Abstract
Background
Dermatophagoides pteronyssinus (DP) and Dermatophagoides farinae (DF) are highly similar disease-associated mites with frequently overlapping geographic distributions. A draft genome of DP was assembled to identify candidate allergens in DP homologous to those in DF, investigate allergen isoforms, and facilitate comparisons with related Acari.
Methods
PacBio and Illumina whole genome sequencing was performed on DP. Assembly and reconstruction of the genomes were optimized for isoform identification in a heterogeneous population. Bioinformatic analyses of Acari genomes were peformed.
Results
The predicted size of the DP nuclear genome is 52.5 Mb. A predicted protein set of 19,368 proteins was identified, including all 19 currently recognized allergens from this species. Orthologs for 12 allergens established for DF were found. The population of DP mites showed a high level of heterozygosity that allowed the identification of 43 new isoforms for both established and candidate allergens in DP, including a new isoform for the major allergen Der p 23. Reanalyzing the previous DF data assuming heterozygosity, 14 new allergen isoforms could be indentified. Some new isoforms were observed in both species suggesting that these isoforms pre-dated speciation. The high quality of both genomes allowed an examination of synteny which showed many allergen orthologs are physically clustered but with species specific exon/intron structures. Comparative genomic analyses with other Acariformes mites showed that most of the allergen homologs are widely conserved within this Superorder.
Conclusions
Candidate allergens in DP were identified to facilitate future serological studies. While DP and DF are highly similar genetically, species-specific allergen isoforms exist to facilitate molecular differentiation.
Keywords: Houst Dust Mite, Genome, Dermatophagoides pteronyssinus, Dermatophagoides farinae, Allergens
Introduction
Sensitization to indoor allergens is often associated with extrinsic asthma [1]. The most common indoor allergens to which individuals are sensitized in the U.S. come from the house dust mites (HDM) Dermatophagoides pteronyssinus (DP) and Dermatophagoides farinae (DF) [2]. Both species have been identified in human dwellings worldwide, although regional patterns of one species predominanting are noted [3]. DP prefers higher relative humidity, and under optimal conditions has a faster growth rate than DF [4]. Determination of the DF genome aided in the identification of 7 new allergens in an Asian population [5]. There are now 31 known proteins comprising the DF allergome, that is, those proteins having been shown to be allergens in patients, while from DP, 19 proteins, all orthologs of a subset of the DF allergome, are officially recognized by the World Health Organization and International Union of Immunological Societies (WHO/IUIS) as allergens. Given this imbalance, the assembly and annotation of the genome of DP provides a valuable resource for allergists to rapidly assess candidate allergens in this species for their relative patient impact geographically, and the potential for species specific identificiation.
The contribution of genetic variation within the population of an allergy causing organism has been studied. For example in birch pollen there are 27 recorded variants of the major allergen Bet v 1, that have been variously assessed for allergic stimulation [6], IgE binding [7], and even natural ligand affinity [8]. Similarly, the polymorphisms in the group 1 and 2 allergens from mites [9,10] have been studied for variations in antibody binding [11–13], and cytokine production in T-cells [14]. Variations in binding can be extremely important for exposure measurements that rely on antibody detection as well as standardizing patient diagnostics and treatments [15]. Our study presents a unique case where allergen isoform variation can be examined at the genomic level, as our genome was generated from a population of randomly breeding diploid dust mites that has been maintained as a colony for many years. While existing variation present in a protein within a population is likely not going to adversely affect the primary function of a given allergen, variation may affect the allergenicity of a protein as that is likely under neutral selection. This offers us the opportunity to examine the naturally occurring variation within a complete collection of allergen genes in an important disease-associated organism.
There exist thousands of common protein domains, although surprisingly, only a few of these are allergenic [16,17]. Within the proteome of DP less than 0.1% of the proteins are allergens [18]. This may be due to the cellular location of a protein within an organism and/or the persistence and stability of a protein in the environment [18]. Many allergens are also members of gene families in which only one of the genes encodes an allergen (e.g. chitin binding domains, cysteine and serine proteases, amylases, and glutathione S transferases). Thus, the presence of a domain is not necessarily predictive of allergenicity. Our genome assembly and annotation allows us to compile collections of gene families of all known allergens. Subsequent analysis of these protein datasets may allow a better understanding of what makes only a small subset of the proteins in a gene family allergenic.
Methods
DP were captured in Ohio, USA and maintained in culture for many years. DP growth conditions were previously described [19]. DNA isolation from whole mite extract is detailed in the Supplemental Material. Explicit methods regarding genome assembly and protein prediction are also described in the Supplemental Material. All original sequence data used herein are deposited to Genbank under Bioproject PRJNA395246.
All multiple sequence alignments were generated using muscle within CLC Genomics Workbench.
Several genomes from Acari suborder, which includes mites and ticks, have recently been published including D. farinae [5], scabies mites Sarcoptes scabiei (SS) [20–22], the honey bee mite Tropilaelaps mercedesae [23], the spider mite Tetranychus urticae [24], which is preyed upon by the mite Metaseiulus occidentalis [25], Ixodes scapularis [26,27], and Varroa destructor [28]. Four mite genomes from the order Oribatida have also been sequenced: Achipteria coleoptrata, Platynothrus peltifer, Steganacarus magnus, and Hypochthonius rufulus [29]. Genomic data for these species was downloaded from GenBank (Table 1).
Table 1.
Genome Statistics for available mite and tick genomes
| Species | Genome size | genome/scaffold N50 | scaffolds | predicted proteins* | Genome NCBI Accession |
|---|---|---|---|---|---|
| Dermatophagoides pteronyssinus | 52.5 Mb | 376 kb | 834 | 19,368 | |
| Dermatophagoides farinae | 53.5 Mb | 187 kb | 515 | 16,376 | GCA_000767015.1 |
| Sarcoptes scabiei | 56.3 Mb | 11.2 kb | 18,860 | 10,644 | GCA_000828355.1 |
| Achipteria coleoptrata | 88.4 Mb | 7.5 kb | 56,345 | ND | GCA_000988765.1 |
| Platynothrus peltifer | 100.5 Mb | 1.6 kb | 105,671 | ND | GCA_000988905.1 |
| Steganacarus magnus | 113.6 Mb | 2.5 kb | 101,545 | ND | GCA_000988885.1 |
| Hypochthonius rufulus | 535 Mb | 4.2 kb | 140,449 | ND | GCA_000988845.1 |
| Tetranychus urticae | 90.8 Mb | 212.8 Mb | 641 | 15,054 | GCA_000239435.1 |
| Ixodes scapularis | 1.76 Gb | 2.94 | 369,492 | 20,467 | GCA_000208615.1 |
| Varroa destructor | 331.9 Mb | 15.6 | 20,448 | ND | GCA_000181155.1 |
| Metaseiulus occidentalis | 151.7 Mb | 200.7 kb | 2,211 | 18,338 | GCA_000255335.1 |
| Tropilaelaps mercedesae | 352.5 Mb | 12.7 kb | 33,764 | 14,342 | GCA_002081605.1 |
The eleven publicly available genomes for the Acari are listed. The five for which publicly available proteomes were available were used for all comparative genomics described herein.
Results
Genome assembly
The genome of DP was assembled from 303,594 PacBio ccs (circular consensus sequencing) reads. After the removal of the microbiome containing contigs, this resulted in a 52.5 Mb genome assembly of 834 contigs with an N50 of 376 kb. A comparison to other available mite and tick genomes is shown in Table 1. This assembly is among the more complete available in terms of the N50 measurement. Judging by the comparison to the extended CEGMA dataset [30] results 97.7% (2686/2748) of these genes were represented in our main assembly, also suggesting a high level of completeness. Analysis with the BUSCO arthropod geneset of 2675 genes [31] found 53.4% complete genes in our genome. A recent summary of nine available arachnid genomes analyzed with BUSCO using this geneset found a mean of 51.4 % complete genes with a range of 13.7–82.3 %[25]. Details regarding the mitochondrial genome [32], microbiome composition, repeat content, and tRNA content and organization can be found in the Supplemental Material.
Allergen identification
Nineteen allergens have been curated from DP by WHO/IUIS, in contrast to 31 from DF. Within our predicted proteome we found orthologs of all 12 DF allergens that have yet to be tested in DP allergic patients (candidate DP groups 16, 22, 25–34). Nearly all were found as full length proteins (Supplemental File 1). Of note, we did identify a full length version of Der p 4, for which only a partial version is listed at WHO/IUIS. We extended our search for conserved allergen orthologs to the other predicted mite proteomes; this is summarized in Supplemental Table 1. For ortholog identification of smaller datasets such as the allergens, we used orthoparahomlist.pl [21]. Twenty of the known allergens in the Dermatophagoides lineage have orthologs in all mites. Of these twenty conserved within mites, 18 were also found in Daphnia pulex and Drosophila melanogaster, suggesting a core group of proteins, that do, in some of these organisims, act as allergens, that are likely to be generally conserved within Arthropods, including the major allergen Der p 1. Only one, Der p 6, a serine protease, appears to be unique to the Dermatophagoides lineage. Also, a Der p 4 ortholog was specifically missing from all three sequenced Sarcoptes scabiei genomes [20–22].
Prediction of novel isoforms of known allergens
Isoforms are defined here as variants from the same genetic locus with one or more amino acid substitutions, insertions, or deletions. Within DP many isoforms have been identified for some of the major allergens such as Der p 1 and Der p 2 that have 24 and 15 isoforms, respectively, but for most others only one isoform has been identified and characterized as an allergen. Only for Der p 5, Der p 9, and Der p 15 have two isoforms been identified. All the sequence data that were generated in this study came from a single laboratory maintained population of randomly breeding DP. Initial characterization of this sequence data by kmer analysis suggested a high level of heterozygosity within this population (Supplemental Figure 1). This was useful for characterization of the genetic variation within this species and the identification of allergen isoforms, but technically presented a challenge in genome assembly (See Supplemental Methods).
To assess this variation within the predicted DP allergome, we searched several of our genome and transcriptome assemblies produced independently from different sequence collections using different algorithms. The assumption was that if new or multiple isoforms of a given protein were present in the population, different assemblies, generated from different sequence preparations and assembled with different algorithms, might differentially predict an isoform, at least for isoforms that are abundant in this population. Furthermore, independently generating these isoforms from different assemblies would give at least in silico validation of their existence. Two transcriptomes from our RNA-seq data (using either Trinity or soapdenovo-trans), two genome assemblies based on the Illumina dataset (using either Phusion or soapdenovo) and the primary genome assembly based on the PacBio data were used to query from the collection of known and candidate allergen proteins and protein predictions were assembled from these. An isoform of an allergen was considered to be new in cases which there is at least one amino acid substitution in any of various data assemblies.
In total, 43 new isoforms were found for 24 known and candidate allergens, including the 12 candidate allergen isoforms in DP related to groups 16, 22 and 24–34 which had not previously identified (Table 2). Only existing isoforms were found for Der p 1 and Der p 2. This might be expected given the depth of the characterization of isoforms for these two major allergens. For 10 of the 19 described DP allergens we did find previously identified isoforms, suggesting the validity of our approach. For the other 9 we found only new isoform(s). Most of these isoforms were found in more than one assembly, giving a higher confidence that their prediction is not due to some anomaly of sequence error or assembly. For 13 allergens, two different isoforms were found in the population and for 6 allergens three or four isoforms were found.
Table 2.
Allergen isoforms identified in the genomes and proteomes of D. pteronyssinus and D. farinae
| Allergen ID | Function | Isoform(s) | Conserved substitutions | Allergen ID | Isoform(s) | Published |
|---|---|---|---|---|---|---|
| Der p 1 | Cysteine protease | Der p 1.0105 | Der f 1 | Derf1.0101 | DEFA_073880 | |
| Der p 2 | Lipid binding | Der p 2.0101, Der p 2.0103 | Der f 2 | Derf2.0102 | DEFA_057430 | |
| Der p 3 | Trypsin | Der p 3.0101, cDerp3b**, cDerp3c** | 17L>W,127N>D, 138A>S | Der f 3 | cDerf3b | DEFA_036500 |
| Der p 4 | alpha amylase | Der p 4.0101, cDerp4b**, cDerp4c** | 523D/G | Der f 4 | cDerf4b* | DEFA_092370 |
| Der p 5 | Structural protein | Der p 5.0101 | Der f 5 | cDer5* | DEFA_009370 | |
| Der p 6 | Chymotrypsin | Der p 6.0101, cDerp6b** | 84L>M | Der f 6 | cDerf6b* | DEFA_160240 |
| Der p 7 | Unknown | cDerp7b, cDerp7c, cDerp7d** | 33I>V | Der f 7 | cDerf7b* | DEFA_012670 |
| Der p 8 | Glutathione transferase | cDerp8b*** | 48Q/E, 162Y/N | Der f 8 | cDerf8b* | DEFA_112610 |
| Der p 9 | Serine protease | cDerp9c*, cDerp9d** | 236S/A | Der f 9 | Derf9.0101 | DEFA_108510 |
| Der p 10 | Tropomyosin | cDerp10b, cDerp10c, cDerp10d* | 258H/Y,267T/A | Der f 10 | Derf10.0101, cDerf10b* | DEFA_012620 |
| Der p 11 | Paramyosin | cDerp11b**, cDerp11c*** | 125Q>H,273E>D,360T>A,382N>D | Der f 11 | cDerf11b* | DEFA_029610 |
| Der p 13 | Fatty acid binding | Der p 13.0101 | Der f 13 | Derf13.0101 | DEFA_016640 | |
| Der p 14 | Vitellogenin | cDerp14b**, cDerp14c**, cDerp14d* | 580S>G,813A>S | Der f 14 | cDerf14b* | DEFA_023480 |
| Der p 15 | Chitinase | Der p 15.0101, Der p 15.0102 | Der f 15 | cDerf15b | DEFA_127470 | |
| CDer p 16 | Gelsolin | cDerp16a**, cDerp16b* | 460D>G | Der f 16 | Derp16.0101, cDerf16b* | DEFA_053360 |
| Der p 18 | Chitinase | cDerp18b**, cDerp18c*** | 457T>A | Der f 18 | Derf18.0101 | DEFA_042810 |
| Der p 20 | Arginine kinase | Der p 20.0101, cDerp20b, cDerp20c | 354I/V,384I/M | Der f 20 | Derf20.0101 | DEFA_122350 |
| Der p 21 | Structural protein | Der p 21.0101, cDerp21b | Der f 21 | Derf21.0101 | DEFA_009360 | |
| CDer p 22 | MD-2–related lipid recognition | cDerp22a*, cDerp22b** | 154H>Q | Der f 22 | Derf22.0101 | DEFA_072800 |
| Der p 23 | Chitin-binding domain type 2 | Der p 23.0101, cDerp23b**, cDerp23c | Der f 23 | Derf23, cDerf23b | DEFA_123860 | |
| Der p 24 | Ubiquinol-cyt C reductase binding | Der p 24.0101 | Der f 24 | Derf24.0101 | DEFA_162130 | |
| CDer p 25 | Triose phosphate isomerase | cDerp25a*, cDerp25b* | Der f 25 | cDerf25b* | DEFA_001450 | |
| CDer p 26 | Myosin, light chain | cDerp26a, cDerp26b*** | 126A>S | Der f 26 | Derf26.0101 | DEFA_126820 |
| CDer p 27 | Serpin | cDerp27 | Der f 27 | cDerf27b* | DEFA_144510 | |
| CDer p 28 | Heat shock protein | cDerp28 | Der f 28 | Derf28.0101 | DEFA_150350 | |
| CDer p 29 | Cyclophilin | cDerp29 | Der f 29 | Derf29.0101 | DEFA_018720 | |
| CDer p 30 | Ferritin | cDerp30a**, cDerp30b*** | 48N/D,58E/R,62D/H,66K/E,74R/K,78L/F | Der f 30 | cDerf30b* | DEFA_057540 |
| CDer p 31 | Cofilin | cDerp31a**, cDerp31b* | insertion | Der f 31 | Derf31.0101 | DEFA_002000 |
| CDer p 32 | Inorganic pyrophosphatase | cDerp32 | Der f 32 | Derf32.0101 | DEFA_053550 | |
| CDer p 33 | alpha tubulin | cDerp33a*, cDerp33b | 125M/L,133A/S | Der f 33 | Derf33.0101 | DEFA_107410 |
| CDer p 34 | Endoribonuclease | cDerp34a**, cDerp34b | 125A>T,144V>I,147A>Q | Der f 34 | cDerf34b* | DEFA_019630 |
Table 2 Footer: The names in bold under the heading Allergen ID are newly identified candidate allergens in DP. Any newly identified candidate allergens/isoforms are noted with the prefix “c”. Newly identified isoforms are noted with an alphabetic suffix of b, c, d, etc. Under Isoforms, those found in the DP or DF population are listed as indicated. One asterisk indicates validation based on prediction in multiple assemblies. Two asterisks indicate validation based on conserved amino acid substitutions between the two species. Three asterisks indicate validation of an isoform based on both of the above criteria. All identified amino acid substitutions seen in both species are listed under Conserved Substitutions. Those for which an ancestral state cannot be inferred are listed in the form of A/B, those for which an ancestral state can be conferred are listed as A>B, with A being the ancestral amino acid. For the DF allergens only confirmation by prediction in another DF transcriptome assembly is noted by an asterisk. The DF Allergen ID contains gene names for the allergens from either the DF predicted proteome (Table 1 of [5]) or those assigned as the ortholog to the appropriate DP allergen. Those listed in bold are also new isoforms that were identified in that paper, but not noted at the time; those not in bold are identical in sequence to the isoforms already defined. For Der f 34, DEFA_019630 is the candidate isoform in the published proteome, the isoform we identified is distinct from this and Der f 34.0101 and is in Supplemental file.
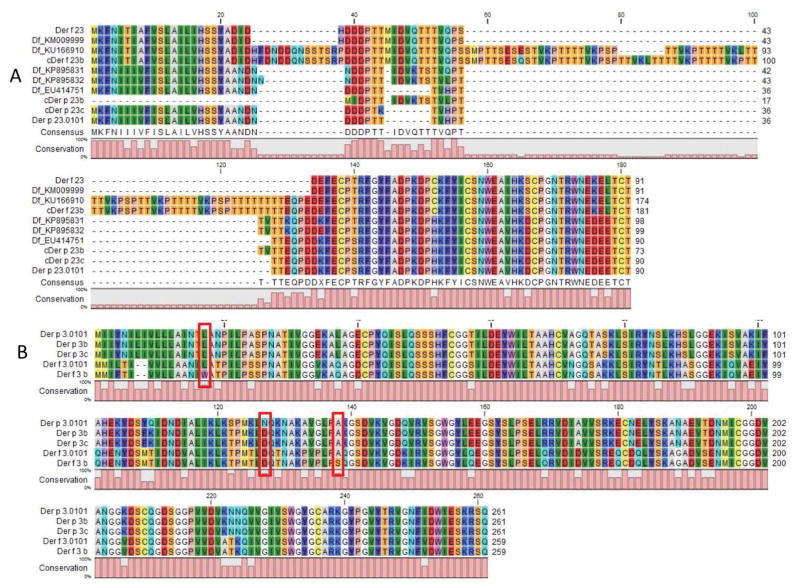
Only for DP groups 5, 13, 15, and 24 were no new isoforms found, suggesting that these proteins are well conserved and possibly subject to more functional selection constraint than the others. Der p 23 is a special case where we found two new candidates (cDer p 23b and cDer p23c). Genbank has five additional Der f 23 variants (Figure 1a), including one, KU166910, with a >60 amino acid insertion with seven copies of a seven amino acid repeating sequence. An examination of the DF genome for a KU166910-like sequence found a new Der f 23 isoform (cDer f 23b) as a single ORF that has a very similer extension containing eight copies of this repeating sequence (Figure 1a). The DP genome contains only the Der p 23 isoforms noted above, all without such extensions. Outside of the group 1 and group 2 allergens, group 23 has the most described isoforms of any Dermatophagoides allergen.
Figure 1. A multiple sequence alignment of the isoforms of groups 23 and 3.
A) All known and newly identified isoforms of group 23 from both D. pteronyssinus and D. farinae are shown aligned. B) Both known and newly identified isoforms of group 3 from both species are included. The three residues boxed in red are examples of amino acid variation conserved between species. As L17, D127, and A138 are observed in both species, the most parsimonious assumption is that these are the ancestral amino acids at these three position and the other is derived after speciation. A list of conserved amino acid substitutions in all allergen isoforms is listed in Table 2.
Novel Isoforms in DF
A more confident validation of the new isoforms found above would be if the sequence variation could also be found in a related species as this would imply a vertical transmission of that isoform, not a confounding assembly or sequence error. A similar search for new isoforms was performed for the previously published DF genome, as heterozygosity was not previously addressed. From WHO/IUIS many isoforms for DF allergen groups 1, 2, 10, and 17, are described, while only two isoforms for three other allergen groups, 20, 25, and 28 are listed. We obtained the DF protein predictions from the authors [5] and independently assembled their RNA-seq data (SRX367593) with Trinity and performed a protein prediction with this and an independent protein prediction with their genome sequence using SNAP [33]. We compared these protein predictions with the set of known DF allergens and with multiple sequence alignments compared the new allergen proteins to the known ones. We again found many examples of new isoform candidates for allergens, suggesting that the mite population used for sequencing the DF genome was from a diverse population also (Table 2). For 11 of the 31 known allergens a new isoform was found, including one for the most recently described Der f 34 allergen [34]. For 8 of these allergens only the novel form was found in any of the protein predictions, and for 6 allergens two isoforms were found. As described for DP, the DF groups 5, 13, and 24 contain only the originally described isoform emphasizing the hypothesis that these three are subject to more functional constraint.
In many cases, a new isoform was found in independent assemblies generated from different sequence preparations. To further validate these new isoforms in both species, multiple alignments were made between the orthologous allergens between the two Dermatophagoides species and for 16 allergens, some of the amino acid substitutions seen in new DP isoforms were observed also in a DF isoform suggesting that many of these differences pre-dated the speciation event from which these two mites resulted. Der p 3 is shown as an example in Figure 1b; L17 is seen in isoforms of both species, while a W17 isoform is seen only in DF. The most parsimonious explanation is that L17 is the ancestral state in both species and that W17 arose in DF after speciation. A similar explanation applies for for A138, but the opposite explanation would apply for D127. In this case D127, present in all Der p 3 isoforms, would be the ancestral amino acid while in DF, one isoform contains a sequence change resulting in N127. For nine of the allergens and candidate allergens, the ancestral state of specific amino acids could be determined (Table 2). In all, for 34 of the 43 newly identified DP allergen isoforms we could validate their existence by one of the above approaches. The 16 allergens for for which we found an isoform conserved between DF and DP are likely genuinely conserved differences as they are found in both species. For those isoforms for which validation is only by confirmation between assemblies, or which are only found in a single assembly, re-sequencing within this population would be an appropriate confirmation.
Genomic organization of allergens in Dermatophagoides
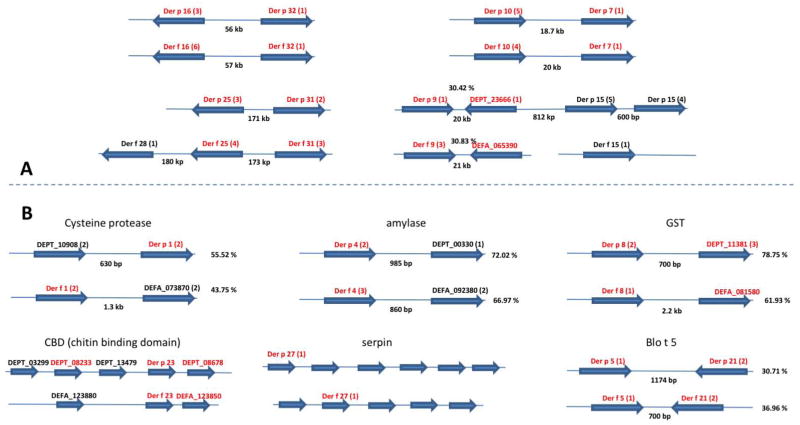
The completeness of the DF and DP genomes allowed a detailed analysis of the conservation of organization of the allergens in both genomes. All of the observed clustering of allergen genes in DP and DF is shown in Figure 2. Many cases of clustering of allergens were found, for instance groups 7 and 10, groups 16 and 32, among others displayed in Figure 2a. In most of these cases, synteny was conserved between the two mites, although this may be underestimated due to gaps in a genome assembly. For instance, Der f 25, Der f 28, and Der f 31 are linked within 350 kb in DF, but only Der p 25 and Der p 31 are linked in the DP assembly.
Figure 2. Conserved genomic organization of allergens.
A) clusters of unrelated allergens that are conserved between D. pteronyssinus and D. farinae. B) Cartoon depiction of the organization of allergens that are part of gene families. In both, orientation of the genes is indicated by an arrow, and the approximate distance between linked genes is shown, not to scale. In parenthesis beside the gene name is the number of exons in a given gene. All chitin binding domain and serpin genes are single exon genes. Orthologous relationships, as defined by the orthoparahomlist.pl script, are highlighted in red. The percent identity between members of a gene family within a species is on the right of a given gene pair. The orthologous relationships between the members of the serpin gene family is inconclusive aside from the Der 27 allergens.
Some of the allergens that exist as part of larger gene families of proteins, were also found to be within clusters of two or more related genes, such as group 1 (cysteine protease), group 4 (amylase), group 8 (glutathione-S transferase), group 23 (chitin binding domain protein, CBD) and group 27 (serpin), Figure 2b. Some of the gene families are extensive. There are 48 CBD (major allergen group 23) proteins in DP (47 in DF) and 5 of these in DP, including Der p 23, are very closely linked within 4 kb while three, including Der f 23, are similarly linked in DF; all three of the latter have orthologous relationships between the two organisms. There are 17 serpins in DP, 6 of these are found within 13 kb, while 5 of 12 serpin proteins are found within 10 kb in DF. Both Der p 1 and Der f 1 genes are part of the larger cysteine protease gene families and are adjacent to related cysteine protease genes in a tandem arrangement. Der p 1 has 55% identity at the amino acid level to DEPT_10908 while Der f 1 has 43 % to DEFA_073870. The allergens of group 5 and 21 are distantly related and present an interesting case of a linkage between related allergens. Each are physically close in both mites, and are in an inverted orientation with similar exon/intron structures. Although the Dermatophagoides groups 5 and 21 proteins have a low identity to each other in both species (30.71 % in DP and 36.96 % in DF) both are clearly related to the Blot 5 allergen of the Blomia tropicalis mite. An examination of the allergen genes alone suggests a high level of conservation of synteny between DP and DF, given the conservation of the orientation of the linked genes and relatively similar distances between them. In the two more extensive gene families, the CBDs and serpins, there does appear to be differentiation between the two species as the cluster of 5 CBD genes in DP is a cluster of three in DF. This could be explained either by gene loss in DF or an expansion of the gene family in DP. While the genomic organization of the above described allergens between DP and DF is well conserved, at the gene level there is little intragenic conservation at the exon/intron level. In only one of the linked pairs, Der 5 and Der 21 (the Blot 5 family) is the exon/intron structure conserved between both species in both allergens.
Comparative genomics of mites and ticks
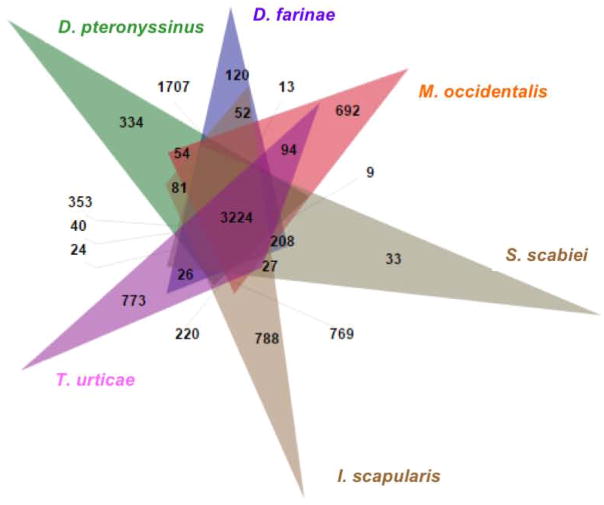
For six of the species in Table 1, genome-wide protein predictions are available, including that of DP presented here. The biology of the Acari has not been well explored at the genomic level and this dataset allows us an opportunity for a variety of important comparisons between these species. We used OrthoVenn to examine the conservation and intersection of the proteomes of these six organisms (Figure 3) [35]. Table 3 shows the total number of proteins, clusters, and singletons in each species. 3224 gene clusters were conserved between all six species. The following summarizes a statistical analysis of GO terms associated with these gene clusters (data not shown). Only one GO term is over-represented in this set by a hypergeometric test at a p-value < 0.05, which is GO: 0051539, 4 iron, 4 sulfur cluster binding protein. A set of 1707 clusters are unique to the Dermatophagoides lineage. Within this set, three GO biological processes are over-represented (p < 0.05): response to nitrosative stress (GO:0051409), cytoplasmic microtubule organization (GO:0031122), and cellular response to nutrient (GO:0031670). In contrast to this, 13 biological processes are over-represented within DP (Supplemental Table 2). All of these involve enzymatic processes, including two peptidase activities, tripeptidyl-peptidase activity (GO:0008240) and exopeptidase activity (GO:0008238) and four different polyphosphate processes. None of these are over-represented in DF in which cell differentiation processes are prominent. Within the central cluster common to all six mites, several types of cytochrome P450s, Glutathione S transferase-1, and an ABC transporter have undergone expansions, suggesting responses to environmental exposure is a common threat to all species. The protein ‘doublesex’ has also undergone an expansion. This and the presence of other insect sexual development proteins in the proteomes of all additional mites and ticks examined (Supplemental Table 3), suggests that there is some conservation in the sexual development strategy between these species and insects. Most conserved expansions are of proteins of known function.
Figure 3. Conserved orthologs in mite and tick genomes.
Predicted proteomes from five publicly available mites and ticks, and D. pteronyssinus, were used. A Venn diagram showing the distribution of conserved ortholog clusters. A cluster is defined as a group of related proteins having a BLAST similarity of at least 10−05. The central overlap of all six species (3224 clusters) represents those proteins that have one or more orthologs in each of the six species at a BLAST cutoff of 10−05, whereas the various other overlapping groups contain clusters representing orthologs conserved between two or more species. The outer spikes (i.e., the D. pteronyssinus spike containing 334 clusters) represent clusters (gene families) of two or more proteins unique to a given species only. The area of the spikes are not proportional to the numbers of genes they contain.
Table 3.
A summary of the ortholog analysis in Figure 3
| Species | Proteins | Clusters | Singletons |
|---|---|---|---|
| D. pteronyssinus | 19368 | 9496 | 8082 |
| D. farinae | 16376 | 8931 | 6505 |
| M. occidentalis | 18338 | 6943 | 4767 |
| S. scabiei | 10473 | 7330 | 2506 |
| I. scapularis | 20486 | 7420 | 9140 |
| T. urticae | 18224 | 6726 | 6042 |
Discussion
The quality of a genome assembly can be judged in a variety of ways. The N50 measurement is often used as a proxy for the completeness of a genome [36]. As noted in Table 1 our assembly compares favorably to other related genomes. A good indication of this is that we found nearly all the syntenic relationships between allergens in both DF and DP to be conserved (Figure 1). Other measurements should also be considered. In our case, it was important to uncover as much information about known allergens as possible. Importantly we found full length versions of all known allergen genes, discovered new candidate isoforms of most, sixteen of which contain variations also seen in DF which thus represents a good evolutionary validation for this subset of isoforms. We also found a set of candidate sexual development genes identical to those found in other mites and ticks (Supplemental Table 3). The extended CEGMA dataset gives a measure of overall eukaryotic gene content, these genes were represented extremely well in the DP assembly. The BUSCO analysis [31], which is a comparison to a collection of conserved arthropod genes, showed only average assembly coverage. Given that the mean BUSCO coverage of all mite and tick genomes was 51.4 % [25] this suggests that a more arachnid-centric dataset may need to be developed. Thus, at the level of specific gene sets of interest, and at an overall level, our genome is a good representation of DP. Other elements of a eukaryotic organisms’ genome were also well represented, including a complement of tRNA genes nearly identical to DF but with a very different organization; 40% of the tRNA genes in DF are within a cluster on a single sequence scaffold; in DP 53% of the tRNAs are clustered but only in small clusters of 2–5 tRNAs. We also found a complete version of a new mitochondrial haplotype for DP and a near full length (5 Mb) genome of Serratia marcescens, the dominant component of the DP microbiome. All data indicators suggest that the sequencing and assembly is high quality and we have accurately identified new isoforms.
Previously we reported that DP allergens represented 0.1% of the DP proteome based on the 19 allergens in WHO/IUIS versus the 25,445 RNAseq transcripts [18]. Using the genome as a guide, we can now revise the total number of proteins from DP to be 19,368, and if we recalculate the percentage of allergens this still rounds to 0.1%. If the 12 DP candidate allergens identified herein have IgE reactivity, that could rise to 0.2%. While the allergens are rare in the genome, some like Der p 1 are highly expressed, which is why exposure measurements have focused on detecting this allergen [37]. However, using the genome as a guide and mass spectrometry techniques, we were able to identify peptides in house dust extract that were unique to DP from proteins that are not currently classified as allergens (data not shown). This indicates that exposure is not limited to allergens. The genome will therefore allow researchers to better catalogue the totality of human exposure which hopefully will help lead to a better understanding of why certain proteins are tolerated and some are allergenic.
Another utility of the genomic analysis is that while DP and DF are closely related organisms, there are hundreds of proteins that are species specific. These proteins may be useful molecular probes to accurately map the geographic range of the species and differentiate exposure.
Intriguingly, a Der p 4 ortholog is missing in SS. This absence of Der p 4 is significant because it has been reported that SS infected patients have a very high anti-Der p 4 IgE titer, on a par with the IgE titer to Der p 1 and Der p 2 [38]. Evidence for Der p 4 exists in either the genome or predicted protein set in all the other 10 mites and ticks and the two outgroups mentioned above, D. pulex and D. melanogaster. Furthermore, Der p 4 was not present in all three available SS genomes that have been sequenced, [20–22] suggesting that its absence is not simply due to the incompleteness of either a single SS genome or its’ protein predictions, but likely a true absence. Der p 4 is an amylase and three other amylases are also predicted in the two Dermatophagoides spp. protein sets; orthologs of these are also found in SS. However these other amylases in SS are low in sequence identity (< 26%) to Der p 4. Without a close homolog to Der p 4 in the SS genome, the reason for this abnormally high response to Der p 4 in scabies patients bears further investigation.
A recent study on the major birch allergen Bet v 1 highlights many of the challenges in creating allergy reference standards [39]. The study demonstrated that in patient products used for immunotherapy from different manufacturers there is a high variability in allergen content [40,41], which has also been noted in cockroach extracts [42]. In addition, a direct comparison of two proposed ELISA standards for Bet v 1 detection showed some discrepancies that were attributed to isoform variations of Bet v 1. At the heart of the isoform variation is genetic diversity. This diversity we were able to describe in some detail from the population of DP mites in this study and a re-evaluation of the DF genomic data [5].
Among the known isoforms of the common Dermatophagoides allergens, many known isoforms have been described of the group 1 and 2 allergens [9,10]. Several studies have corroborated the Bet v 1 study above indicating that the isoform variations affect antibody binding to mite allergens [11–13]. We found the existence of multiple isoforms of many of the known allergens in DP. A more detailed analysis of the existing genomic data for DF found a similar level of variation. As both these HDM genomes were assembled from isolated populations of laboratory-maintained mites, this may underestimate variations in existing population structure in dust mite allergens worldwide, undermining the hope for a suitable reference standard. However, the data presented in Table 2 shows that at least some of the isoform variation predates the speciation event in Dermatophagoides spp. This suggests that with proper characterization and knowledge of existing isoforms, standardization should be possible.
The human genome has been analyzed for new drug targets for treating human disease and improving human health [43]. Arthropod genomes are also analysed for drug or pesticide targets, i.e. for essential mite molecules or mechanisms that can be targeted with lethal compounds [44]. In the same way the genetic revolution has also expanded to include the development of acaricides, for example in ticks [45] and spider mites [46]. Patients sensitized to mite allergens are encouraged to reduce allergen exposure in the home with multiple integrated strategies [47]. Current acaricide treatment using tannic acid or benzyl benzoate alone is generally regarded as ineffective in reducing allergen levels [48,49]. However, the singletons in Table 3 could be promising selective acaricide targets for mites because there are no relatives in current protein databases. This study of the DP genome may also be useful in the developement of acaricides with improved effectiveness.
Knowledge of the genomes and proteomes of multiple mite species will allow molecular allergologists to rapidly make comparisons of cross-reacitivty, species specificity, and exposure measurements as allergens are characterized. We identified multiple isoforms of many allergens within a single population of both DP and DF which suggests there is much more allergen variation yet to be discovered. The likelihood is that this genomic variation could contribute to differences in allergenicity between mite populations. This study should highlight the need to understand the interplay between human genetic variation and the genetic variation of the allergy causing organisms as being important in understanding allergies in general.
Supplementary Material
Acknowledgments
The authors thanks S. Dean Rider and Larry Arlian for providing the genomic mite DNA and helpful suggestions. The authors thank David Fargo and Jian-Liang Li for a critical reading of the manuscript. This study was supported by the Intramural Research Program of the National Institute of Environmental Health Sciences (Z01- ES102885-01).
Footnotes
Conflicts of Interest- All authors are U.S. Government employees or contractors and have no conflicts of interest or competing interests to disclose.
Ethical Statement- No human patients were studied.
Author Contributions
TR analyzed genomic data. JM and NISC performed genome sequencing and quality analysis. TR and GM analyzed data and wrote the paper.
Conflicts of Interest
The authors affirm there are no conflicts of interest to report.
References
- 1.Salo PM, Arbes SJ, Crockett PW, Thorne PS, Cohn RD, Zeldin DC. Exposure to multiple indoor allergens in US homes and its relationship to asthma. J Allergy Clin Immun. 2008;121:678–684. doi: 10.1016/j.jaci.2007.12.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salo PM, Arbes SJ, Jaramillo R, Calatroni A, Weir CH, Sever ML, Hoppin JA, Rose KM, Liu AH, Gergen PJ, Mitchell HE, Zeldin DC. Prevalence of allergic sensitization in the United States: Results from the National Health and Nutrition Examination Survey (NHANES) 2005–2006. J Allergy Clin Immun. 2014;134:350–359. doi: 10.1016/j.jaci.2013.12.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas WR. Geography of house dust mite allergens. Asian Pac J Allergy. 2010;28:211–224. [PubMed] [Google Scholar]
- 4.Arlian LG, Confer PD, Rapp CM, Vyszenski-Moher DL, Chang JCS. Population dynamics of the house dust mites Dermatophagoides farinae, D-pteronyssinus, and Euroglyphus maynei (Acari: Pyroglyphidae) at specific relative humidities. J Med Entomol. 1998;35:46–53. doi: 10.1093/jmedent/35.1.46. [DOI] [PubMed] [Google Scholar]
- 5.Chan TF, Ji KM, Yim AK, Liu XY, Zhou JW, Li RQ, Yang KY, Li J, Li M, Law PT, Wu YL, Cai ZL, Qin H, Bao Y, Leung RK, Ng PK, Zou J, Zhong XJ, Ran PX, Zhong NS, Liu ZG, Tsui SK. The draft genome, transcriptome, and microbiome of Dermatophagoides farinae reveal a broad spectrum of dust mite allergens. J Allergy Clin Immunol. 2015;135:539–548. doi: 10.1016/j.jaci.2014.09.031. [DOI] [PubMed] [Google Scholar]
- 6.Smole U, Balazs N, Hoffmann-Sommergruber K, Radauer C, Hafner C, Wallner M, Ferreira F, Grossinger R, de Jong EC, Wagner S, Breiteneder H. Differential T-cell responses and allergen uptake after exposure of dendritic cells to the birch pollen allergens Bet v 1.0101, Bet v 1.0401 and Bet v 1. 1001. Immunobiology. 2010;215:903–909. doi: 10.1016/j.imbio.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Ferreira F, Hirtenlehner K, Jilek A, GodnikCvar J, Breiteneder H, Grimm R, HoffmannSommergruber K, Scheiner O, Kraft D, Breitenbach M, Rheinberger HJ, Ebner C. Dissection of immunoglobulin E and T lymphocyte reactivity of isoforms of the major birch pollen allergen Bet v 1: Potential use of hypoallergenic isoforms for immunotherapy. Journal of Experimental Medicine. 1996;183:599–609. doi: 10.1084/jem.183.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Loetzen CS, Jacob T, Hartl-Spiegelhauer O, Vogel L, Schiller D, Sporlein-Guttler C, Schobert R, Vieths S, Hartl MJ, Rosch P. Ligand Recognition of the Major Birch Pollen Allergen Bet v 1 is Isoform Dependent. Plos One. 2015:10. doi: 10.1371/journal.pone.0128677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeong KY, Lee IY, Yong TS, Lee JH, Kim EJ, Lee JS, Hong CS, Park JW. Sequence polymorphisms of Der f 1, Der p 1, Der f 2 and Der p 2 from Korean house dust mite isolates. Exp Appl Acarol. 2012;58:35–42. doi: 10.1007/s10493-012-9553-x. [DOI] [PubMed] [Google Scholar]
- 10.Piboonpocanun S, Malainual N, Jirapongsananuruk O, Vichyanond P, Thomas WR. Genetic polymorphisms of major house dust mite allergens. Clinical and Experimental Allergy. 2006;36:510–516. doi: 10.1111/j.1365-2222.2006.02464.x. [DOI] [PubMed] [Google Scholar]
- 11.Christensen LH, Riise E, Bang L, Zhang CQ, Lund K. Isoallergen Variations Contribute to the Overall Complexity of Effector Cell Degranulation: Effect Mediated through Differentiated IgE Affinity. J Immunol. 2010;184:4966–4972. doi: 10.4049/jimmunol.0904038. [DOI] [PubMed] [Google Scholar]
- 12.Park JW, Kim KS, Jin HS, Kim CW, Kang DB, Choi SY, Yong TS, Oh SH, Hong CS. Der p 2 isoallergens have different allergenicity, and quantification with 2-site ELISA using monoclonal antibodies is influenced by the isoallergens. Clinical and Experimental Allergy. 2002;32:1042–1047. doi: 10.1046/j.1365-2222.2002.01421.x. [DOI] [PubMed] [Google Scholar]
- 13.Tanyaratsrisakul S, Jirapongsananuruk O, Kulwanich B, Hales BJ, Thomas WR, Piboonpocanun S. Effect of Amino Acid Polymorphisms of House Dust Mite Der p 2 Variants on Allergic Sensitization. Allergy Asthma Immun. 2016;8:55–62. doi: 10.4168/aair.2016.8.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hales BJ, Hazell LA, Smith W, Thomas WR. Genetic variation of Der p 2 allergens: effects on T cell responses and immunoglobulin E binding. Clinical and Experimental Allergy. 2002;32:1461–1467. doi: 10.1046/j.1365-2745.2002.01500.x. [DOI] [PubMed] [Google Scholar]
- 15.van Ree R. Indoor allergens: Relevance of major allergen measurements and standardization. J Allergy Clin Immun. 2007;119:270–277. doi: 10.1016/j.jaci.2006.10.033. [DOI] [PubMed] [Google Scholar]
- 16.Radauer C, Breiteneder H. Pollen allergens are restricted to few protein families and show distinct patterns of species distribution. J Allergy Clin Immun. 2006;117:141–147. doi: 10.1016/j.jaci.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 17.Radauer C, Breiteneder H. Evolutionary biology of plant food allergens. J Allergy Clin Immun. 2007;120:518–525. doi: 10.1016/j.jaci.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 18.Ogburn RN, Randall TA, Xu Y, Roberts JH, Mebrahtu B, Karnuta JM, Rider SD, Kissling GE, London RE, Pomes A, Arlian L, Fitzgerald MC, Mueller GA. Are dust mite allergens more abundant and/or more stable than other Dermatophagoides pteronyssinus proteins? J Allergy Clin Immunol. 2017;139:1030–1032. e1031. doi: 10.1016/j.jaci.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avula-Poola S, Morgan MS, Arlian LG. Diet Influences Growth Rates and Allergen and Endotoxin Contents of Cultured Dermatophagoides farinae and Dermatophagoides pteronyssinus House Dust Mites. Int Arch Allergy Imm. 2012;159:226–234. doi: 10.1159/000336026. [DOI] [PubMed] [Google Scholar]
- 20.Mofiz E, Holt DC, Seemann T, Currie BJ, Fischer K, Papenfuss AT. Genomic resources and draft assemblies of the human and porcine varieties of scabies mites, Sarcoptes scabiei var. hominis and var. suis. Gigascience. 2016;5:23. doi: 10.1186/s13742-016-0129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rider SD, Morgan MS, Arlian LG. Draft genome of the scabies mite. Parasite Vector. 2015;8:585. doi: 10.1186/s13071-015-1198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arlian LG, Morgan MS, Rider SD. Sarcoptes scabiei: genomics to proteomics to biology. Parasite Vector. 2016;9:380. doi: 10.1186/s13071-016-1663-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong X, Armstrong SD, Xia D, Makepeace BL, Darby AC, Kadowaki T. Draft genome of the honey bee ectoparasitic mite, Tropilaelaps mercedesae, is shaped by the parasitic life history. Gigascience. 2017;6:1–17. doi: 10.1093/gigascience/gix008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grbic M, Van Leeuwen T, Clark RM, Rombauts S, Rouze P, Grbic V, Osborne EJ, Dermauw W, Ngoc PC, Ortego F, Hernandez-Crespo P, Diaz I, Martinez M, Navajas M, Sucena E, Magalhaes S, Nagy L, Pace RM, Djuranovic S, Smagghe G, Iga M, Christiaens O, Veenstra JA, Ewer J, Villalobos RM, Hutter JL, Hudson SD, Velez M, Yi SV, Zeng J, Pires-daSilva A, Roch F, Cazaux M, Navarro M, Zhurov V, Acevedo G, Bjelica A, Fawcett JA, Bonnet E, Martens C, Baele G, Wissler L, Sanchez-Rodriguez A, Tirry L, Blais C, Demeestere K, Henz SR, Gregory TR, Mathieu J, Verdon L, Farinelli L, Schmutz J, Lindquist E, Feyereisen R, Van de Peer Y. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479:487–492. doi: 10.1038/nature10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoy MA, Waterhouse RM, Wu K, Estep AS, Ioannidis P, Palmer WJ, Pomerantz AF, Simao FA, Thomas J, Jiggins FM, Murphy TD, Pritham EJ, Robertson HM, Zdobnov EM, Gibbs RA, Richards S. Genome Sequencing of the Phytoseiid Predatory Mite Metaseiulus occidentalis Reveals Completely Atomized Hox Genes and Superdynamic Intron Evolution. Genome Biol Evol. 2016;8:1762–1775. doi: 10.1093/gbe/evw048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gulia-Nuss M, Nuss AB, Meyer JM, Sonenshine DE, Roe RM, Waterhouse RM, Sattelle DB, de la Fuente J, Ribeiro JM, Megy K, Thimmapuram J, Miller JR, Walenz BP, Koren S, Hostetler JB, Thiagarajan M, Joardar VS, Hannick LI, Bidwell S, Hammond MP, Young S, Zeng QD, Abrudan JL, Almeida FC, Ayllon N, Bhide K, Bissinger BW, Bonzon-Kulichenko E, Buckingham SD, Caffrey DR, Caimano MJ, Croset V, Driscoll T, Gilbert D, Gillespie JJ, Giraldo-Calderon GI, Grabowski JM, Jiang D, Khalil SMS, Kim D, Kocan KM, Koci J, Kuhn RJ, Kurtti TJ, Lees K, Lang EG, Kennedy RC, Kwon H, Perera R, Qi YM, Radolf JD, Sakamoto JM, Sanchez-Gracia A, Severo MS, Silverman N, Simo L, Tojo M, Tornador C, Van Zee JP, Vazquez J, Vieira FG, Villar M, Wespiser AR, Yang YL, Zhu JW, Arensburger P, Pietrantonio PV, Barker SC, Shao RF, Zdobnov EM, Hauser F, Grimmelikhuijzen CJP, Park Y, Rozas J, Benton R, Pedra JHF, Nelson DR, Unger MF, Tubio JMC, Tu ZJ, Robertson HM, Shumway M, Sutton G, Wortman JR, Lawson D, Wikel SK, Nene VM, Fraser CM, Collins FH, Birren B, Nelson KE, Caler E, Hill CA. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat Commun. 2016:7. doi: 10.1038/ncomms10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Zee JP, Geraci NS, Guerrero FD, Wikel SK, Stuart JJ, Nene VM, Hill CA. Tick genomics: The Ixodes genome project and beyond. Int J Parasitol. 2007;37:1297–1305. doi: 10.1016/j.ijpara.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 28.Cornman SR, Schatz MC, Johnston SJ, Chen YP, Pettis J, Hunt G, Bourgeois L, Elsik C, Anderson D, Grozinger CM, Evans JD. Genomic survey of the ectoparasitic mite Varroa destructor, a major pest of the honey bee Apis mellifera. Bmc Genomics. 2010;11:602. doi: 10.1186/1471-2164-11-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bast J, Schaefer I, Schwander T, Maraun M, Scheu S, Kraaijeveld K. No Accumulation of Transposable Elements in Asexual Arthropods. Mol Biol Evol. 2016;33:697–706. doi: 10.1093/molbev/msv261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genornes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 31.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 32.Dermauw W, Van Leeuwen T, Vanholme B, Tirry L. The complete mitochondrial genome of the house dust mite Dermatophagoides pteronyssinus (Trouessart): a novel gene arrangement among arthropods. Bmc Genomics. 2009:10. doi: 10.1186/1471-2164-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korf I. Gene finding in novel genomes. BMC bioinformatics. 2004:5. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ElRamlawy KG, Fujimura T, Baba K, Kim JW, Kawamoto C, Isobe T, Abe T, Hodge-Hanson K, Downs DM, Refaat IH, Al-Azhary DB, Aki T, Asaoku Y, Hayashi T, Katsutani T, Tsuboi S, Ono K, Kawamoto S. Der f 34, a Novel Major House Dust Mite Allergen Belonging to a Highly Conserved Rid/YjgF/YER057c/UK114 Family of Imine Deaminases. Journal of Biological Chemistry. 2016;291:21607–21615. doi: 10.1074/jbc.M116.728006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Coleman-Derr D, Chen GP, Gu YQ. OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Research. 2015;43:W78–W84. doi: 10.1093/nar/gkv487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–327. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tovey ER, Chapman MD, Platts-Mills TA. Mite faeces are a major source of house dust allergens. Nature. 1981;289:592–593. doi: 10.1038/289592a0. [DOI] [PubMed] [Google Scholar]
- 38.Walton SF, Slender A, Pizutto S, Mounsey KE, Opresecu F, Thomas WR, Hales BJ, Currie BJ. Analysis of IgE binding patterns to house dust mite allergens in scabies-endemic communities: insights for both diseases. Clinical and Experimental Allergy. 2015;45:1868–1872. doi: 10.1111/cea.12553. [DOI] [PubMed] [Google Scholar]
- 39.Kaul S, Zimmer J, Dehus O, Costanzo A, Daas A, Buchheit KH, Asturias JA, Barber D, Carnes J, Chapman M, Dayan-Kenigsberg J, Doring S, Fuhrer F, Hanschmann KM, Holzhauser T, Ledesma A, Moingeon P, Nony E, Pini C, Plunkett G, Reese G, Sandberg E, Sander I, Strecker D, Valerio C, van Ree R, Vieths S. Standardization of allergen products: 3. Validation of candidate European Pharmacopoeia standard methods for quantification of major birch allergen Bet v 1. Allergy. 2016;71:1414–1424. doi: 10.1111/all.12898. [DOI] [PubMed] [Google Scholar]
- 40.Larenas-Linnemann D, Cox LS, Diagnostics IA. European allergen extract units and potency: review of available information. Ann Allerg Asthma Im. 2008;100:137–145. doi: 10.1016/S1081-1206(10)60422-X. [DOI] [PubMed] [Google Scholar]
- 41.Larenas-Linnemann D, Esch R, Plunkett G, Brown S, Maddox D, Barnes C, Constable D. Maintenance dosing for sublingual immunotherapy by prominent European allergen manufacturers expressed in bioequivalent allergy units. Ann Allerg Asthma Im. 2011;107:448–458. doi: 10.1016/j.anai.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 42.Patterson ML, Slater JE. Characterization and comparison of commercially available German and American cockroach allergen extracts. Clinical and Experimental Allergy. 2002;32:721–727. doi: 10.1046/j.1365-2222.2002.01397.x. [DOI] [PubMed] [Google Scholar]
- 43.Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, Karlsson A, Al-Lazikani B, Hersey A, Oprea TI, Overington JP. A comprehensive map of molecular drug targets. Nat Rev Drug Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ffrench-Constant RH. The Molecular Genetics of Insecticide Resistance. Genetics. 2013;194:807–815. doi: 10.1534/genetics.112.141895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lees K, Woods DJ, Bowman AS. Transcriptome analysis of the synganglion from the brown dog tick, Rhipicephalus sanguineus. Insect Mol Biol. 2010;19:273–282. doi: 10.1111/j.1365-2583.2009.00968.x. [DOI] [PubMed] [Google Scholar]
- 46.Kwon DH, Clark JM, Lee SH. Toxicodynamic mechanisms and monitoring of acaricide resistance in the two-spotted spider mite. Pestic Biochem Phys. 2015;121:97–101. doi: 10.1016/j.pestbp.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 47.Krieger J, Jacobs DE, Ashley PJ, Baeder A, Chew GL, Dearborn D, Hynes HP, Miller JD, Morley R, Rabito F, Zeldin DC. Housing Interventions and Control of Asthma-Related Indoor Biologic Agents: A Review of the Evidence. J Public Health Man. 2010;16:S11–S20. doi: 10.1097/PHH.0b013e3181ddcbd9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gotzsche PC, Hammarquist C, Burr M. House dust mite control measures in the management of asthma: meta-analysis. Brit Med J. 1998;317:1105–1110. doi: 10.1136/bmj.317.7166.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodfolk JA, Hayden ML, Couture N, Plattsmills TAE. Chemical Treatment of Carpets to Reduce Allergen - Comparison of the Effects of Tannic-Acid and Other Treatments on Proteins Derived from Dust Mites and Cats. J Allergy Clin Immun. 1995;96:325–333. doi: 10.1016/s0091-6749(95)70051-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.