Abstract
Background & Aims
Patients with severe alcoholic hepatitis (AH) have a high risk of death within 90 days. Corticosteroids, which can cause severe adverse events, are the only treatment that increases short-term survival. It is a challenge to predict outcomes of patients with severe AH. Therefore, we developed a scoring system to predict patient survival, integrating baseline molecular and clinical variables.
Methods
We obtained fixed liver biopsy samples from 71 consecutive patients diagnosed with severe AH and treated with corticosteroids from July 2006 through December 2013 in Brussels, Belgium (derivation cohort). Gene expression patterns were analyzed by microarrays and clinical data were collected for 180 days. We identified gene expression signatures and clinical data that are associated with survival without liver transplantation at 90 and 180 days after initiation of corticosteroid therapy. Findings were validated using liver biopsies from 48 consecutive patients with severe AH treated with corticosteroids, collected from March 2010 through February 2015 at hospitals in Belgium and Switzerland (validation cohort 1) and in liver biopsies from 20 patients (9 received corticosteroid treatment), collected from January 2012 through May 2015 in the United States (validation cohort 2).
Results
We integrated data on expression patterns of 123 genes and the model for end-stage liver disease (MELD) score to assign patients to groups with poor survival (29% survived 90 days and 26% survived 180 days) and good survival (76% survived 90 days and 65% survived 180 days) (P<.001) in the derivation cohort. We named this assignment system the gene signature-MELD (gs-MELD) score. In the validation cohort 1, the gs-MELD score discriminated patients with a poor survival (43% survived 90 days) from those with a good survival (96% survived 90 days) (P<.001). The gs-MELD score also discriminated between patients with a poor survival at 180 days (34% survived) and a good survival at 180 days (84% survived) (P<.001). The time-dependent area under the receiver operator characteristic curve for the score was 0.86 (95% CI, 0.73–0.99) for survival at 90 days, and 0.83 (95% CI, 0.71–0.96) for survival at 180 days. This score outperformed other clinical models to predict survival of patients with severe AH in validation cohort 1. In validation cohort 2, the gs-MELD discriminated patients with a poor survival at 90 days (12% survived) from those with a good survival at 90 days (100% survived) (P<.001).
Conclusions
We integrated data on baseline liver gene expression pattern and MELD score to create the gs-MELD scoring system, which identifies patients with severe AH, treated or not with corticosteroids, most and least likely to survive for 90 and 180 days.
Keywords: MELD, transcription, ethanol, cirrhosis
Introduction
Alcoholic hepatitis (AH) is a syndrome characterized by a recent onset of jaundice in individuals who chronically abuse alcohol.1 Severe AH is commonly defined by a Maddrey’s discriminant function of 32 or higher.2 It is a highly lethal disease with a 90-day mortality rate of up to 50%.1, 3 Corticosteroids have been the only reproducible medical treatment option improving the short-term survival of severe AH patients over the few past decades.4 Nevertheless, a significant fraction of AH patients do not respond to this treatment, and frequently experience severe adverse effects, such as infection, that often occur within a few days of treatment initiation.5, 6 Furthermore, corticosteroids non-responders have currently no available therapeutic options.7, 8 Thus, early prognostic prediction prior to corticosteroid treatment is critical in the management of severe AH.
To date, several pre-treatment clinical/histological prognostic scores, have been proposed.2, 3, 9–11 The performance of these static scores at the individual level is extremely variable between studies and none of these indices have shown to reproducibly outperformed the others. Recently, the combination of a pre-treatment index, the model for end stage liver disease (MELD), and a dynamic score, the Lille model,12 calculated 7 days after initiation of corticosteroid therapy, was suggested as a very efficient predictive algorithm of short-term survival.13 However, using a dynamic score may expose the non-responder subgroup to a high risk of infection without any benefit. This clearly indicates an urgent unmet need for better pre-treatment predictors of survival. To address this challenge, we aimed to develop and establish a prognostic score, combining pre-treatment clinical variables and a molecular signature and to implement this score in a Food and Drug Administration (FDA)-approved platform ready for clinical deployment.
Methods
Patients
Patients included in the current study had the same following criteria. All of them were chronic alcohol consumers (>60 g/day for males and >40 g/day for females), had onset of jaundice within 90 days of enrollment, a serum bilirubin level of more than 5 mg/dL, and less than 60 days of abstinence before the onset of jaundice.7 All patients had a Maddrey’s discriminant function of 32 or more.2 Histological confirmation of AH was performed in all patients by transjugular liver biopsy. Catheterization of the right jugular vein was performed under ultrasound guidance. An introducer sheath was inserted according to Seldinger’s technique and advanced to the hepatic vein under fluoroscopic control. A 18-gauge biopsy needle was used for tissue sampling.14 Histological criteria included the presence of steatosis, ballooned hepatocytes, Mallory bodies, with infiltration of polymorphonuclear neutrophils,7, 8 Needle biopsy specimens of the liver were obtained from all patients prior to potential corticosteroid therapy and archived as formalin-fixed, paraffin-embedded (FFPE) tissue blocks. Patients with viral or autoimmune hepatitis, hemochromatosis, or co-infection with human immunodeficiency virus were excluded. The derivation cohort included a total of 73 consecutive patients recruited between July 2006 and December 2013 at C.U.B. Hôpital Erasme, Brussels, Belgium (Figure 1A). Validation cohort 1 included 48 consecutive patients recruited between March 2010 and February 2015 at Hôpital de Jolimont, Haine-Saint-Paul, Belgium (n=26), Centre Hospitalier Universitaire Vaudois, University of Lausanne, Lausanne, Switzerland (n=12), and University Hospitals of Geneva, Geneva, Switzerland (n=10).15 All patients in the derivation cohort and validation cohort 1 received methylprednisolone orally at a dose of 32 mg/day, after confirmation of the histological diagnosis of AH, for a maximum of 28 days.7 Eleven patients in the derivation cohort had a controlled infection at baseline that did not preclude the administration of corticosteroids, none were infected in the validation cohort 1.16 The clinical utility of the NanoString assay-based prognostic score was further validated in another independent validation cohort 2 (n=20), including clinically high-risk patients with histologically-confirmed severe AH who were referred from January 2012 through May 2015, to Mount Sinai Medical Center, United States, for evaluation of early liver transplantation.17 These patients either had a contraindication for the use of steroids due to comorbidities such as hepatorenal syndrome (HRS° and/or uncontrolled infection (n=11) or were non-responders to corticosteroids after 7 days of treatment according to the Lille score (n=9).
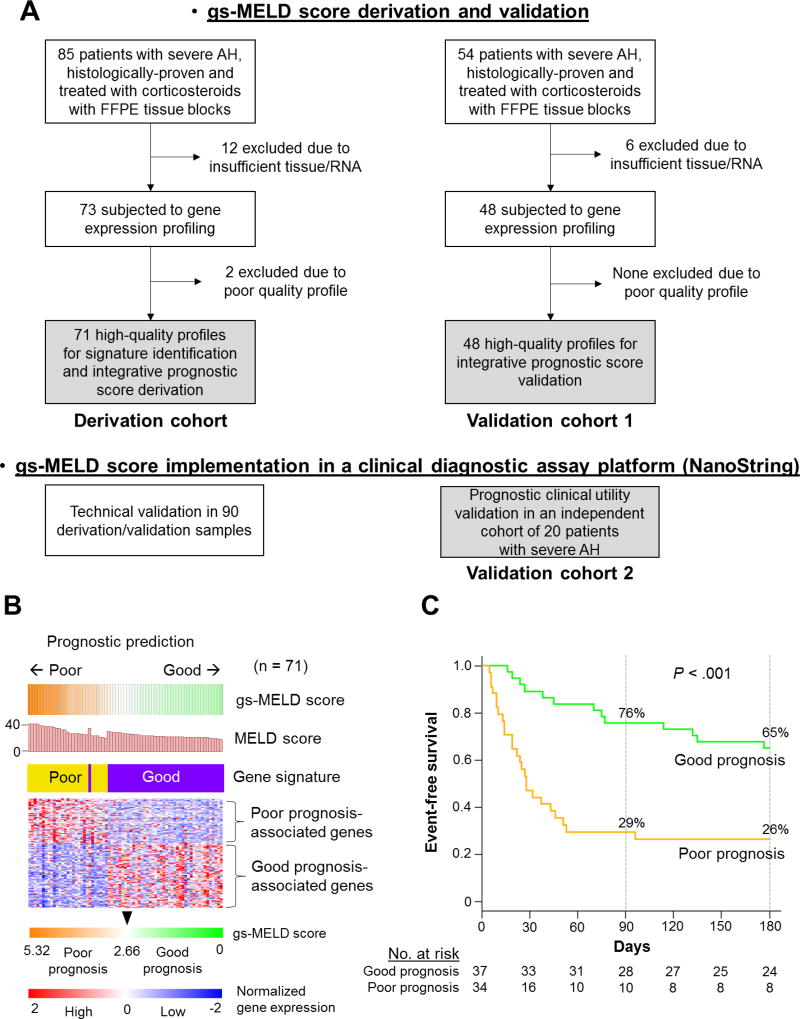
Figure 1. Development of the gs-MELD score.
Panel A shows the selection of patients included in the study. The gray background indicates the final number of patients in each cohort. Panel B shows expression patterns of the 123-gene signature in the derivation cohort. Red and blue colors indicate high and low gene expression, respectively. The level of expression of the 123 genes and the MELD score in the derivation cohort, defined the gs-MELD score. Patients were assigned to a good- or poor-prognosis group according to the median (2.66) of the gs-MELD score. Thirty-seven (52%) of the patients had a gs-MELD score equal to or lower than 2.66, and 34 (48%) patients had a score greater than 2.66 and were classified as good, and poor prognosis, respectively. Panel C shows event-free survival curves for the gs-MELD score in the derivation cohort. In the good (green line) and poor-prognosis (orange line) groups, 76% and 29% of patients were free of events at 90 days vs 65% and 26% at 180 days, in the good and poor-prognosis groups respectively (P < .001). AH: alcoholic hepatitis, FFPE: Formalin-fixed, paraffin-embedded, MELD: Model for End-Stage Liver Disease, gs: gene signature.
We also included 67 patients with alcoholic cirrhosis and no histological signs of AH, 13 with non-severe AH defined as biopsy-documented AH and a Maddrey’s discriminant function lower than 32, and 6 with alcoholic steatosis without fibrosis from C.U.B. Hôpital Erasme, Brussels, Belgium. Written informed consent was obtained from all individuals, and Ethical Committees at C.U.B. Hôpital Erasme, Hôpital de Jolimont, Centre Hospitalier Universitaire Vaudois, University Hospitals of Geneva, and Mount Sinai Medical Center approved the study.
Studied clinical variables and endpoints
The following values were collected or calculated at baseline (day 0, date of corticosteroid therapy initiation) unless otherwise specified: age, sex, prothrombin time, international normalized ratio (INR), bilirubin (day 0 and day 7), albumin, serum creatinine, Maddrey’s discriminant function, and MELD scores.2, 11 The Lille score was calculated at day 7 and a score of 0.45 or greater defines non-responders to corticosteroids.12
The primary endpoint was to develop an integrative prognostic score of 90- and 180-day survival without death or liver transplantation defined as event-free survival. This composite outcome was chosen since LT is considered to be a salvage therapy in severe AH and involves patients with very severe disease.18 These time points were also chosen based on recommendations from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) Alcoholic Hepatitis Consortia.19 Furthermore, survival before 180 days is mainly driven by liver injury and not significantly impacted by alcohol-relapse.20 Follow-up time was defined as the period from the first day of initiation of therapy to 180 days after treatment started in the derivation cohort and in the validations cohorts. When patients were not treated (55% of patients in the validation cohort 2), the follow-up started at the day of liver biopsy. Data for patients who had not died including those lost to follow-up were censored at the date of the last follow-up visit. The secondary outcome was to implement this score in an FDA-approved platform ready for clinical deployment. Finally, we tested whether other gene signature could be associated with the occurrence of severe infection (defined as infection causing death) and type 1 HRS.21
Transcriptome profiling
Using five 10-micron-thick FFPE liver biopsy tissue sections, total RNA was extracted using the AllPrepDNA/RNA Mini Kit (Qiagen), 250 ng of total RNA with sufficient quality (RPL13A qPCR Ct value <30) was assayed using Affymetrix GeneTitan Human Genome U219 microarray (Affymetrix). Raw scan data were normalized by Robust Multiarray Average algorithm,22 and poor quality transcriptome profiles were determined by inter-sample correlation <0.70.23 Less variable genes across the samples, based on coefficient of variation <0.4, were excluded; and a 123-gene prognostic gene signature was defined in the derivation cohort as previously described.23 One hundred fourteen genes with the largest absolute Cox scores (to fit the assay format) were implemented in an nCounter Elements assay (NanoString), and 200 to 500 ng of total RNA samples was assayed.24 Gene signature-based outcome prediction was performed using the Nearest Template Prediction algorithm,25 based on a prediction confidence of p<0.05 defining the poor-prognosis group, while the rest of the subjects were designated to the good-prognosis group. Prognostic molecular pathways in derivation cohort were determined by surveying the Molecular Signature Database (MSigDB, www.broadinstitute.org/msigdb) using Gene Set Enrichment Analysis.26 Regulatory gene networks (modules) in the transcriptome profiles of the derivation cohort were determined as previously described.27, 28 All bioinformatics analyses were performed by using GenePattern (www.broadinstitute.org/genepattern) and R statistical language (www.r-project.org). Transcriptome data are available at NCBI Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo, accession number GSE94417) and EBI ArrayExpress (www.ebi.ac.uk/arrayexpress, E-MTAB-2664) for the 10 patients from Geneva in validation cohort 1. We also added liver gene expression data from 21 healthy individuals from publicly available datasets (n=7 from GSE28619, and n=14 from GSE48452).29, 30
Statistical analysis
Inter-group difference for continuous and categorical variables was tested using the Wilcoxon rank-sum test and Fisher’s exact test, respectively. Prognostic associations of the clinical and molecular variables were assessed using Kaplan-Meier curves (for entire follow-up period), Fisher’s exact test (for each time point), log-rank test, and Cox regression modeling. Prognostic performance of the scores was assessed by hazard ratio (HR) and 95% confidence intervals (CI), area under the ROC curve (AUC) and time-dependent AUC with higher values demonstrating improved diagnostic performance of a given model as a function of time,31 cindex with higher values indicating better predictive model ability,32 Akaike’s Information Criterion (AIC) and Bayesian information criterion (BIC), with lower AIC and BIC values indicating better model fit.33, 34. An integrative clinical and molecular prognostic score was developed in the derivation cohort based on the regression coefficient of a multivariable Cox regression model including the gene signature and a baseline prognostic indicator, the MELD score.11 Prognostic performances of the integrative score, were evaluated in validation cohort 1 and compared with its individual components, the Lille model,12 and the combination of the MELD and the Lille scores,13 all assessed as continuous variables. The integrative score was further validated in a different platform (NanoString) in validation cohort 2. A two-tailed p<0.05 was considered statistically significant. All data analyses were performed using the R statistical language.
Results
Patients
Among the 73 patients included in the derivation cohort, two were excluded due to low-quality transcriptome profiles (Figure 1, panel A). Clinical demographics of the patients at the time of enrollment were comparable between the derivation cohort (n=71) and validation cohort 1 (n=48) cohorts except higher INR, and Maddrey’s discriminant function in the derivation cohort (Table 1). Conversely, patients in validation cohort 2 (n=20) had a worse liver function compared to those in the derivation cohort (Table 1). All patients were initially hospitalized and followed after discharge. At 90 days, no patients in the derivation cohort, 1 (2.1 %) patient in the validation cohort 1, and no patients in the validation 2 cohort were lost to follow-up. At 180 days, 1 (1.4%) patient in the derivation cohort, 2 (4.2%) in the validation cohort 1, and none in the validation 2 cohort were lost to follow-up.
Table 1.
Characteristics of patients and clinical outcomes
| Characteristics | Derivation cohort (n = 71) |
Validation cohort 1 (n = 48) |
P* | Validation cohort 2 (n = 20) |
P** |
|---|---|---|---|---|---|
| Age, y | 53 (47–60) | 53 (46–60) | 0.92 | 44 (41–48) | 0.001 |
| Male sex, n | 44 (62%) | 25 (52%) | 0.35 | 8 (40%) | 0.12 |
| INR | 1.9 (1.7–2.2) | 1.7 (1.5–2.0) | 0.003 | 2.3 (2.0–2.7) | 0.031 |
| Bilirubin, mg/dL | 13 (7.3–24.0) | 13 (7.8–18) | 0.54 | 26.1 (23.5–35.3) | <0.001 |
| Creatinine, mg/dL | 0.8 (0.6–1.0) | 0.8 (0.6–0.9) | 0.37 | 1.7 (1.3–2.6) | <0.001 |
| Albumin, g/L | 27 (23–29) | 26 (24–28) | 0.96 | 28 (26–31) | 0.076 |
| Maddrey score | 57.5 (45–82) | 50 (41–69) | <0.001 | 85 (69–103) | 0.003 |
| MELD score | 23 (21–28) | 22 (20–25) | 0.074 | 34 (32–38) | <0.001 |
| Lille score | |||||
| - median score | 0.32 (0.13–0.67) | 0.30 (0.12–0.46)¥ | 0.54 | 0.99 (0.93–1.00)‡ | <0.001 |
| - non-responders | 28 (39%) | 14 (30%) | 0.43 | 9 (100%)‡ | <0.001 |
| Death or liver transplantation | |||||
| - 90 days | 33 (46%) | 13 (27%) | 0.037 | 13 (65%) | 0.21 |
| - 180 days | 38 (54%) | 18 (38%) | 0.095 | 15 (75%) | 0.12 |
Derivation cohort vs. validation cohort 1.
Derivation cohort vs. validation cohort 2.
INR: international normalized ratio; MELD: model for end-stage liver disease. Continuous variables are presented as median (interquartile range).
Lille score could not be calculated in 2 patients in the validation cohort 1 since they died before 7 days of treatment.
In validation cohort 2, 9 patients (45%) were treated with corticosteroids.
Prognostic gene signature in severe AH
In the derivation cohort, a 123-gene prognostic signature was defined (Supplementary Table 1), designating 28 patients (39%) as having a poor prognostic signature pattern. Patients assigned to the poor-prognosis group had a significantly lower event-free survival at 90 and 180 days compared to patients with a good prognosis (Supplementary Figure 1). Molecular pathway analysis revealed association of inflammatory and oxidative stress response pathways with poor prognosis, and association of cell proliferation and hepatic stellate cell gene signature,35 presumably reflecting a fibrogenic wound healing response, with good prognosis (Supplementary Table 2). Regulatory gene network analysis revealed association of lipid, bile acid, and xenobiotic metabolism with better prognosis (Supplementary Figure 2 and Supplementary Table 3). The poor prognosis signature pattern was also assessed in other patients with alcoholic liver disease (ALD) [Supplementary Table 4, Supplementary Figure 3A] and healthy liver individuals (Supplementary Figure 3B). The poor-prognosis pattern of the 123-gene signature was absent in individuals with a healthy liver and alcoholic steatosis, whereas partial signature gene dysregulation was observed in 23% of non-severe AH and 18% of alcoholic cirrhosis patients (Supplementary Table 5). Among the 123 signature genes, 51 genes were specifically dysregulated in severe AH with poor prognosis, whereas 72 genes were also dysregulated in the subsets of alcoholic cirrhosis and non-severe AH patients (Supplementary Figure 3C). Each gene subset showed inferior prognostic capability compared to the full 123-gene signature (c-index of 0.65, 0.57 and 0.70 for the 72, 51 and 123-gene signatures respectively), indicating that the both components are necessary to maintain the prognostic performance of the signature.
Derivation of an integrative clinical and molecular prognostic score: gene signature-MELD (gs-MELD) score
A prognostic score, integrating the gene signature and the MELD score (gene signature-MELD [gs-MELD] score), was developed by using regression coefficients from the multivariable Cox regression model in the derivation cohort as follows:
Patients with a gs-MELD greater than 2.66 (the median in the derivation cohort) were considered to have a poor prognosis (Figure 1B). This group had an event-free probability of survival of 29%, and 26% at 90 and 180 days, respectively (P < .001, Figure 1C). The gs-MELD remained strongly associated with the occurrence of mortality or liver transplantation even after exclusion of these 11 infected patients in the derivation cohort (P < .001, Supplementary Figure 4).
Validation of the gs-MELD score
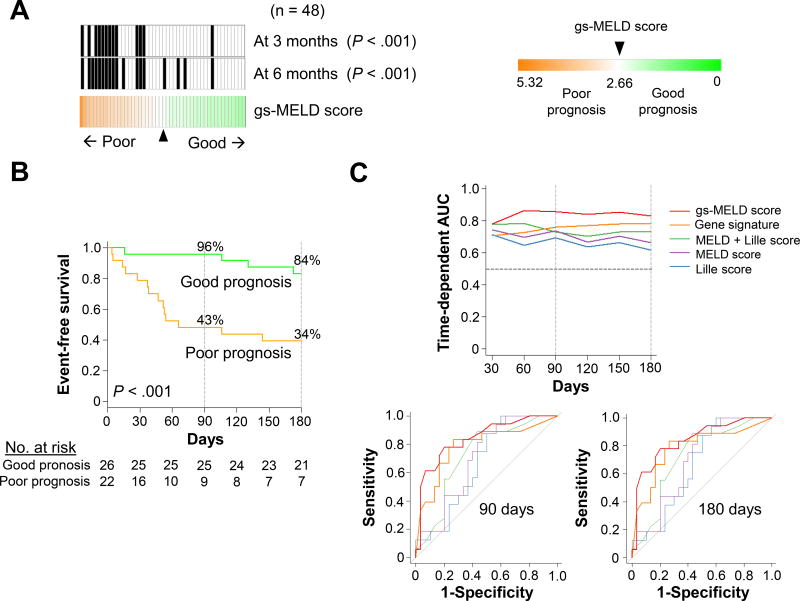
Using the cut-off defined in the derivation cohort, the prognostic performance of the gs-MELD score was evaluated in the validation cohort 1, similarly profiled by genome-wide DNA microarray (Figure 2A). Patients assigned to the poor prognosis group (gs-MELD score > 2.66) had a significantly lower event-free probability of survival compared to those classified as good prognosis at 90 (43% vs 96%, P<.001) and 180 days (34% vs 84% P<.001, Figure 2B). The gs-MELD score was superior to the MELD score alone for 90-day (AUC, time-dependent AUC and c-index were increased by 0.15, 0.13 and 0.12, and AIC was decreased by 8) and 180-day prognosis (increased by 0.11, 0.10 and 0.09, and decreased by 3, respectively) (Table 2). Of note,, prognostic association of the score was superior to a recently published combined model integrating the MELD and Lille scores.13 The prognostic performance of the gs-MELD score remained stable over time compared to other clinical scores (Figure 2C).
Figure 2. Prognostic performances of the gs-MELD score in the validation cohort 1.
Panel A shows the distribution of the gs-MELD score among the 48 subjects of the validation cohort 1. The cut-off (2.66) was defined in the derivation cohort. The black vertical bars represent subjects who experienced death or liver transplantation at 90 or 180 days. Twenty-six and 22 patients were classified as good and poor prognosis, respectively. Thirteen (27%) and 18 (38%) died or were transplanted at 90 (P < .001) and 180 days (P<.001), respectively. Panel B shows event-free survival curves for the gs-MELD score. In the good (green line) and poor-prognosis (orange line) groups, 96% and 43% of patients were free of events at 90 days vs 84% and 34% at 180 days, in the good and poor-prognosis groups respectively (P < .001). Panel C, upper part, shows time-dependent area under the ROC curve (AUC) corresponding to the evolution of AUC during follow-up. The horizontal black dotted line indicates the generally used cut-off (0.70) of clinically meaningful utility. In the lower part, ROC curves at 90 and 180 days are displayed. MELD: Model for End-Stage Liver Disease.
Table 2.
Prognostic association of continuous scores with 90- and 180-day probability of death or liver transplantation in validation cohort 1.
| Variables | HR (95% CI) |
P | 90 days | 180 days | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC | Time- dependent AUC (95% CI) |
C-index (95% CI) |
AIC¥ | BIC¥ | AUC | Time-dependent AUC (95% CI) |
C-index (95% CI) |
AIC¥ | BIC¥ | |||
| gs-MELD score | 4.19 (2.23–7.89) | <0.001 | 0.86 (0.73–0.99) | 0.86 (0.73–0.99) | 0.83 (0.66–0.99) | 72 | 72 | 0.84 (0.72–0.96) | 0.83 (0.71–0.96) | 0.80 (0.66–0.94) | 105 | 106 |
| 123-gene signature | 1.57 (1.23–2.00) | <0.001 | 0.76 (0.59–0.94) | 0.76 (0.59–0.93) | 0.73 (0.57–0.89) | 78 | 78 | 0.79 (0.64–0.93) | 0.78 (0.64–0.93) | 0.73 (0.60–0.87) | 108 | 109 |
| MELD score | 1.16 (1.03–1.30) | 0.010 | 0.71 (0.55–0.88) | 0.73 (0.57–0.89) | 0.71 (0.55–0.87) | 80 | 80 | 0.73 (0.58–0.87) | 0.73 (0.58–0.89) | 0.71 (0.57–0.85) | 108 | 109 |
| Lille score‡* | 1.13 (0.97–1.32) | 0.13 | 0.70 (0.53–0.86) | 0.69 (0.53–0.85) | 0.68 (0.51–0.86) | 80 | 80 | 0.67 (0.51–0.82) | 0.62 (0.44–0.79) | 0.65 (0.51–0.80) | 115 | 116 |
| Lille and MELD scores | 1.84 (0.99–3.41) | 0.055 | 0.73 (0.57–0.88) | 0.74 (0.59–0.88) | 0.71 (0.53–0.88) | 78 | 78 | 0.70 (0.55–0.85) | 0.66 (0.50–0.83) | 0.68 (0.53–0.82) | 113 | 114 |
MELD: model for end-stage liver disease; HR: hazard ratio; CI: confidence interval; AUC: area under the curve; AIC: Akaike information criterion. BIC: Bayesian information criterion. Scores are presented as median (interquartile range).
HR for the Lille score was calculated per increase of 0.1 unit.
Lille score could not be calculated in 2 patients since they died before 7 days of treatment.
Since the Lille score could not be calculated in two patients AIC and BIC were calculated for 46 patients out of 48 to allow meaningful comparisons between all scores.
Evaluation of other gene signatures
First, we tested various signatures including fewer genes in replication cohort 1 using HRs and their 95% CI as a surrogate and observed that the number of genes could be reduced to 32 while maintaining the prognostic performances (Supplementary Figure 5). Using a similar method to integrate the gene signature with the MELD score, we developed a 32-gene signature-MELD (gs32-MELD) score in the derivation cohort and evaluate the performance in the validation 1 cohort (Supplementary Figure 6). The prognostic performances of the gs32-MELD were slightly inferior to the gs-MELD (HR, 4.04; 95% CI, 1.56–10.46; P = .002 vs HR, 4.19; 95% CI, 2.23–7.89 P < .001 for the gs32-MELD and gs-MELD respectively) in the validation 1 cohort. In addition, we developed a gene signature in the derivation cohort for the occurrence of severe infection (Supplementary Figure 7A). Patients harboring the poor prognosis signature had a significantly higher probability of severe infection (P = .019) in the validation cohort 1 (Supplementary Figure 7B). Similarly, we defined a signature for the occurrence of HRS in the derivation cohort (Supplementary Figure 7C). However, the incidence of this outcome did not differ significantly between the poor and good prognosis signature (P = .81) [Supplementary Figure 7D].
Implementation of the prognostic gene signature in a clinical assay platform
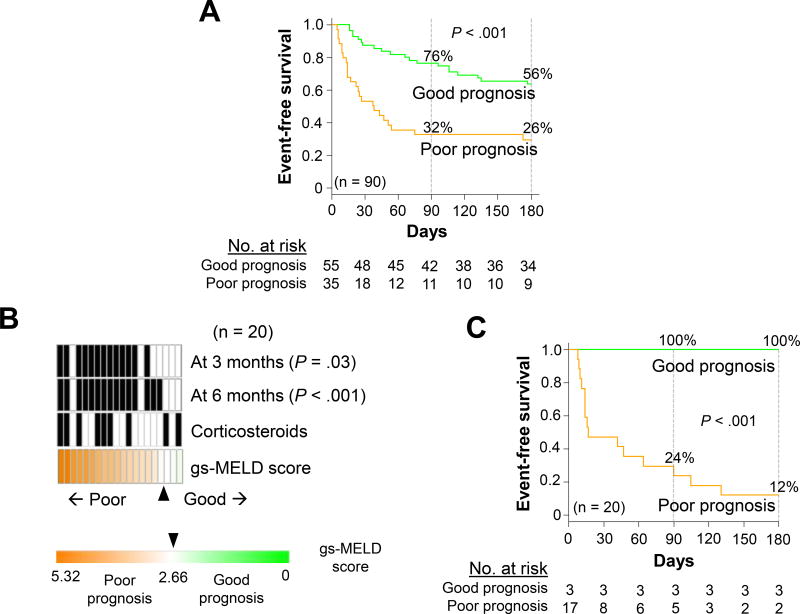
Subsequently, the prognostic gene signature was implemented in an FDA-approved clinical diagnostic platform (NanoString) using the 114 top prognostic genes with the largest absolute Cox scores to fit onto the platform (120-gene platform, including 6 normalization genes, Supplementary Table 2). For technical validation of the assay, 90 patients from the derivation and validation cohorts with remaining RNA were re-analyzed. The gs-MELD score-based prognostic prediction was compared between the original microarray and the NanoString platforms. The NanoString assay-based score using the predefined cut-off value yielded highly reproducible prognostic predictions (accuracy = 89%, p<0.001, Supplementary Figure 3) with retained prognostic association (p<0.001, Figure 3A). To assess whether the gs-MELD score would be useful in very high-risk subjects, we further evaluated this score in another independent cohort of 20 patients with severe AH, referred as potential candidates for early liver transplantation (validation cohort 2) (Table 1). Even in this high-risk cohort of patients representing more advanced disease, the gs-MELD score identified a subset of patients with striking prognostic differences (Figure 3B and 3C), further supporting the broad clinical utility of the gs-MELD score across the whole spectrum of severity of severe AH.
Figure 3. Implementation of the gs-MELD score in an FDA-approved clinical diagnostic platform (NanoString).
Panel A shows event-free survival curves for the gs-MELD score in these 90 patients. At 90 days, 78% and 35% of patients were free of events vs 67% and 30% of patients at 180 days in the good (green line) and poor-prognosis (orange line) groups, respectively (P< .001). Panel B shows the distribution of the gs-MELD score in validation cohort 2 including very high-risk patients (n = 20). Nine patients (45%) were treated with corticosteroids. Twelve (60%) and 15 (75%) died or were transplanted at 90 (P < .001) and 180 days (P<.001), respectively. Panel C shows survival curves for the gs-MELD score in validation cohort 2. At 90 days, 100% and 24% of patients were free of events vs 100% and 12% of patients at 180 days in the good (green line) and poor-prognosis (orange line) groups, respectively (P < .001). MELD: Model for End-Stage Liver Disease, gs: gene-signature.
Discussion
This study shows for the first time that a gene expression signature is associated with survival in biopsied patients with severe AH treated with corticosteroids or not due to uncontrolled infection and evaluated for liver transplantation. It also highlights the high predictive value of the gs-MELD, a new score combining a gene signature and the MELD score calculated at baseline in a real-world population of severe AH patients. In addition, we also demonstrate that this score can be implemented in an assay platform.
Corticosteroids are the only standard-of-care treatment that have reproducibly been shown to provide short-term survival benefits in patients with severe AH.36 However, a common side effect of this therapy is an increased susceptibility to infection one of the most frequent causes of mortality in severe AH patients.16 Although the gs-MELD cannot be advocated as a predictor of response to corticosteroids per se as the population studied did not receive corticosteroids based on a predefined protocol like in randomized controlled trials, the score identified a subgroup of patients with very poor prognosis despite the use of corticosteroids. These patients could be selected for alternative management strategies including early evaluation for liver transplantation,18 or inclusion in clinical trials for new pharmaceutical drugs. Importantly, in the second validation cohort, the gs-MELD also predicted survival in patients at very high risk of mortality referred for liver transplantation evaluation in the context of severe AH of whom some were contradicted to receive corticosteroid therapy. This indicates that the gs-MELD score may be considered as a more general prognostic predictor of outcome in subjects with severe AH, and is prognostic in a wide variety of clinical settings, on different continents and at different stages of severe AH severity although future studies will need to further characterize these findings. The score could also improve the design of future clinical trials by stratifying patients according to their prognosis and thus help selecting those suitable for new therapeutic strategies and/or treatments.
A potential limitation of this study was its relatively small sample size, compared to others aiming to identify gene signatures associated with clinical outcomes in liver disease patients.23, 37 However, research in the field of severe AH faces several challenges. First, although severe AH is one of the most life threatening liver diseases, it remains a rare complication in heavy drinkers.1 Second, it is also a heterogeneous disease, and both recruiting and retaining patients with alcohol use disorders is challenging.19 Third, many centers rely on clinical criteria and do not perform transjugular liver biopsy as a routine practice for patients with decompensated cirrhosis and suspicion of severe AH.7, 8 Overall, we believe that the patient population included in this study represents a unique cohort in light of the aforementioned issues and to the best of our knowledge, it is the largest population to date in the field of gene based biomarker prognosis prediction in severe AH. The performance of the Lille score in validation cohort 1 was inferior to the one reported in some randomized controlled trials.38, 39 However, it remains unclear whether this clinical model has similar prognostic performance in real-world cohorts especially in non-French centers”.
Interestingly, we observed that poor-prognosis pattern of the signature gene expression was partially observed in a subset of non-severe AH (23%) and alcoholic cirrhosis (18%) patients; 72 signature genes were already dysregulated at the stage of non-severe AH and alcoholic cirrhosis, and 51 genes were additionally dysregulated uniquely in severe AH (Supplementary Figure 3C). This observation suggests that dysregulation of the 72 genes underlie non-severe AH- or alcoholic cirrhosis-affected livers, and additional dysregulation of 51 genes confer manifestation of lethal severe AH. Of note, each subset of the gene signature showed inferior prognostic capability compared to the full 123-gene signature, indicating that both components of the signature are necessary for prognostic prediction. Histological confirmation of AH may not be mandatory in many cases with typical presentation.8, 19 Nevertheless, European Association for the Study of the Liver guidelines recommend the use of histologic analysis, notably in clinical trials, as relying only on clinical criteria is associated with a significant risk of misclassification.7 Despite considerations regarding diagnosis, we have shown the added value of liver biopsy in the understanding of underlying molecular signaling pathways. We have also confirmed the feasibility of needle biopsy profiling in FFPE tissue which generate gene expression data comparable to their fresh frozen counterpart,40 and the ability of the gs-MELD to remain predictive when implemented in an independent assay platform is a strong sign of its robustness. Of note, this score can be calculated in two days using this FDA-approved platform, which can accommodate up to 800 genes in a single reaction,41 including one day for RNA extraction and another day for gene signature assay and score calculation. Nevertheless, we acknowledge that the gs-MELD may not be available for a widespread use in clinical practice since, as mentioned above, liver biopsy may only be required in situations of uncertainty,8, 19 some centers do not have the capacities to perform systematic transjugular liver biopsy in this setting,7, 8 and the gene signature assay is not routinely available. Therefore, we are actively pursuing the development of candidate blood surrogates that have already been shown to be effective in cancer research.42
In conclusion, the gs-MELD, a combined score including genome-based and biological information, predicts survival in a real-world population of severe AH patients managed according to current standards of care including corticosteroid therapy when not contraindicated. Although these findings should be further validated in independent prospective cohorts, the gs-MELD could serve as a reference for the development of serum surrogate biomarkers that certainly represent the future of prognosis prediction in severe AH.
Supplementary Material
Acknowledgments
We are grateful to Dr. Olivier Le Moine, and Dr. Arnaud Lemmers for the identification of patients, obtaining consent and collection of samples at C.U.B. Hôpital Erasme, Université Libre de Bruxelles, Brussels, Belgium. We also thank Eric Quertinmont at the Laboratory of Experimental Gastroenterology, Université Libre de Bruxelles, Brussels, Belgium for his help with RNA extraction.
Grant support: The study was supported by the Fonds Erasme (Edouard and Suzanne Jacobs Convention), the Belgian Association for the Study of the Liver, and a Research Project (T100914F) from the Fund for Scientific Research-FNRS (F.R.S.-FNRS). ET is a Research Associate of the F.R.S.-FNRS and DF is a Research Director of the F.R.S.-FNRS. NG received funding from the FLAGS and Nuovo-Soldati foundations. YH is supported by NIH/NIDDK DK099558, European Union ERC-2014-AdG-671231HEPCIR, Irma T. Hirschl Trust, and US Department of Defense (W81XWH-16-1-0363). AC and GB are supported by the BridgeIRIS project funded by INNOVIRIS, Brussels-Capital Region, Brussels, Belgium. The funders had no role in the study design, data collection, data analysis, data interpretation, or writing of the report.
Writing Assistance: We acknowledge the contribution of a medical writer, Sandy Field, PhD, for her assistance concerning English-language editing. Dr. Field was compensated by the Université Libre de Bruxelles for her work.
Abbreviations
- AH
alcoholic hepatitis
- MELD
model for end-stage liver disease
- FDA
Food and Drug Administration
- FFPE
Formalin-fixed, paraffin-embedded
- HRS
hepatorenal syndrome
- HR
hazard ratio
- CI
confidence interval
- AUC
area under the receiver operating characteristic curve
- AIC
Akaike’s Information Criterion
- BIC
Bayesian information criterion
- ALD
alcoholic liver disease
- gs
gene signature
- gs-MELD
gene signature-MELD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure: The authors declare no competing interest in the present study.
Transcript Profiling: Transcriptome data are available at NCBI Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo, accession number GSE94417) and EBI ArrayExpress (www.ebi.ac.uk/arrayexpress, E-MTAB-2664)
Author Contributions: ET recruited patients, collected data, drafted and revised the manuscript. NG performed statistical analyses, drafted and revised the manuscript. NF performed statistical analyses and revised the manuscript. WMS performed the regulatory gene modules analysis and bioinformatics support, AC gave conceptual advice and bioinformatics support. AM and LS recruited subjects. PDem, CS and JS reviewed histological slides. TG recruited patients and collected data. SNT reviewed histological slides. JD and GYI recruited patients and collected data. CMi provided valuable technical support with transcriptome experiments. TS recruited patients and collected data. GB gave conceptual advice and bioinformatics support. KA, JH, DD, VL recruited patients. LRB reviewed histological slides. VDN provided valuable technical support for NanoString experiments. CMo and PDel recruited patients, collected data, and revised the manuscript. YH performed statistical analyses, conceptualized and supervised the study, drafted and revised the manuscript. DF obtained the funding, conceived and supervised the study, drafted and revised the manuscript. All authors were involved in the critical revision of the manuscript for important intellectual content.
References
- 1.Lucey MR, Mathurin P, Morgan TR. Alcoholic hepatitis. N Engl J Med. 2009;360:2758–2769. doi: 10.1056/NEJMra0805786. [DOI] [PubMed] [Google Scholar]
- 2.Maddrey WC, Boitnott JK, Bedine MS, et al. Corticosteroid therapy of alcoholic hepatitis. Gastroenterology. 1978;75:193–199. [PubMed] [Google Scholar]
- 3.Altamirano J, Miquel R, Katoonizadeh A, et al. A histologic scoring system for prognosis of patients with alcoholic hepatitis. Gastroenterology. 2014;146:1231–1239. e1–6. doi: 10.1053/j.gastro.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thursz M, Morgan TR. Treatment of Severe Alcoholic Hepatitis. Gastroenterology. 2016;150:1823–1834. doi: 10.1053/j.gastro.2016.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thursz MR, Richardson P, Allison M, et al. Prednisolone or Pentoxifylline for Alcoholic Hepatitis. N Engl J Med. 2015;372:1619–1628. doi: 10.1056/NEJMoa1412278. [DOI] [PubMed] [Google Scholar]
- 6.Gustot T, Maillart E, Bocci M, et al. Invasive aspergillosis in patients with severe alcoholic hepatitis. J Hepatol. 2014;60:267–274. doi: 10.1016/j.jhep.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 7.EASL clinical practical guidelines: management of alcoholic liver disease. Journal of hepatology. 2012;57:399–420. doi: 10.1016/j.jhep.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 8.O'Shea RS, Dasarathy S, McCullough AJ. Alcoholic liver disease. Hepatology. 2010;51:307–328. doi: 10.1002/hep.23258. [DOI] [PubMed] [Google Scholar]
- 9.Dominguez M, Rincon D, Abraldes JG, et al. A new scoring system for prognostic stratification of patients with alcoholic hepatitis. Am J Gastroenterol. 2008;103:2747–2756. doi: 10.1111/j.1572-0241.2008.02104.x. [DOI] [PubMed] [Google Scholar]
- 10.Forrest EH, Evans CD, Stewart S, et al. Analysis of factors predictive of mortality in alcoholic hepatitis and derivation and validation of the Glasgow alcoholic hepatitis score. Gut. 2005;54:1174–1179. doi: 10.1136/gut.2004.050781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn W, Jamil LH, Brown LS, et al. MELD accurately predicts mortality in patients with alcoholic hepatitis. Hepatology. 2005;41:353–358. doi: 10.1002/hep.20503. [DOI] [PubMed] [Google Scholar]
- 12.Louvet A, Naveau S, Abdelnour M, et al. The Lille model: a new tool for therapeutic strategy in patients with severe alcoholic hepatitis treated with steroids. Hepatology. 2007;45:1348–1354. doi: 10.1002/hep.21607. [DOI] [PubMed] [Google Scholar]
- 13.Louvet A, Labreuche J, Artru F, et al. Combining Data From Liver Disease Scoring Systems Better Predicts Outcomes of Patients With Alcoholic Hepatitis. Gastroenterology. 2015;149:398–406. e8. doi: 10.1053/j.gastro.2015.04.044. quiz e16-7. [DOI] [PubMed] [Google Scholar]
- 14.Dohan A, Guerrache Y, Boudiaf M, et al. Transjugular liver biopsy: indications, technique and results. Diagn Interv Imaging. 2014;95:11–15. doi: 10.1016/j.diii.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Lanthier N, Rubbia-Brandt L, Lin-Marq N, et al. Hepatic cell proliferation plays a pivotal role in the prognosis of alcoholic hepatitis. J Hepatol. 2015;63:609–621. doi: 10.1016/j.jhep.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Louvet A, Wartel F, Castel H, et al. Infection in patients with severe alcoholic hepatitis treated with steroids: early response to therapy is the key factor. Gastroenterology. 2009;137:541–548. doi: 10.1053/j.gastro.2009.04.062. [DOI] [PubMed] [Google Scholar]
- 17.Im GY, Kim-Schluger L, Shenoy A, et al. Early Liver Transplantation for Severe Alcoholic Hepatitis in the United States--A Single-Center Experience. Am J Transplant. 2016;16:841–849. doi: 10.1111/ajt.13586. [DOI] [PubMed] [Google Scholar]
- 18.Mathurin P, Moreno C, Samuel D, et al. Early liver transplantation for severe alcoholic hepatitis. The New England journal of medicine. 2011;365:1790–800. doi: 10.1056/NEJMoa1105703. [DOI] [PubMed] [Google Scholar]
- 19.Crabb DW, Bataller R, Chalasani NP, et al. Standard Definitions and Common Data Elements for Clinical Trials in Patients With Alcoholic Hepatitis: Recommendation From the NIAAA Alcoholic Hepatitis Consortia. Gastroenterology. 2016;150:785–790. doi: 10.1053/j.gastro.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louvet A, Labreuche J, Artru F, et al. Main drivers of outcome differ between short and long-term in severe alcoholic hepatitis: A prospective study. Hepatology. 2017 doi: 10.1002/hep.29240. (in press) [DOI] [PubMed] [Google Scholar]
- 21.Salerno F, Gerbes A, Gines P, et al. Diagnosis, prevention and treatment of hepatorenal syndrome in cirrhosis. Gut. 2007;56:1310–1318. doi: 10.1136/gut.2006.107789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 23.Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King LY, Canasto-Chibuque C, Johnson KB, et al. A genomic and clinical prognostic index for hepatitis C-related early-stage cirrhosis that predicts clinical deterioration. Gut. 2015;64:1296–1302. doi: 10.1136/gutjnl-2014-307862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoshida Y. Nearest template prediction: a single-sample-based flexible class prediction with confidence assessment. PLoS One. 2010;5:e15543. doi: 10.1371/journal.pone.0015543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song WM, Zhang B. Multiscale Embedded Gene Co-expression Network Analysis. PLoS Comput Biol. 2015;11:e1004574. doi: 10.1371/journal.pcbi.1004574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa S, Wei L, Song WM, et al. Molecular Liver Cancer Prevention in Cirrhosis by Organ Transcriptome Analysis and Lysophosphatidic Acid Pathway Inhibition. Cancer Cell. 2016;30:879–890. doi: 10.1016/j.ccell.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Affo S, Dominguez M, Lozano JJ, et al. Transcriptome analysis identifies TNF superfamily receptors as potential therapeutic targets in alcoholic hepatitis. Gut. 2013;62:452–460. doi: 10.1136/gutjnl-2011-301146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahrens M, Ammerpohl O, von Schonfels W, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18:296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 31.Blanche P, Dartigues JF, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med. 2013;32:5381–5397. doi: 10.1002/sim.5958. [DOI] [PubMed] [Google Scholar]
- 32.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 33.Akaike H. Fitting autoregressive models for prediction. Ann Inst Stat Math. 1969;21:243–247. [Google Scholar]
- 34.Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6:461–464. [Google Scholar]
- 35.Zhang DY, Goossens N, Guo J, et al. A hepatic stellate cell gene expression signature associated with outcomes in hepatitis C cirrhosis and hepatocellular carcinoma after curative resection. Gut. 2016;65:1754–1764. doi: 10.1136/gutjnl-2015-309655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh S, Murad MH, Chandar AK, et al. Comparative Effectiveness of Pharmacological Interventions for Severe Alcoholic Hepatitis: A Systematic Review and Network Meta-analysis. Gastroenterology. 2015;149:958–70. e12. doi: 10.1053/j.gastro.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Hoshida Y, Villanueva A, Sangiovanni A, et al. Prognostic gene expression signature for patients with hepatitis C-related early-stage cirrhosis. Gastroenterology. 2013;144:1024–1030. doi: 10.1053/j.gastro.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathurin P, O'Grady J, Carithers RL, et al. Corticosteroids improve short-term survival in patients with severe alcoholic hepatitis: meta-analysis of individual patient data. Gut. 2011;60:255–260. doi: 10.1136/gut.2010.224097. [DOI] [PubMed] [Google Scholar]
- 39.Mathurin P, Louvet A, Duhamel A, et al. Prednisolone with vs without pentoxifylline and survival of patients with severe alcoholic hepatitis: a randomized clinical trial. JAMA. 2013;310:1033–1041. doi: 10.1001/jama.2013.276300. [DOI] [PubMed] [Google Scholar]
- 40.Mittempergher L, de Ronde JJ, Nieuwland M, et al. Gene expression profiles from formalin fixed paraffin embedded breast cancer tissue are largely comparable to fresh frozen matched tissue. PLoS One. 2011;6:e17163. doi: 10.1371/journal.pone.0017163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kulkarni MM. Digital multiplexed gene expression analysis using the NanoString nCounter system. Curr Protoc Mol Biol. 2011;Chapter 25(Unit25B):10. doi: 10.1002/0471142727.mb25b10s94. [DOI] [PubMed] [Google Scholar]
- 42.Vathipadiekal V, Wang V, Wei W, et al. Creation of a Human Secretome: A Novel Composite Library of Human Secreted Proteins: Validation Using Ovarian Cancer Gene Expression Data and a Virtual Secretome Array. Clin Cancer Res. 2015;21:4960–4969. doi: 10.1158/1078-0432.CCR-14-3173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.