Abstract
BACKGROUND
Hyperhemolysis syndrome (HHS) is an uncommon, but life-threatening, transfusion-related complication of red blood cell transfusion. HHS has predominantly been described in patients with sickle cell disease (SCD) and is difficult to diagnose and treat. The pathogenesis of HHS, including its occurrence in only a subset of apparently susceptible individuals, is poorly understood. We undertook whole-exome sequencing (WES) of 12 SCD-HHS patients to identify shared genetic variants that might be relevant to the development of HHS.
STUDY DESIGN AND METHODS
DNA from adults with SCD having at least one previous episode of HHS were subject to WES. High-quality variants were passed through a series of bioinformatics filters to identify variants that were uncommon among African populations represented in public databases. Recurrent, putative loss-of-function variants occurring in biologically plausible genes were prioritized and then genotyped in a larger, ancestry-matched cohort of non-HHS controls.
RESULTS
A rare, heterozygous stop-gain variant (p.Glu210Ter) in MBL2 was significantly enriched among HHS cases (p = 0.002). This variant is predicted to result in a premature termination codon that escapes nonsense-mediated mRNA decay, potentially leading to a novel phenotype. We also observed a complex insertion-deletion variant in the final exon of KLRC3 that was enriched among cases (p = 0.0019), although neither variant was found among seven pediatric SCD-HHS patients.
CONCLUSION
Our results suggest a potential role for rare genetic defects in the development of HHS among adult SCD patients. Such enriched variants may ultimately be useful for identifying high-risk individuals and informing therapeutic approaches in HHS.
Individuals with sickle cell disease (SCD) frequently require therapeutic red blood cell (RBC) transfusions1 for acute complications—such as aplastic crisis or acute chest syndrome—or to prevent primary or secondary stroke.2,3 Nonetheless, the benefits of RBC transfusion therapy are not without cost; compared to other transfused patients, patients with SCD have an increased risk of alloimmunization, in which antibodies (alloantibodies) are directed against the donor RBC antigens. If not suspected before transfusion, alloimmunization can result in delayed hemolytic transfusion reactions (DHTRs),1 in which donor RBCs are destroyed.
Another consequence of recurrent transfusions is a life-threatening condition known as hyperhemolysis syndrome (HHS). This is a relatively uncommon reaction that has been observed in a small subset of patients receiving antigen-negative, cross-match compatible RBCs. Patients with HHS break down both transfused and autologous RBCs, resulting in severe vasoocclusive crises, characterized by pain, fever, and occasionally, acute chest syndrome, as well as markers of acute hemolysis such as hemoglobinuria and jaundice.4 The breakdown of both autologous and donor RBCs is supported by the presence of markers for both sickle hemoglobin (HbS) and normal hemoglobin (HbA) in the urine, mirroring observations of steroid-responsive reticulocytopenia in which the destruction of reticulocytes cannot be explained by suppression of erythropoiesis.5 HHS, therefore, is sometimes viewed as a different clinical entity from DHTRs (in which only the donor RBCs are broken down), although the two often coexist in patients.
The symptoms of HHS mimic other complications of SCD, and hemolysis can occur even in the absence of detectable RBC alloantibodies. This makes the diagnosis of HHS particularly challenging. Subsequent transfusions may further aggravate the condition and negate the therapeutic value of transfusions. Difficulties in diagnosis also mean that data on the incidence and prevalence of HHS are not readily available; however, one retrospective review of pediatric and adult sickle cell patients followed over a 10-year period reported an incidence of 5.1% in the pediatric population and 1.6% in the adult population.6 The prevalence of HHS in our own sickle cell transfusion cohort was approximately 3% to 5%7 in keeping with this and previous reports;8 however, these figures are potentially an underestimate, given the high likelihood of underreporting and/or misdiagnosed cases.
Although HHS is well described in the literature,5,9-15 its etiology is poorly understood. Hypotheses have focused on SCD pathophysiology as a chronic disease characterized by a state of heightened inflammation, and have sought to explain HHS in the context of activated macrophages,5 oxidative stress,16 and hypercoagulability.17 It has been proposed that transfusion-induced suppression of erythropoiesis may contribute to HHS. This is supported by one study in which patients with HHS exhibited a significant decrease in the absolute reticulocyte counts (reticulocytopenia);12 however, it is unclear whether this is a primary cause or secondary effect of transfusion and does not readily explain the concomitant autologous hemolysis. Another prevailing theory is that of “bystander hemolysis” in which there is the destruction of antigen-negative RBCs during immune hemolysis of antigen-positive RBCs,14 although there is not presently compelling evidence for this. Yet another theory is of macrophage-induced hemolysis, which is underscored by the fact that HbS cells readily adhere to macrophages, and RBC antibodies are not always detected during acute HHS; indeed, intravenous immunoglobulins and steroids both suppress macrophage activity and have been recommended for the treatment of HHS.18 Finally, Chadebech and colleagues15 posited that the oxidative nature of the bloodstream of SCD patients can result in donor RBCs exposing more phosphatidylserine on the outer surface of the RBC. Phosphatidylserine is a signal for eryptosis (suicidal RBC death) and its increased exposure may result in the subsequent destruction of both donor and autologous RBCs. Few of these hypotheses have been studied in detail, however, and none answer the question of why only some SCD patients get HHS, while the vast majority do not. There is thus a lingering mechanistic gap that hinders therapeutic progress.
Given that SCD patients receiving transfusions are exposed to similar environmental factors, but only a small percentage (approx. 5%) go on to develop HHS, there is a potential role for recipient genetics in the pathogenesis of HSS. Recent advances in genomics technologies have heralded new approaches to investigating the role of genetic factors in HHS. One such approach is whole-exome sequencing (WES), which has become the mainstay of uncovering DNA sequence variants underlying Mendelian diseases.19-21 WES has since been extended to the discovery of variants that underlie complex traits such as fasting glucose22 and increased risk for myocardial infarction.23
Understanding the molecular underpinnings of HHS could offer fresh insights into the early detection of HHS susceptibility and usher in the development of new therapies, not only for HHS patients with SCD, but also for those with other hematologic conditions. This would have strong parallels to similarly uncommon reactions mediated by gene–environment interactions, such as the increased sensitivity to the HIV antiretroviral drug abacavir among persons carrying HLA-B*5701.24 We undertook WES of 12 adult SCD individuals, all with a confirmed history of HHS, and compared the frequency of recurrent, predicted protein-damaging variants identified in these individuals with that observed in public genetic databases and an adult non-HHS cohort of similar ancestry.
MATERIALS AND METHODS
Sample population
Details of the full cohort have been previously published;7 briefly, individuals with SCD were recruited through Life-Share Blood Services. All included samples had at least one hyperhemolysis event, with at least two of these patients having more than one episode of HHS (J.M. Moulds, unpublished data). Also of note is that we used a conservative set of criteria for diagnosis of HHS, which excluded some other borderline patients. Participants had peripheral blood collected, deidentified, and genomic DNA extracted from buffy coats or whole blood. Within this cohort (Table 1), 17 individuals met criteria for having HHS, defined as “a posttransfusion decrease in Hb to less than pretransfusion levels without evidence of bleeding”;7 of these, 12 individuals had sufficient DNA available for WES and were used as cases in a case-only discovery, in which variants in public databases were utilized as external references. The remaining non-HHS individuals in the cohort (n = 202) were used as geographic and ancestral controls for our discovery cohort.
TABLE 1.
Demographic variables and HbS status in HHS and nonhyperhemolysis groups*
| HHS (n = 12) | Non-HHS (n = 365) | |
|---|---|---|
| Sex | ||
| Male | 3 | 158 |
| Female | 9 | 199 |
| NA | 0 | 8 |
| Age group | ||
| Adult | 11 | 331 |
| Child | 1 | 34 |
| HbS status | ||
| SS | 12 | 354 |
| SC | 0 | 11 |
| Alloimmune category | ||
| Responder | 12 | 179 |
| Nonresponder | 0 | 186 |
| Responder alloantibodies, median (min, max, IQR) | 6.5 (1, 8, 2.5) |
HbS status includes patients that are homozygous for HbS (SS) or compound heterozygous for HbS and HbC (SC).
IQR = interquartile range.
WES
WES was carried out at the Human Genome Sequencing Center, Baylor College of Medicine, as previously described.20,25,26 Briefly, DNA samples were processed and quantified to meet quality control (QC) criteria before WES (HiSeq 2500, Illumina) using the sequencing-by-synthesis kits (TruSeq, Illumina) per the manufacturer’s protocol. Exon enrichment was performed using an exome capture reagent (NimbleGen VCRome 2.1(rebal), Roche Sequencing). Indexed adapters were used to allow multiplexed sequencing of six samples and sequencing was performed using paired-end 100-bp read lengths.
Sequence alignment and variant calling
Base calling and QC were performed using real-time analysis software sequence pipeline (Illumina). Alignment and annotation were undertaken using the Mercury pipeline.27 In brief, 100-bp end reads were aligned to the NCBI human reference genome (UCSC hg19)28 using Burrows-Wheeler Aligner,29 and duplicates marked with Picard,30 followed by local realignment around likely short insertions-deletions (indels) and quality score recalibration using genome analysis toolkit.31
Variant annotation and filtering
Atlas232 was used to identify single-nucleotide variants (SNVs) and indels from the sequence data. VCFtools v0.1.9.033 was used to filter for variants with depth-of-coverage of at least 10× and a “PASS” FILTER flag. Variant Tools v2.6.134 was used to annotate and filter variants using dbNSFP (database for nonsynonymous SNPs’ functional predictions),35 which compiles protein prediction scores from four algorithms: SIFT,36 PolyPhen,37 MutationTaster,38 and likelihood ratio test.39 Variants were also removed based on the following criteria: variants with a minor allele frequency (MAF) greater than or equal to 5% in the 1000 Genomes Project,40 Exome Variant Server,41 and The Exome Aggregation Consortium (ExAC);42 variants with allele read ratios of less than 0.3 or found on sex chromosomes; and variants in genes in a list suspected to have a high false-positive rate due to high polymorphism, assembly misalignment, or misleading reference genome information.43
Sanger validation and genotyping of candidate variants
Dideoxy Sanger sequencing was used to validate candidate variants and to genotype candidate variants in the larger non-HHS control cohort (Table 1). Polymerase chain reaction (PCR) primers (Sigma-Aldrich) were designed using Primer-blast44 targeting 100- to 200-bp region flanking target SNVs, and primer quality was tested using OligoAnalyzer 3.1 (https://www.idtdna.com/calc/analyzer). Details of the primers utilized are given in Table S1 (available as supporting information in the online version of this paper). PCR amplification was done using DNA polymerase (PrimeSTAR GXL, Clontech Laboratories) with patients’ DNA as template, according to manufacturer’s instructions (annealing temperature 638C). PCR products were visualized (1% agarose gel stained with ethidium bromide) and then sequenced (Genewiz).
Replication in a secondary data set
To determine the generalizability of our findings, we interrogated WES data from an independent SCD pediatric cohort (n = 1238), aged 2 to 21 years of age, obtained from several US sickle cell centers. SCD genotypes included HbSs and HbSβ0. This cohort included seven children with a history of HHS, defined in the same way as the primary cohort; however, the transfusion history (either having had a transfusion or having had a number of transfusions) was unavailable for the remainder of the cohort. WES was carried out at the Human Genome Sequencing Center using the VCRome 2.1 capture reagent (Nimblegen); details of sequencing alignment, variant calling, and QC have been previously published.20,25,26,45 Variant sites were evaluated to ensure that they were captured at a similar read depth as the discovery cohort; all of the candidate variants had good coverage with more than 25× median coverage for all samples. The frequency of the candidate variants in the secondary cohort was then stratified by phenotype and reported.
Statistical analysis
Allele frequencies in the HHS and non-HHS cohorts were compared using a two-tailed Fisher’s exact test (p value), alongside odds ratios (OR) and 95% confidence intervals (CIs). p values were subjected to a Bonferroni correction significance threshold of p = 0.01 based on the number of rare, recurrent, putative loss-of-function (pLOF) variants (n = 6) observed in the data set. Margin of error for proportions was computed using a 95% CI.
Ethics statement
All samples evaluated in this study were deidentified clinical samples left over from pretransfusion genetic testing performed at LifeShare Blood Services; a waiver of informed consent was obtained from the institutional review board of Baylor College of Medicine and no additional clinical data apart from transfusion (alloimmunization/alloantibody) status and basic demographic information was available. The study was also approved by the ethics board of St Luke’s Episcopal Hospital (Houston, TX).
RESULTS
Whole-exome sequencing identified a total of 3,007,766 variants of which 1,544,050 variants “PASS”ed our QC FILTER. Further filtering (summarized in Fig. S1, available as supporting information in the online version of this paper) for nonsynonymous variants (n = 46,488) in dbNSFP and uncommon variants (MAF ≤ 0.05 in the African population) with a SIFT score of less than 0.05 resulted in a total of 8986 SNVs and indels.
Identification of candidate variants
We first focused on the class of variants predicted to have the most damaging effect on the resulting protein—so-called LOF variants, which included stop-gains, frame-shifts, nonsense, and splicing (within 2 bp of intron-exon boundaries) variants. Rare singleton variants are likely to represent private familial variants;46 thus we focused on recurrent variants (occurring more than once) that might be enriched in our HHS discovery cohort. This yielded a total of six variants (Table 2), all of which were heterozygous stop-gains predicted to result in a premature termination codon (PTC) in the last exon, except for C1orf194, which had a PTC in Exon 2/5. PTC-generating mutations usually give rise to a C-terminal truncated polypeptide by escaping nonsense-mediated decay and so result in either a dominant negative or gain of function phenotype47,48 (Fig. S2, available as supporting information in the online version of this paper).
TABLE 2.
Candidate LOF variants in cohort
| Public database frequencies*
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Biologic processes (http://www.uniprot. org) |
dbSNP ID | Position | Codon change |
Allele count |
Amino acid change |
1000 Genomes† |
ExAC‡ | ESP§ |
| MBL2 (mannose-binding protein C) | Calcium-dependent lectin involved in innate immune defense. Binds mannose, fucose, and N-acetylglucosamine on different microorganisms and activates the lectin complement pathway. | rs74754826 | chr10: 54528016 | GAG>TAG | 3 | E210X | 0.010 | 0.007 | 0.006 |
| KLRC3 (NKG2-E type II integral membrane protein killer cell lectin-like receptor subfamily C, Member 3) | Plays a role as a receptor for the recognition of MHC class I HLA-E molecules by NK cells and some cytotoxic T cells. | rs145456037 | chr12: 10568251 | AAA>TAA | 5 | K244X | 0.046 | 0.058 | NA |
| PCSK5 (proprotein convertase subtilisin/kexin type 5) | Establishment by proteolytic activation of a number of important factors such as alpha-integrins. | rs77068135 | chr9: 78790143 | TGG>TGA | 4 | W666X | NA | 0.036 | NA |
| IDI2 (isopentenyl-diphosphate delta-isomerase 2) | Involved in the biosynthesis of isoprenoids. | rs1044261 | chr10: 1065710 | TGG>TAG | 4 | W144X | 0.038 | 0.046 | 0.047 |
| EPHB2 (ephrin type-B receptor 2) | Receptor tyrosine kinase which binds transmembrane ephrin-B family ligands residing on adjacent cells, leading to contact-dependent bidirectional signaling into neighboring cells. | rs76826147 | chr1: 23240250 | AAA>TAA | 3 | K1019X | 0.043 | 0.046 | 0.040 |
| C1orf194 (uncharacterized protein) | Unknown | rs62623709 | chr1: 109650558 | TAC>TAA | 3 | Y61X | 0.031 | 0.035 | 0.040 |
African population MAFs are shown. Global MAFs for all variants were lower than African MAFs except PCSK5 (global MAF = 0.060).
The 1000 Genomes database is the largest public catalog of human variation and genotype data.
Exome Aggregation Consortium (ExAC) comprises of exome sequencing data from 60,706 unrelated individuals participating in various disease-specific and population genetic studies. Individuals affected by severe pediatric disease are excluded so this data set should serve as a useful reference set of allele frequencies for severe disease studies.
NHLBI GO Exome Sequencing Project (ESP) comprises of exome sequencing data from various studies of heart, lung, and blood disorders.
We did not observe any significantly enriched candidate variants in the HHS group under a homozygous recessive model. A more relaxed model of recurrent, rare, likely damaging variants identified 163 missense mutations with a SIFT score of less than 0.05 (where a lower SIFT score implies a higher probability that the variant is damaging). Of these, 41 (Table S3, available as supporting information in the online version of this paper) variants were predicted to be damaging by four or more algorithms: SIFT,36 PolyPhen,37 MutationTaster,38 likelihood ratio test,39 FATHMM,49 MetaLR,50 and PROVEAN;51 however, none of the affected genes were considered to be strong biologic candidates for HHS and together with the known uncertainty of prediction algorithms,39,52 we opted to focus on the above pLOF variants in this analysis.
Comparison of variant frequencies
We then prioritized pLOF variants for replication in the larger cohort based on the known biologic function of the gene and the relative frequency of the variant in public databases (Table 2). Variants in MBL2, KLRC3, and PCSK6 were deemed the strongest biologic candidates. These were validated using dideoxy Sanger sequencing (Fig. 1) and then genotyped in the remainder of the study cohort (n = 202), which included individuals with SCD from the same geographical region and of similar ethnicity ancestry.7 The rs74754826 variant in MBL2 (mannose-binding lectin 2; transcript ID ENST00000373968: c.628G>T, p.Glu210Ter), which had a frequency of less than 1% in public databases, was observed in three of 12 HHS samples (25 ± 24.5%) and this was significantly more than in non-HHS controls (three of 202 heterozygous individuals; p = 0.003; OR, 22.1; 95% CI, 3.9-125.2; Table 3).
Fig. 1.
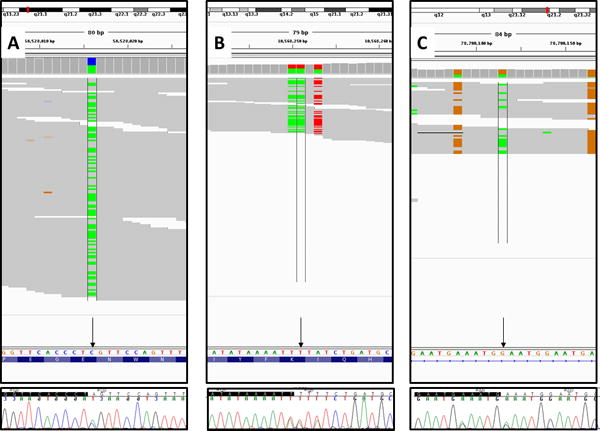
Sanger sequencing validation of three candidate variants (arrows)—rs74754826 (average ref/alt read ratio, 59:58) in MBL2 gene (A); rs145456037 (average ref/alt read ratio = 30:35) in the KLRC3 gene (B), for which the reverse complement sequence is shown; and rs77068135 (average ref/alt read ratio, 13:20) in the PCSK5 gene (C). For each variant, reads were visualized using Integrative Genomics Viewer/IGV (top panel), where colored bars represent nucleotide substitutions: green for adenine, red for thymine, orange for guanine, and blue for cytosine. Sanger trace files were visualized using Sequencher v5.3 (bottom panel).
TABLE 3.
Comparison of candidate variants in HHS versus non-HHS in the discovery cohort
| Allele frequencies
|
Genotype counts*
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| HHS (n = 12)
|
Non-HHS (n = 365)
|
||||||||
| Gene | Variant | p value | HHS | Non-HHS | OR (95% CI) | Alt/Ref | Ref/Ref | Alt/Ref | Ref/Ref |
| MBL2 | rs74754826 | 0.0025 | 0.125 | 0.007 | 22.11 (3.90–125.24) | 3 | 9 | 3 (202) | 199 (202) |
| KLRC3 | rs145456037 | 0.0019 | 0.167 | 0.034 | 9.93 (2.67–36.98) | 5 | 7 | 10 (149) | 139 (149) |
| PCSK5 | rs77068135 | 0.0623 | 0.167 | 0.061 | 3.60 (0.99–13.05) | 4 | 8 | 20 (164) | 144 (164) |
No homozygous variants (Alt/Alt) were observed.
Alt = alternate allele; Ref = reference allele.
The rs145456037 (transcript ID ENST00000381903: c.730A>T, p.Lys244Ter) in KLCR3 (killer cell lectin-like receptor subfamily C, member 3) was observed in five of 12 SCD-HHS (41.7 ± 27.9%) and this was also significantly enriched in comparison to non-HHS individuals (10 of 149; Table 3) with a slightly lower OR (p = 0.002; OR, 9.9; 95% CI, 2.3-37.0). On closer inspection, however, this variant was always (and only) observed with two other adjacent variants, suggestive of a complex indel resulting from a dinucleotide insertion (10,568,249-10,568,250insAA) followed by a dinucleotide deletion (10,568,252-10,568,253delTA); this would be anticipated to result in two predicted-damaging adjacent missense amino acid changes in the last exon rather than yielding a stop-gain mutation (details in supplementary data, available as supporting information in the online version of this paper).
Finally, rs77068135 in PCSK5 (proprotein convertase subtilisin/kexin type 5; transcript ID ENST00000376767: c.1998G>A, p.Trp666Ter) was found in four HHS individuals (33.3 ± 26.7%); however, this was not significantly enriched in comparison with non-HHS controls (n = 164; p = 0.062; Table 3). After comparing HHS patients, all of whom were responders, with controls, who were also responders, our results remained consistent with our original findings—MBL2 (p = 0.003; OR, 33.7; 95% CI, 3.2-357.9), KLRC3 (p = 0.007; OR, 8.9; 95% CI, 1.9-41.4), and PCSK5 (p = 0.118; OR, 3.0; 95% CI, 0.8-11.5).
Replication and transferability
To evaluate the transferability of our findings to other SCD cohorts, we evaluated the frequency of our candidate pLOF variants in an independent pediatric SCD cohort that comprised seven HHS and 1231 non-HHS individuals; of note, transfusion history for non-HHS was unavailable for this study. Consistent with public databases and our internal control, we observed similar MAFs in the non-HHS group; however, we did not observe any significant enrichment of the variants among those with HHS (Table S4, available as supporting information in the online version of this paper).
DISCUSSION
Although there is limited knowledge on the etiology of HHS, the confluence of environmental, donor, and recipient factors thus far surmised suggests a complex interplay between various factors.4,53 Using WES, we identified a rare, stop-gain SNV in MBL2 and a rare damaging indel in KLCR3 that are enriched among DHTR-positive, adult SCD individuals from Louisiana and Southeast Texas with a history of HHS.
MBL2 encodes a protein of the same name (MBL) that belongs to the collectin family of proteins. It is known to play a key role in the innate immune system through recognition of microorganism cell wall components mannose and N-acetylglucosamine and subsequently activating complement-mediated lysis via the lectin pathway.54 Deficiencies in this gene have been associated with susceptibility to autoimmune and infectious diseases,55 and previous SNVs in the promoter regions of MBL2 have been associated with altered serum MBL levels. Common missense variants in Exon 1 of MBL254 have been described to affect the collagen helical domain56 resulting in disruption of the assembly of MBL multimers and a functional MBL deficiency. The rare, nonsense variant found to be significantly enriched in our cohort occurs in the last exon of MBL2, which codes for the carbohydrate-recognition domain that is involved in the binding of carbohydrates in a calcium-dependent manner.57 Given that nonsense mutations in the last exon usually result in either a dominant negative or gain of function phenotype via escape of nonsense-mediated decay,47,48 we posit that the predominant role of this variant is an abrogation of MBL function rather than a deficiency of serum levels. Unlike the classical pathway of complement activation, the lectin-mediated pathway is not antibody dependent,58 which would allow for destruction of autologous RBCs in the absence of alloantibodies—the so-called “bystander hypothesis.”14 The role of complement in bystander hemolysis is supported by observations of a strong positive direct agglutination test, regardless of alloimmunization, in 26 DHTR patients; this complement sensitization was apparent even months after the transfusion.59 Thus, MBL2 makes for an interesting candidate for functional studies of HHS.
We also observed evidence for association with a presumed indel in KLRC3. Reduced expression of KLRC3 has been associated with autoimmune diseases60 and the resulting KLRC3 protein is thought to act as a receptor for the recognition of HLA-E molecules by NK cells and some cytotoxic T cells and may also regulate their activity.61 Given a proposed roll for NK cells in hemolytic transfusion reactions,62 KLRC3 is also an interesting HHS candidate.
The variants in MBL2 and KLRC3, taken together, account for, at most, six of the 12 HHS cases in our cohort, suggesting that there may be other contributing variants, perhaps similarly related to innate immunity. Tacit in this observation is that two of the heterozygous MBL2 variant carriers also carried the KLRC3 variant (Fig. S4, available as supporting information in the online version of this paper). This biallelic carriage was not observed among non-HHS controls and suggests that there may be a role for more complex epistatic effects in which variants in multiple genes might synergistically contribute to the phenotype.63 Alternately, predisposition to HHS may be mediated by more complex combinations of variants in a single gene; for example, one major limitation was our small sample size, which together with an exon-based assessment, did not allow for extended evaluation of common MBL2 promoter variants that might be acting in tandem with our rare MBL2 variant. Genetically, we chose to evaluate the “lowest hanging fruit” among SCD-HHS individuals—rare, recurrent, pLOF coding variants—leaving other tractable disease models such as damaging singleton variation or noncoding regulatory variation unevaluated.
Our efforts to generalize these findings to an independent pediatric cohort were also insightful. We were unable to replicate any significant associations between these variants and HHS. While this could be an example of a false-positive “winner’s curse,” it should be noted that, unlike the discovery cohort, our replication cohort was composed largely of children, who temporally differ from their adult counterparts in immune development64 and environmental exposure; this may partially explain our lack of replication. Second, the penetrance of our variants is necessarily incomplete (explaining their appearance in the primary control cohort); it may be that additional environmental, genetic, or transfusion-related factors, not yet evident in the pediatric cohort, are required to trigger HHS4,53 among carriers of our candidate variants. Similarly, although the non-HHS patients (n = 1231) were used as controls, one caveat is the current unavailability of the transfusion history of this group, leaving the possibility that they may not have been sufficiently exposed to develop HHS. The allele frequencies observed in the replication cohort established the rarity of the MBL2 variant (MAF < 0.01) in the general population; conversely, the variants in KLRC3 and PCSK5 were relatively more common. This places the greatest weight on MBL2 as the most promising candidate emerging from this study.
HHS is an uncommon phenomenon; consequently, there are few established cohorts through which to contextualize our findings. This is compounded by the difficulty in robustly diagnosing HHS, for which the hallmark symptoms may be attributed to other comorbidities of SCD; this is a particular problem for post hoc assemblies of HHS cases within existing cohorts. In our study, for instance, although the primary diagnosis of HHS was secure among the identified cases, it remains unclear whether individuals in the non-HHS SCD cohort may have had HHS diagnosed as something else or may have developed HHS later. These observations underscore the challenges inherent to developing the large, well-phenotyped cohorts that are needed for future genetic studies of HHS. Such studies will need to include both qualitative and quantitative clinical data on RBC transfusions and alloantibodies and span the breadth of age and severity to definitively evaluate the extent of recipient genetic susceptibility to HHS.
In conclusion, our foray into the genetic predisposition to HHS among transfusion recipients with SCD, although preliminary, opens new avenues for future studies of HHS. Larger studies that utilize newer “omics” technologies at genome scale to robustly identify candidate variants in transfusion recipients, combined with suitably powered and well-phenotyped cohorts and functional assays of candidate genes, are an attractive way to begin to understand more about this unusual syndrome. The candidate genes identified here hint at a potential role for defects in innate immunity in HHS and support hypotheses of complement- and macrophage-mediated hemolysis in HHS. Our results may also have diagnostic and therapeutic implications; ultimately, it may be possible to use recipient genetic profiling to inform the need for pretransfusion prophylaxis and provide this in a pathogenesis-targeted fashion to reduce the incidence of an unusual, but potentially devastating, transfusion response.
Supplementary Material
Table S1. Candidate variants in HHS replication cohort.
Table S2. Primers used for PCR and Sanger sequencing.
Table S3. Summary of variants that were predicted to be damaging by four or more algorithms.
Fig. S1. Schematic of variant filtering pipeline after whole exome sequencing. Twelve individuals with clinical definitions of SCD and HHS were sequenced. The top-to-bottom variant filtering workflow shows the variants remaining after each filtering step together with the software tools used.
Fig. S2. Protein domains of candidate genes. Protein topology highlighting position of the mutations (red arrows) and protein domains for MBL2 (Top), KLRC3 (Middle), and PCSK5 (Bottom). The PCSK5 transcript of interest (ENST00000376767) lacks a portion of the CRM domain (shaded) (http://www.uniprot.org/).
Fig. S3. Histogram of the frequency distribution of filtered variants. Non-synonymous variants in dbNSFP, with a “PASS” flag, SIFT score < 0.05, MAF ≤5% (utilizing data in the 1000 Genomes Project Exome Variant Server, Exome Sequencing Project (ESP) and The Exome Aggregation Consortium, ExAC) were selected. Variants with allelic ratios < 0.3 and variants found in sex chromosomes and potential false positive genes were filtered out. The histogram shows the frequency distribution of the final set of uncommon, stop-gain variants that were enriched in the HHS cohort.
Fig. S4. Distribution of candidate gene variant carriers. Rare damaging variants in 3 genes (MBL2, KLRC3 and PCSK5) were found in 7 of the 12 patients with HHS. 5 of the 12 patients did not have any of the 3 candidate variants. Venn diagram shows distribution and intersection of the pLOF variants in the 3 candidate genes across the 12 HHS patients.
Acknowledgments
We are grateful to Dr John Belmont for his valuable input in the study design and for his insightful feedback and Nancy Hall for her outstanding technical assistance with the Sanger genotyping.
This work is primarily supported by funding from the National Blood Foundation to NH (NBF Grant 1103112502). Part of the analyses was supported by Grant U54 HG003273 from the National Human Genome Research Institute awarded to JF and VS. NH is partly supported by Grant 2013096 from the Doris Duke Charitable Foundation.
ABBREVIATIONS
- DHTR(s)
delayed hemolytic transfusion reaction(s)
- HHS
hyperhemolysis syndrome
- MAF
minor allele frequency
- pLOF
putative loss-of-function
- PTC
premature termination codon
- SCD
sickle cell disease
- SNV(s)
single-nucleotide variant(s)
- WES
whole-exome sequencing
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
AUTHOR CONTRIBUTIONS
NH and AC designed the study; JMM ascertained and distinguished the clinical samples; SM carried out genotyping and data analyses; JF, VS, and AG carried out analysis on replication cohort; and all authors contributed to writing the manuscript.
CONFLICT OF INTEREST
The authors have disclosed no conflicts of interest.
References
- 1.Wayne AS, Kevy SV, Nathan DG. Transfusion management of sickle cell disease [abstract] Blood. 1993;81:1109–23. [PubMed] [Google Scholar]
- 2.Adams RJ, McKie VC, Hsu L, et al. Prevention of a first stroke by transfusions in children with sickle cell anemia and abnormal results on transcranial Doppler ultrasonography. N Engl J Med. 1998;339:5–11. doi: 10.1056/NEJM199807023390102. [DOI] [PubMed] [Google Scholar]
- 3.Lee MT, Piomelli S, Granger S, et al. Stroke Prevention Trial in Sickle Cell Anemia (STOP): extended follow-up and final results. Blood. 2006;108:847–52. doi: 10.1182/blood-2005-10-009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Habibi A, Mekontso-Dessap A, Guillaud C, et al. Delayed hemolytic transfusion reaction in adult sickle-cell disease: presentations, outcomes, and treatments of 99 referral center episodes. Am J Hematol. 2016;91:989–94. doi: 10.1002/ajh.24460. [DOI] [PubMed] [Google Scholar]
- 5.Win N. Hyperhemolytic transfusion reaction in sickle cell disease. Transfusion. 2001;41:323–8. doi: 10.1046/j.1537-2995.2001.41030323.x. [DOI] [PubMed] [Google Scholar]
- 6.Aygun B, Padmanabhan S, Paley C, et al. Clinical significance of RBC alloantibodies and autoantibodies in sickle cell patients who received transfusions. Transfusion. 2002;42:37–43. doi: 10.1046/j.1537-2995.2002.00007.x. [DOI] [PubMed] [Google Scholar]
- 7.Hanchard NA, Moulds JM, Belmont JW, et al. A genome-wide screen for large-effect alloimmunization susceptibility loci among red blood cell transfusion recipients with sickle cell disease. Transfus Med Hemother. 2014;41:453–61. doi: 10.1159/000369079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox JV, Steane E, Cunningham G, et al. Risk of alloimmunization and delayed hemolytic transfusion reactions in patients with sickle cell disease. Arch Intern Med. 1988;148:2485–9. [PubMed] [Google Scholar]
- 9.Talano JA, Hillery CA, Gottschall JL, et al. Delayed hemolytic transfusion reaction/hyperhemolysis syndrome in children with sickle cell disease. Pediatrics. 2003;111:e661–5. doi: 10.1542/peds.111.6.e661. [DOI] [PubMed] [Google Scholar]
- 10.de Montalembert M, Dumont MD, Heilbronner C, et al. Delayed hemolytic transfusion reaction in children with sickle cell disease. Haematologica. 2011;96:801–7. doi: 10.3324/haematol.2010.038307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eleng N. Delayed hemolytic transfusion reaction in sickle cell disease. Am J Clin Med Res. 2013;1:40–4. [Google Scholar]
- 12.Petz LD, Calhoun L, Shulman IA, et al. The sickle cell hemolytic transfusion reaction syndrome. Transfusion. 1997;37:382–92. doi: 10.1046/j.1537-2995.1997.37497265338.x. [DOI] [PubMed] [Google Scholar]
- 13.Vidler JB, Gardner K, Amenyah K, et al. Delayed haemolytic transfusion reaction in adults with sickle cell disease: a 5-year experience. Br J Haematol. 2015;169:746–53. doi: 10.1111/bjh.13339. [DOI] [PubMed] [Google Scholar]
- 14.King KE, Shirey RS, Lankiewicz MW, et al. Delayed hemolytic transfusion reactions in sickle cell disease: simultaneous destruction of recipients’ red cells. Transfusion. 1997;37:376–81. doi: 10.1046/j.1537-2995.1997.37497265337.x. [DOI] [PubMed] [Google Scholar]
- 15.Chadebech P, Habibi A, Nzouakou R, et al. Delayed hemolytic transfusion reaction in sickle cell disease patients: evidence of an emerging syndrome with suicidal red blood cell death. Transfusion. 2009;49:1785–92. doi: 10.1111/j.1537-2995.2009.02199.x. [DOI] [PubMed] [Google Scholar]
- 16.Kato GJ, Hebbel RP, Steinberg MH, et al. Vasculopathy in sickle cell disease: biology, pathophysiology, genetics, translational medicine, and new research directions. Am J Hematol. 2009;84:618–25. doi: 10.1002/ajh.21475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ataga KI. Hypercoagulability and thrombotic complications in hemolytic anemias. Haematologica. 2009;94:1481–4. doi: 10.3324/haematol.2009.013672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Win N. Hyperhemolysis syndrome in sickle cell disease. Expert Rev Hematol. 2009;2:111–5. doi: 10.1586/ehm.09.2. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–9. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med. 2013;369:1502–11. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valencia CA, Husami A, Holle J, et al. Clinical impact and cost-effectiveness of whole exome sequencing as a diagnostic tool: a pediatric center’s experience. Front Pediatr. 2015;3:67. doi: 10.3389/fped.2015.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li AH, Morrison AC, Kovar C, et al. Analysis of loss-of-function variants and 20 risk factor phenotypes in 8,554 individuals identifies loci influencing chronic disease. Nat Genet. 2015;47:640–2. doi: 10.1038/ng.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Do R, Stitziel NO, Won HH, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–6. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallal S, Nolan D, Witt C, et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359:727–32. doi: 10.1016/s0140-6736(02)07873-x. [DOI] [PubMed] [Google Scholar]
- 25.Lupski JR, Gonzaga-Jauregui C, Yang Y, et al. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med. 2013;5:57. doi: 10.1186/gm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMichael G, Bainbridge MN, Haan E, et al. Whole-exome sequencing points to considerable genetic heterogeneity of cerebral palsy. Mol Psychiatry. 2015;20:176–82. doi: 10.1038/mp.2014.189. [DOI] [PubMed] [Google Scholar]
- 27.Reid JG, Carroll A, Veeraraghavan N, et al. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15:30. doi: 10.1186/1471-2105-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picard [Internet] Cambridge (MA): Broad Institute; [cited 2016 Sep 24]. Available from: http://broadinstitute.github.io/picard/ [Google Scholar]
- 31.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Challis D, Yu J, Evani US, et al. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–8. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.San Lucas FA, Wang G, Scheet P, et al. Integrated annotation and analysis of genetic variants from next-generation sequencing studies with variant tools. Bioinformatics. 2012;28:421–2. doi: 10.1093/bioinformatics/btr667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32:894–9. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lubeck E, Coskun AF, Zhiyentayev T, et al. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2012;9:743–8. [Google Scholar]
- 39.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–61. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.1000 Genomes Project Consortium. Auton A, Abecasis GR, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.NHLBI GO Exome Sequencing Project (ESP) Exome Variant Server [Internet] Seattle (WA): University of Washington; [cited 2013 Feb 1]. Available from: http://evs.gs.washington.edu/EVS/ [Google Scholar]
- 42.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuentes Fajardo KV, Adams D, Mason CE, et al. Detecting false-positive signals in exome sequencing. Hum Mutat. 2012;33:609–13. doi: 10.1002/humu.22033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye J, Coulouris G, Zaretskaya I, et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheehan VA, Crosby JR, Sabo A, et al. Whole exome sequencing identifies novel genes for fetal hemoglobin response to hydroxyurea in children with sickle cell anemia. PLoS One. 2014;9:1–8. doi: 10.1371/journal.pone.0110740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lupski JR, Belmont JW, Boerwinkle E, et al. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 48.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 49.Shihab HA, Gough J, Cooper DN, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong C, Wei P, Jian X, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24:2125–37. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi Y, Sims GE, Murphy S, et al. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walters-Sen LC, Hashimoto S, Thrush DL, et al. Variability in pathogenicity prediction programs: impact on clinical diagnostics. Mol Genet Genomic Med. 2015;3:99–110. doi: 10.1002/mgg3.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narbey D, Habibi A, Chadebech P, et al. Incidence and predictive score for delayed hemolytic transfusion reaction in adult patients with sickle cell disease. Am J Hematol. 2017;92:1340–8. doi: 10.1002/ajh.24908. [DOI] [PubMed] [Google Scholar]
- 54.Heitzeneder S, Seidel M, Förster-Waldl E, et al. Mannan-binding lectin deficiency—good news, bad news, doesn’t matter? Clin Immunol. 2012;143:22–38. doi: 10.1016/j.clim.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 55.Thiel S, Frederiksen PD, Jensenius JC. Clinical manifestations of mannan-binding lectin deficiency. Mol Immunol. 2006;43:86–96. doi: 10.1016/j.molimm.2005.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wallis R, Drickamer K. Molecular determinants of oligomer formation and complement fixation in mannose-binding proteins. J Biol Chem. 1999;274:3580–9. doi: 10.1074/jbc.274.6.3580. [DOI] [PubMed] [Google Scholar]
- 57.Hartshorn KL, White MR, Crouch EC. Contributions of the N- and C-terminal domains of surfactant protein D to the binding, aggregation, and phagocytic uptake of bacteria. Infect Immun. 2002;70:6129–39. doi: 10.1128/IAI.70.11.6129-6139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stowell SR, Winkler AM, Maier CL, et al. Initiation and regulation of complement during hemolytic transfusion reactions. Clin Dev Immunol. 2012;2012:307093. doi: 10.1155/2012/307093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salama A, Mueller-Eckhardt C. Delayed hemolytic transfusion reactions. Evidence for complement activation involving allogeneic and autologous red cells. Transfusion. 1984;24:188–93. doi: 10.1046/j.1537-2995.1984.24384225018.x. [DOI] [PubMed] [Google Scholar]
- 60.Nakata S, Imagawa A, Miyata Y, et al. Low gene expression levels of activating receptors of natural killer cells (NKG2E and CD94) in patients with fulminant type 1 diabetes. Immunol Lett. 2013;156:14–155. doi: 10.1016/j.imlet.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 61.Orbelyan GA, Tang F, Sally B, et al. Human NKG2E is expressed and forms an intracytoplasmic complex with CD94 and DAP12. J Immunol. 2014;193:610–6. doi: 10.4049/jimmunol.1400556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gilsanz F, De La Serna J, Moltó L, et al. Hemolytic anemia in chronic large granular lymphocytic leukemia of natural killer cells: cytotoxicity of natural killer cells against autologous red cells is associated with hemolysis. Transfusion. 1996;36:463–6. doi: 10.1046/j.1537-2995.1996.36596338025.x. [DOI] [PubMed] [Google Scholar]
- 63.Posey JE, Harel T, Liu P, et al. Resolution of disease phenotypes resulting from multilocus genomic variation. N Engl J Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simon AK, Hollander GA, McMichael A. Evolution of the immune system in humans from infancy to old age. Proc Biol Sci. 2015;282:20143085. doi: 10.1098/rspb.2014.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Candidate variants in HHS replication cohort.
Table S2. Primers used for PCR and Sanger sequencing.
Table S3. Summary of variants that were predicted to be damaging by four or more algorithms.
Fig. S1. Schematic of variant filtering pipeline after whole exome sequencing. Twelve individuals with clinical definitions of SCD and HHS were sequenced. The top-to-bottom variant filtering workflow shows the variants remaining after each filtering step together with the software tools used.
Fig. S2. Protein domains of candidate genes. Protein topology highlighting position of the mutations (red arrows) and protein domains for MBL2 (Top), KLRC3 (Middle), and PCSK5 (Bottom). The PCSK5 transcript of interest (ENST00000376767) lacks a portion of the CRM domain (shaded) (http://www.uniprot.org/).
Fig. S3. Histogram of the frequency distribution of filtered variants. Non-synonymous variants in dbNSFP, with a “PASS” flag, SIFT score < 0.05, MAF ≤5% (utilizing data in the 1000 Genomes Project Exome Variant Server, Exome Sequencing Project (ESP) and The Exome Aggregation Consortium, ExAC) were selected. Variants with allelic ratios < 0.3 and variants found in sex chromosomes and potential false positive genes were filtered out. The histogram shows the frequency distribution of the final set of uncommon, stop-gain variants that were enriched in the HHS cohort.
Fig. S4. Distribution of candidate gene variant carriers. Rare damaging variants in 3 genes (MBL2, KLRC3 and PCSK5) were found in 7 of the 12 patients with HHS. 5 of the 12 patients did not have any of the 3 candidate variants. Venn diagram shows distribution and intersection of the pLOF variants in the 3 candidate genes across the 12 HHS patients.


