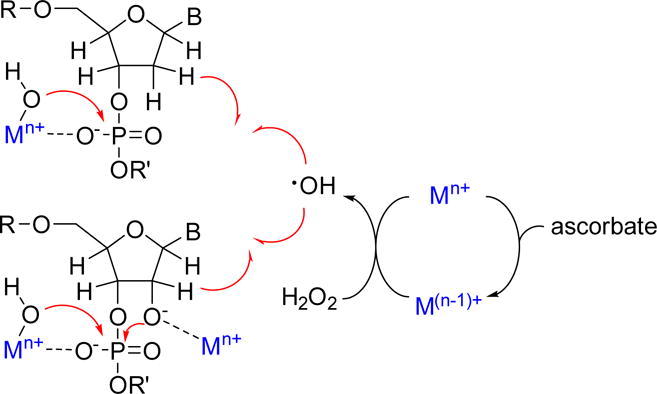
Summary of hydrolytic and oxidative cleavage pathways for nucleic acids, promoted by metal complexes.
Oxidative cleavage, utilizing redox active metals, represents an alternative approach to degrade nucleic acids, often by pathways involving intermediate formation of reactive oxygen species (ROS) following electron transfer from the metal to molecular oxygen or peroxide [18]. The oxidized metal can subsequently be reduced by ascorbate or DTT, to continue formation of ROS, which typically abstract hydrogen from the deoxyribose/ribose ring followed by spontaneous cleavage of C–C and C–O bonds [18] (Figure 1). The formation of different 3′ overhang products through different C–H abstractions can be detected by use of mass spectroscopy [19].
Due to the lack of a hydrophobic environment in the substrate binding pocket of natural nucleases, small-molecule metal complexes typically exhibit slower DNA cleavage rates in solution, therefore, some strategies to enhance cleavage activities have been developed. A recent study reported the design of amphiphilic Zn-cyclen complexes conjugated to phosphocholine derivatives that, in turn, allow the formation of Zn-cyclen surface modified micelles [20]. Theses Zn-containing micelle/vehicles exhibit ~103–106 fold increase in catalytic activity for DNA hydrolysis, relative to the isolated Zn-cyclen. In addition to hydrophobicity, facilitating direct interaction with DNA or RNA can also play an important role in promoting effective nucleic acid cleavage. For example, Zn and Cu complexes that are linked to a cationic peptide show improved binding to DNA through electrostatic interactions and more efficient DNA cleavage [9]. A Zn complex of a diaza-crown ether was reported to exhibit more robust DNA hydrolysis relative to its copper analogues, since Zn(II) is more Lewis acidic than Cu(II) [12]. For oxidative cleavage promoted by redox active metals, an appropriate reduction potential for the M(n−1)+/Mn+ couple, relative to that of the redox active coreagents (such as O2/O2−, H2O2/·OH) can establish more efficient redox cycling and promote rapid formation of ROS [21]. Overall, improvements in hydrophobicity, promoting contacts with the substrate, enhanced Lewis acidity (for hydrolysis) or optimizing reduction potentials (for oxidative cleavage) are all factors that have been shown to influence the catalytic cleavage reactivity of metal complexes.
Potential Applications
In contrast to protein-based nucleases that often display some measure of substrate selectivity, artificial nucleases based on metal complexes usually disregard the nucleic acid sequence and the ability to differentiate between ribose and deoxyribose rings. Consequently, such catalysts are capable of degrading a broader spectrum of substrates. The versatility of artificial nucleases has enabled development of novel molecular tools that complement conventional nucleases. Fe-EDTA and other metal complexes have been widely used as molecular probes to provide information on biological activities associated with nucleic acids, including protein-DNA/RNA interactions [22,23], the structure of DNA/RNA [24], and footprinting of nucleic acids [25] (Figure 2). For example, methidiumpropyl-EDTA-Fe has been used in the mapping of chromatin structure [26]. In these cases, the metal complex is typically used alone, to cleave the solvent-exposed sites of nucleic acids. Alternatively, metal complexes can also be linked to protein or nucleic acids to introduce substrate selectivity and promote targeted cleavage of specific sites on nucleic acids [27]. For example, the interaction between the transcription elongation factor, NusA, and nascent RNA was studied by use of Fe-EDTA-linked NusA, formed by coupling p-bromoacetamidobenzyl-EDTA (BABE) to the cysteine side chain of NuaA [28]. Similarly, Fe-EDTA has been conjugated to BvgAT194C or BvgAV148C to examine formation of the transcription complex formed by template DNA, phosphorylated BvgA, and RNA polymerase [29]. In addition to proteins, a protocol has also been reported for the site-specific incorporation of Fe-EDTA into 4-chlorophenyl-uridine of a pre-mRNA to study spliceosome assembly of pre-mRNA through RNA cleavage by the Fe-EDTA motif [24,30]. Use of metal complexes as artificial nucleases is not only complementary to other protein-based nucleases, but in some cases can also be more feasible than protein-based nucleases. Both DNA and RNA can be cleaved by artificial nucleases, while DNase and RNase, respectively, would be required if protein-based nucleases were applied. Solvent-exposed sites of nucleic acids are more accessible to these small-molecule metal complexes, relative to bulky protein-based nucleases. Metal complexes, such as Fe-BABE, can be conveniently conjugated to a specific residue of a protein or nucleic acid through chemical coupling to cysteine or nucleobases (Figure 2a), while a protein-based nuclease can only be tethered to the N- or C- terminus of a protein and may potentially have a negative impact on protein/nucleic acid interactions and protein folding.
Figure 2.
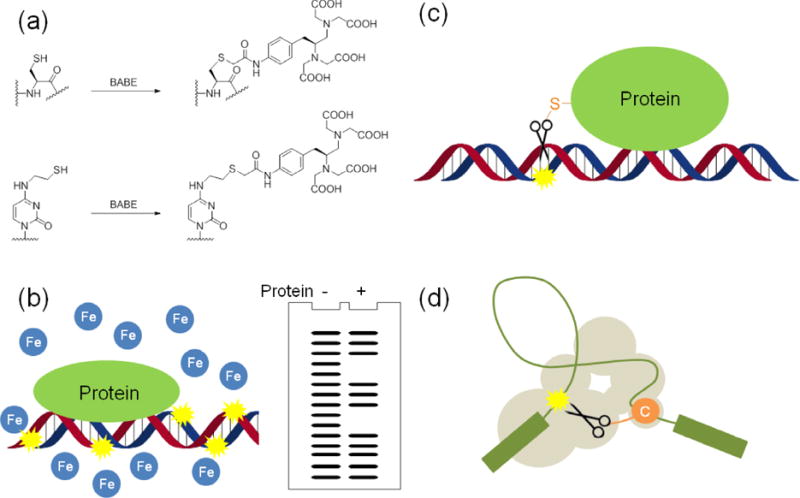
Examples of the application of metal complexes as molecular tools to study DNA/RNA. (a) Incorporation of BABE into cysteine or N4-modified cytosine. (b) Solvent-exposed sites of DNA studied by metal complexes. (c) DNA-protein interaction revealed by site-selective cleavage. (d) Structure of RNA mapped by metal complexes (modified from reference [24]).
In addition to in vitro biochemical studies, metal complexes that target nucleic acids can also be developed as therapeutics [31]. In fact, DNA cleavage can activate the DNA damage response signaling pathway in cells, while unrepaired DNA damage further activates the apoptotic pathway and cell death [31]. As a result, artificial nucleases that promote DNA damage can be used as anticancer agents. In fact, bleomycin, a natural product of S. verticillus, can promote DNA cleavage when bound to Fe or Cu, and has been approved as an anticancer agent since 1973. The mechanism of cellular selectivity of bleomycin is not well understood, but in part may reflect a defective DNA repair machinery in cancer cells, making it more difficult to identify and repair DNA lesions induced by bleomycin [32]. Similarly, a number of synthetic metal complexes have been designed that promote DNA cleavage and their anticancer activity has been broadly tested [33]. DNA breaks induced in cancer cells by metal complexes are typically confirmed by use of the TUNEL assay and/or comet assay. However, most reported examples of metal nuclease complexes exhibit very little cleavage selectivity. In contrast to other metal complexes that lack selectivity, metal complexes targeting specific DNA structures and sequences may improve drug efficacy and mitigate side effects, such as the design of a Cu complex that targets G-quadruplex telomeric DNA (Figure 3a) [34]. The incorporation of a G4 targeting motif to a Cu complex can enhance cleavage selectivity against G-quadruplex structures, relative to duplex structures. In turn, G-quadruplex-targeting metal complexes should exhibit improved cell selectivity due to the increased formation of G-quadruplex during S-phase DNA synthesis and the up-regulated synthesis of DNA in cancer cells [35]. In fact, the designed G-quadruplex-targeting Cu complexes have been shown to shorten telomere length in cancer cells through DNA cleavage, representing a novel mode of drug action that differs from bleomycin and other nonselective metal complexes [34].
Figure 3.
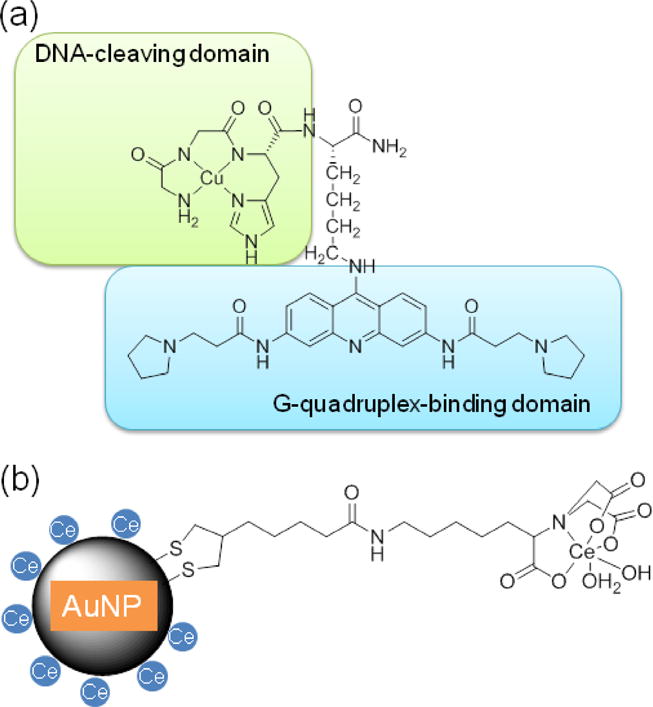
(a) A G-quadruplex-targeting Cu complex as an anticancer agent. (b) Ce-coated gold nano particles inhibiting biofilm formation
In addition to anticancer therapeutics, DNA damage to other microbes may be developed into novel antibiotics or agents that inhibit biofilm formation [36]. Drug resistance against conventional antibiotics is an increasing challenge in clinical practice [37]. The main classes of antibiotics include beta-lactams, tetracycline derivatives, quinolones that target DD-transpeptidases, 16S rRNA, and DNA helicases, respectively. Therefore, DNA cleavage by metal complexes may provide an alternative target and mechanism that differs from those identified with conventional antibiotics and provide new opportunities to circumvent drug resistance. Nanoparticles surface-coated with Ce(IV) have been reported to degrade extracellular DNA (eDNA) and inhibit the growth of biofilm (Figure 3b) [38].
Nucleic acids of viruses represent a novel drug target susceptible to artificial nucleases [39]. Metal complexes that function as selective artificial nucleases, targeting HIV RRE (Rev response element) RNA, HCV SLIIB RNA, and HCV SLIV RNA have been described. To design a selective metal complex, the RNA cleaving motif is incorporated into a selective RNA binding motif, the TRQARRNRRRRWRERQR peptide for RRE [40,41], a YrFK peptide for SLIIB [42,43], and LaR2C peptides derived from La Protein for SLIV [44]. Selective cleavage of RNA is confirmed by use of MS, implying a cognate interaction between metal complex and substrate RNA that should be ascribed to RNA recognition by the targeting domain. Moreover, cellular replicon assays support the decrease of HCV RNA in cells following treatment with such complexes, consistent with intracellular RNA cleavage as the mode of action [42–44].
Challenge and future directions
Metal complexes have seen wide application as molecular tools for in vitro studies of a variety of biochemical problems. In vivo applications are less evident and represent an area for further exploration. Application of metal complexes that target disease-associated DNA/RNA as potential therapeutic agents should enjoy a promising future [45]. However, most metal complexes reported in current literature exhibit low substrate selectivity, and the potential for off-target activity, low efficiency in mediating chemistry, and the potential for damage to other biomolecules will result in low efficacy and side effects unless significant improvements in, and consideration of, substrate targeting are made. Approaches have been illustrated in recent works that describe applications of metal complexes to promote oxidative damage/cleavage to proteins and other metabolites, such as saccharides [46,47]. Therefore, the need for substrate selectivity becomes critically important, and the introduction of a substrate-binding moiety to designed metal complexes may improve substrate selectivity. Aside from the issue of cellular delivery and lifetime, other problems more pertinent to the coordination chemistry of such metal complexes will also require attention for cellular or in vivo application. The potential for metal dissociation (kinetic and thermodynamic lability) has also engendered anxiety concerning metal toxicity and has led to disfavor for therapeutic application of metal complexes. In addition to stability, chelate ligands used to develop metal complexes should also exhibit selectivity for the desired metal, since biologically-relevant metals, such as Ca (1.2–1.43 mM, serum) [48] and Mg (0.75–0.95 mM, serum) [49], may competitively replace the metal of choice. These problems are not insignificant and will provide ample opportunity for creativity in the design and study of the next generation of metal complexes that promote catalytic cleavage of nucleic acids.
Highlights.
New catalytic metal complexes as synthetic nucleases
Surface adhesion in micelles and nanoparticles enhances nuclease activity
Incorporation of a targeting motif improves selectivity and catalytic activity
Applications in biotechnology and as therapeutics are summarized
Acknowledgments
This work was supported by grants from the National Institutes of Health [HL093446]. Z.Y. was supported by the Pelotonia Fellowship Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Lipfert J, Doniach S, Das R, Herschlag D. Understanding Nucleic Acid-Ion Interactions. Annu Rev Biochem. 2014;83:813–841. doi: 10.1146/annurev-biochem-060409-092720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allentoft ME, Collins M, Harker D, Haile J, Oskam CL, Hale ML, Campos PF, Samaniego JA, Gilbert MTP, Willerslev E, et al. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proc R Soc B Biol Sc. 2012;279:4724–4733. doi: 10.1098/rspb.2012.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang W. Nucleases: diversity of structure, function and mechanism. Q Rev Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mancin F, Scrimin P, Tecilla P. Progress in artificial metallonucleases. Chem Commun. 2012;48:5545–5559. doi: 10.1039/c2cc30952a. [DOI] [PubMed] [Google Scholar]
- 6.Mancin F, Scrimin P, Tecilla P, Tonellato U. Artificial metallonucleases. Chem Commun. 2005:2540–2548. doi: 10.1039/b418164f. [DOI] [PubMed] [Google Scholar]
- 7.Dupureur CM. Roles of metal ions in nucleases. Curr Opin Chem Biol. 2008;12:250–255. doi: 10.1016/j.cbpa.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Baum DA, Silverman SK. Deoxyribozymes: useful DNA catalysts in vitro and in vivo. Cell Mol Life Sci. 2008;65:2156–2174. doi: 10.1007/s00018-008-8029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soler M, Figueras E, Serrano-Plana J, Gonzalez-Bartulos M, Massaguer A, Company A, Martinez MA, Malina J, Brabec V, Feliu L, et al. Design, Preparation, and Characterization of Zn and Cu Metallopeptides Based On Tetradentate Aminopyridine Ligands Showing Enhanced DNA Cleavage Activity. Inorg Chem. 2015;54:10542–10558. doi: 10.1021/acs.inorgchem.5b01680. [DOI] [PubMed] [Google Scholar]
- 10.Salvio R, Volpi S, Cacciapaglia R, Sansone F, Mandolini L, Casnati A. Upper Rim Bifunctional cone-Calix[4]arenes Based on a Ligated Metal Ion and a Guanidinium Unit as DNAase and RNAase Mimics. J Org Chem. 2016;81:4728–4735. doi: 10.1021/acs.joc.6b00644. [DOI] [PubMed] [Google Scholar]
- 11.Zhou WH, Yu TM, Vazin M, Ding JS, Liu JW. Cr3+ Binding to DNA Backbone Phosphate and Bases: Slow Ligand Exchange Rates and Metal Hydrolysis. Inorg Chem. 2016;55:8193–8200. doi: 10.1021/acs.inorgchem.6b01357. [DOI] [PubMed] [Google Scholar]
- 12.Xie Li FZ, Li JQ, Feng FM. Copper and zinc complexes of a diaza-crown ether as artificial nucleases for the efficient hydrolytic cleavage of DNA. New J Chem. 2015;39:5654–5660. [Google Scholar]
- 13.Yang JW, Lin YL, Dong C, Zhou CQ, Chen JX, Wang B, Zhou ZZ, Sun B, Chen WH. Synthesis, hydrolytic DNA-cleaving activities and cytotoxicities of EDTA analogue-tethered pyrrole-polyamide dimer-based Ce(IV) complexes. Eur J Med Chem. 2014;87:168–174. doi: 10.1016/j.ejmech.2014.09.057. [DOI] [PubMed] [Google Scholar]
- 14.Luong TKN, Govaerts I, Robben J, Shestakova P, Parac-Vogt TN. Polyoxometalates as artificial nucleases: hydrolytic cleavage of DNA promoted by a highly negatively charged Zr-IV-substituted Keggin polyanion. Chem Commun. 2017;53:617–620. doi: 10.1039/c6cc08555e. [DOI] [PubMed] [Google Scholar]
- 15.Muxel AA, Neves A, Camargo MA, Bortoluzzi AJ, Szpoganicz B, Castellano EE, Castilho N, Bortolotto T, Terenzi H. New La(III) Complex Immobilized on 3-Aminopropyl-Functionalized Silica as an Efficient and Reusable Catalyst for Hydrolysis of Phosphate Ester Bonds. Inorg Chem. 2014;53:2943–2952. doi: 10.1021/ic402705r. [DOI] [PubMed] [Google Scholar]
- 16.Piovezan C, Jovito R, Bortoluzzi AJ, Terenzi H, Fischer FL, Severino PC, Pich CT, Azzolini GG, Peralta RA, Rossi LM, et al. Heterodinuclear (FeZnII)-Zn-III-Bioinspired Complex Supported on 3-Aminopropyl Silica. Efficient Hydrolysis of Phosphate Diester Bonds. Inorg Chem. 2010;49:2580–2582. doi: 10.1021/ic902489j. [DOI] [PubMed] [Google Scholar]
- 17.Massoud SS, Perkins RS, Louka FR, Xu W, Le Roux A, Dutercq Q, Fischer RC, Mautner FA, Handa M, Hiraoka Y, et al. Efficient hydrolytic cleavage of plasmid DNA by chloro-cobalt(II) complexes based on sterically hindered pyridyl tripod tetraamine ligands: synthesis, crystal structure and DNA cleavage. Dalton Trans. 2014;43:10086–10103. doi: 10.1039/c4dt00615a. [DOI] [PubMed] [Google Scholar]
- 18.Jiang Q, Xiao N, Shi PF, Zhu YG, Guo ZJ. Design of artificial metallonucleases with oxidative mechanism. Coord Chem Rev. 2007;251:1951–1972. [Google Scholar]
- 19.Joyner JC, Keuper KD, Cowan JA. Analysis of RNA cleavage by MALDI-TOF mass spectrometry. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gks811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20••.Gruber B, Kataev E, Aschenbrenner J, Stadlbauer S, Konig B. Vesicles and Micelles from Amphiphilic Zinc(II)-Cyclen Complexes as Highly Potent Promoters of Hydrolytic DNA Cleavage. J Am Chem Soc. 2011;133:20704–20707. doi: 10.1021/ja209247w. Vesicles and micelles coated with surface Zn complexes promote more efficient DNA cleavage than Zn-cyclen alone. [DOI] [PubMed] [Google Scholar]
- 21.Joyner JC, Reichfield J, Cowan JA. Factors Influencing the DNA Nuclease Activity of Iron, Cobalt, Nickel, and Copper Chelates. J Am Chem Soc. 2011;133:15613–15626. doi: 10.1021/ja2052599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dey B, Thukral S, Krishnan S, Chakrobarty M, Gupta S, Manghani C, Rani V. DNA-protein interactions: methods for detection and analysis. Mol Cell Biochem. 2012;365:279–299. doi: 10.1007/s11010-012-1269-z. [DOI] [PubMed] [Google Scholar]
- 23.James T, Hsieh ML, Knipling L, Hinton D. Determining the Architecture of a Protein-DNA Complex by Combining FeBABE Cleavage Analyses, 3-D Printed Structures, and the ICM Molsoft Program. Methods Mol Biol. 2015;1334:29–40. doi: 10.1007/978-1-4939-2877-4_3. [DOI] [PubMed] [Google Scholar]
- 24••.Grewal CS, Kent OA, MacMillan AM. Radical probing of spliceosome assembly. Methods. 2017;125:16–24. doi: 10.1016/j.ymeth.2017.06.030. A reported protocol decribing incorportion of FeEDTA into RNA. [DOI] [PubMed] [Google Scholar]
- 25.Yang ZY, Price NE, Johnson KM, Wang YS, Gates KS. Interstrand cross-links arising from strand breaks at true abasic sites in duplex DNA. Nucleic Acids Res. 2017;45:6275–6283. doi: 10.1093/nar/gkx394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Ishii H, Kadonaga JT, Ren B. MPE-seq, a new method for the genome-wide analysis of chromatin structure. Proc Natl Acad Sci U S A. 2015;112:E3457–3465. doi: 10.1073/pnas.1424804112. An example studying solvent-exposed sites of genomic DNA with Fe-EDTA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duval M, Marenna A, Chevalier C, Marzi S. Site-Directed Chemical Probing to map transient RNA/protein interactions. Methods. 2017;117:48–58. doi: 10.1016/j.ymeth.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 28•.Mishra S, Sen R. N protein from lambdoid phages transforms NusA into an antiterminator by modulating NusA-RNA polymerase flap domain interactions. Nucleic Acids Res. 2015;43:5744–5758. doi: 10.1093/nar/gkv479. An example studying protein/RNA interaction with Fe-EDTA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Boulanger A, Moon K, Decker KB, Chen Q, Knipling L, Stibitz S, Hinton DM. Bordetella pertussis fim3 gene regulation by BvgA: Phosphorylation controls the formation of inactive vs. active transcription complexes. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E526–E535. doi: 10.1073/pnas.1421045112. An example studying protein/RNA interaction with Fe-EDTA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kent OA, MacMillan AM. Early organization of pre-mRNA during spliceosome assembly. Nat Struct Biol. 2002;9:576–581. doi: 10.1038/nsb822. [DOI] [PubMed] [Google Scholar]
- 31.Mjos KD, Orvig C. Metallodrugs in Medicinal Inorganic Chemistry. Chem Rev. 2014;114:4540–4563. doi: 10.1021/cr400460s. [DOI] [PubMed] [Google Scholar]
- 32.Chen JY, Stubbe J. Bleomycins: Towards better therapeutics. Nature Reviews Cancer. 2005;5:102–112. doi: 10.1038/nrc1547. [DOI] [PubMed] [Google Scholar]
- 33.Santini C, Pellei M, Gandin V, Porchia M, Tisato F, Marzano C. Advances in Copper Complexes as Anticancer Agents. Chem Rev. 2014;114:815–862. doi: 10.1021/cr400135x. [DOI] [PubMed] [Google Scholar]
- 34••.Yu Z, Han ML, Cowan JA. Toward the Design of a Catalytic Metallodrug: Selective Cleavage of G-Quadruplex Telomeric DNA by an Anticancer Copper-Acridine-ATCUN Complex. Angew Chem Int Ed. 2015;54:1901–1905. doi: 10.1002/anie.201410434. A G-quadruplex-targeting Cu complex that can promote cleavage of G-quadruplex telomeric DNA and telomere reduction in cancer cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nature Chemistry. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joyner JC, Hodnick WF, Cowan AS, Tamuly D, Boyd R, Cowan JA. Antimicrobial metallopeptides with broad nuclease and ribonuclease activity. Chem Commun. 2013;49:2118–2120. doi: 10.1039/c3cc38977d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38••.Chen ZW, Ji HW, Liu CQ, Bing W, Wang ZZ, Qu XG, et al. A Multinuclear Metal Complex Based DNase-Mimetic Artificial Enzyme: Matrix Cleavage for Combating Bacterial Biofilms. Angew Chem Int Ed. 2016;55:10732–10736. doi: 10.1002/anie.201605296. An example describing Ce-containing nanopartiple that can inhbit biofilm formation throught promoting eDNA cleavage. [DOI] [PubMed] [Google Scholar]
- 39.Joyner JC, Cowan JA. Target-directed catalytic metallodrugs. Braz J Med Biol Res. 2013;46:465–485. doi: 10.1590/1414-431X20133086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Joyner JC, Cowan JA. Targeted Cleavage of HIV RRE RNA by Rev-Coupled Transition Metal Chelates. J Am Chem Soc. 2011;133:9912–9922. doi: 10.1021/ja203057z. Metal complexes that selectively cleave HIV RRE RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joyner JC, Keuper KD, Cowan JA. Kinetics and mechanisms of oxidative cleavage of HIV RRE RNA by Rev-coupled transition metal-chelates. Chemical Science. 2013;4:1707–1718. doi: 10.1039/C3SC22135K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bradford S, Cowan JA. Catalytic metallodrugs targeting HCV IRES RNA. Chem Commun (Camb) 2012;48:3118–3120. doi: 10.1039/c2cc17377h. [DOI] [PubMed] [Google Scholar]
- 43•.Bradford SS, Ross MJ, Fidai I, Cowan JA. Insight into the Recognition, Binding, and Reactivity of Catalytic Metallodrugs Targeting Stem LoopIIb of HepatitisC IRES RNA. ChemMedChem. 2014;9:1275–1285. doi: 10.1002/cmdc.201400070. Substrate-selective metallopeptides that degrades HCV SLIIB IRES RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Ross MJ, Bradford SS, Cowan JA. Catalytic metallodrugs based on the LaR2C peptide target HCV SLIV IRES RNA. Dalton Trans. 2015;44:20972–20982. doi: 10.1039/c5dt02837j. Substrate-selective metallopeptides that degrades HCV SLIV IRES RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Z, Cowan JA. Catalytic Metallodrugs: Substrate-Selective Metal Catalysts as Therapeutics. Chem Eur J. 2017;23:14113–14127. doi: 10.1002/chem.201701714. [DOI] [PubMed] [Google Scholar]
- 46.Yu Z, Cowan JA. Design of Artificial Glycosidases: Metallopeptides that Remove H Antigen from Human Erythrocytes. Angew Chem Int Ed. 2017;56:2763–2766. doi: 10.1002/anie.201612079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fidai I, Hocharoen L, Bradford S, Wachnowsky C, Cowan JA. Inactivation of sortase A mediated by metal ATCUN complexes. J Biol Inorg Chem. 2014;19:1327–1339. doi: 10.1007/s00775-014-1190-x. [DOI] [PubMed] [Google Scholar]
- 48.Peacock M. Calcium metabolism in health and disease. Clin J Am Soc Nephrol. 2010;5(Suppl 1):S23–30. doi: 10.2215/CJN.05910809. [DOI] [PubMed] [Google Scholar]
- 49.Musso CG. Magnesium metabolism in health and disease. Int Urol Nephrol. 2009;41:357–362. doi: 10.1007/s11255-009-9548-7. [DOI] [PubMed] [Google Scholar]


