Abstract
Consumption of fermentable dietary fibers (DFs), which can induce growth and/or activity of specific beneficial populations, is suggested a promising strategy to modulate the gut microbiota and restore health in microbiota-linked diseases. Until today, inulin and fructo-oligosaccharides (FOS) are the best studied DFs, while little is known about the gut microbiota-modulating effects of β-glucan, α-galactooligosaccharide (α-GOS) and xylo-oligosaccharide (XOS). Here, we used three continuous in vitro fermentation PolyFermS model to study the modulating effect of these DFs on two distinct human adult proximal colon microbiota, independently from the host. Supplementation of DFs, equivalent to a 9 g daily intake, induced a consistent metabolic response depending on the donor microbiota. Irrespective to the DF supplemented, the Bacteroidaceae-Ruminococcaceae dominated microbiota produced more butyrate (up to 96%), while the Prevotellaceae-Ruminococcaceae dominated microbiota produced more propionate (up to 40%). Changes in abundance of specific bacterial taxa upon DF supplementation explained the observed changes in short-chain fatty acid profiles. Our data suggest that the metabolic profile of SCFA profile may be the most suitable and robust read-out to characterize microbiota-modulating effects of a DF and highlights importance to understand the inter-individual response to a prebiotic treatment for mechanistic understanding and human application.
Introduction
The human gut microbiota is composed of around 1014 bacterial cells that belong to more than 1000 species1, dominated by members belonging to the two phyla, Firmicutes and Bacteroidetes1,2. Diet is known to strongly influence the composition of the gut microbiota as well as metabolites, dominated by the canonical short chain fatty acids (SCFAs) acetate, propionate and butyrate3,4. Among healthy individuals, compositional and functional properties of the microbiome vary substantially, leading to highly variable responses to dietary interventions5–9. Baseline bacterial composition of the host microbiome has repeatedly been observed to be a key factor to explain responses of the gut microbiota to different dietary interventions10–12. Within the collective genome of several millions genes, the microbiome harbors the capacity of primary degradation of substrates by specialized bacteria, cross-feeding and competition, making the stratification of microbiota response profiles a major challenge in the field1,10.
Around 40 g of complex carbohydrates reach the colon each day after escaping breakdown by host enzymes9,13,14. Endogenous enzymes are unable to degrade numerous complex carbohydrates and plant polysaccharides4. Non-digestible dietary fibers have been shown to have a beneficial effect on intestinal wellbeing acting as bulking agent and substrates for growth and activity of specific endogenous bacterial populations within the gastrointestinal tract (GIT)15. The breakdown of the complex carbohydrates by the gut microbiota is a key factor for the stability and diversity of the intestinal ecosystem yielding energy not only for the host, but also for its microbiota. The presence of end metabolites such as the SCFAs acetate, propionate and butyrate, and absence of intermediate metabolite accumulation, such as for lactate, formate and succinate, are generally recognized as markers for a healthy microbiome16. Metabolism of fibers is occurring in the colon, especially in the proximal colon resulting in an increased production of organic acids and a decrease in luminal pH of 5.5–5.917. Acidification but more importantly the production of intermediate and end-metabolites have important consequences for the microbial composition, the establishment of key bacterial interactions and the proper functioning of host physiology18,19. Microbial SCFAs impact on gut health, as energy source for the intestinal epithelium and epigenetic factor influencing immune response, epithelial integrity, electrolytes re-absorption and gut motility. Butyrate is an energy substrate used by colonocytes, while acetate and propionate reach systemic circulation and affect metabolism and function of peripheral organs (e.g. liver, pancreas, brain, muscle)4,20,21. Therefore fiber modulation of microbiota composition and functions has crystallized as a promising strategy to promote gut and host health.
Dietary fibers (DFs) as main substrate for the gut microbiota are key factors of the microbial network in the gut. In particular inulin and fructo-oligosaccharides (FOS) have repeatedly been shown to selectively modulate the gut microbiota in vitro and in vivo with benefits for host health (“prebiotic effect”)22–27. However, the lack of specific responses of bacterial groups has posed an important challenge in understanding the prebiotic mechanisms of most fibers12,28. Multiple studies aiming at identification of novel prebiotics, such as β-glucans, galacto-oligosaccharides (GOS) and xylo-oligosaccharides (XOS), have encountered the same challenge of functional redundancy within phylogenetically diverse bacterial groups and limited understanding of the metabolite functions.
In vitro fermentation models are powerful approaches to investigate gut microbiota functionality without host effects in a highly controlled environment29. These models allow the strict control of physiologic parameters, such as retention time, pH, temperature and anaerobiosis, and medium composition used to mimic the diet. Colonic models from simple short-term batch fermentations to multistage long-term continuous flow models were developed30–32. Continuous models further control the medium flow rate, for culturing of microbiota in steady-state conditions, allowing a fiber to develop its full effect along the entire trophic chain, thereby increasing the physiological relevance of the experiment. In particular, continuous fermentation systems with immobilized gut microbiota were shown to simulate the high-cell density, biodiversity and long-term stability of the intestinal microbiota32. This prevents washout of less competitive bacteria and ensures the repeated exposure of a single microbiota to different fibers29,30,32. The PolyFermS model allows the parallel testing of different treatments on singular microbiota.
The aim of this study was therefore to investigate the effects of novel potential prebiotics on healthy adult gut microbiota using PolyFermS in vitro continuous colonic fermentation model mimicking the adult proximal colon microbiota. Immobilization of two distinct fecal microbiota obtained from two healthy adult donors was performed. The two microbiota were propagated and stabilized in an inoculum reactor (IR) seeded with microbiota immobilized in polymer gel beads allowing continuous and prolonged culture of the microbiome31,33. The model design allowed parallel testing of four different dietary fiber supplementations (β-glucan, XOS, α-GOS and inulin) compared to a control reactor with no supplemented fiber, all inoculated with the same microbiota produced in IR. The effect of dietary fibers derived from plants was tested at a physiological concentration mimicking a daily intake of 9 g fibers in test reactors for 5–7 days to reach pseudo-steady state conditions. Microbiota composition and diversity was monitored with 16S rRNA gene amplicon sequencing, and SCFAs analysis by HPLC.
Results
Donor selection and transfer to PolyFermS
Fecal microbiota of 30 healthy individuals were screened using 16S rRNA gene amplicon sequencing to select two fecal donors with distinct taxonomic profile for dietary fiber supplementation study. Fecal microbiota variation across the donors was stratified into enterotypes Ruminococcus, Bacteroides and Prevotella as decribed by Arumugam et al.34 (Fig. 1A). Two donors, D3 and D4, were selected based on the difference in their dominant microbial taxa. The microbiota composition of both donors was representative for a healthy human microbiota (Supplementary Table S2), with Firmicutes (D3: 51%; D4: 57%) and Bacteroidetes (D3: 26%; D4: 32%) as the dominant bacterial phyla. Donors differed from each other on family level with D3 fecal microbiota characterized by higher levels of Bacteroidaceae (D3: 17%; D4: 8%) and presence of Verrucomicrobiaceae (D3: 6%; D4: < 0%), and archaeal family Methanobacteriaceae (D3: 9%; D4: 2%). Whereas D4 fecal microbiota was characterized by higher levels of Prevotellaceae (D4: 17%; D3: 0.1%) and Lachnospiraceae (D4: 13%; D3: 7%) compared to D3 fecal microbiota (Supplementary Table S2B). For assessing the α-diversity, the Shannon index for both fecal microbiota was calculated and both donors had a comparable Shannon index (D3: 5.8 ± 0.9; D4: 5.8 ± 0.9; Supplementary Table S3).
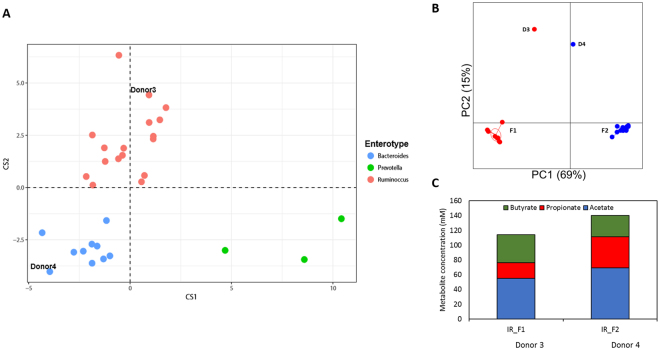
Figure 1.
(A) Variation in fecal microbiota among the 30 healthy individuals represented in an enterotype plot for 16S rRNA gene amplicon sequence dataset generated as described on http://enterotyping.embl.de/. (B) Principle Coordinate Analysis based on the weighted UniFrac distance matrix on OTU level generated from fecal and non-treated PolyFermS microbiota of fermentation 1 (F1 with donor 3 microbiota) and fermentation 2 (F2 with donor 4 microbiota), showing conservation of the two distinct donor microbiota profiles in PolyFermS model. Microbiota included from inoculum reactor (IR) and control reactor (CR) from F1 on day 12, day 19, day 24 and day 31 and for F2 on day 8, day 14, day 19, day 25, day 31 and day 37. (C) Bar-plot representation of mean acetate (blue), propionate (red) and butyrate (green) concentrations (mM) of PolyFermS effluent samples of IR during stable operation phase and connection with experimental reactors (IR_F1: day 1–day 36; IR_F2: day 1–day 42).
Fecal microbiota of both donors were studied in the PolyFermS continuous intestinal fermentation model operated with conditions mimicking the proximal colon microbiota. The model is composed of an inoculum reactor (IR), containing immobilized human adult fecal microbiota used to continuously inoculate several second stage control and experimental reactors mounted in parallel. The second stage reactors with the same complete microbiota allow comparing different treatments with a control. During 49 days of continuous operation of IRs inoculated with different donor microbiota, the microbial stability was assessed on phylogenetic and metabolic levels. The Shannon diversity, taking in account both abundance and evenness of species present in a sample, was reduced in the in vitro model compared with fecal microbiota (Supplementary Table S3). The mean Shannon index of IR and second stage untreated microbiota (CR) was 4.3 ± 0.1 for F1, containing D3 microbiota (Shannon index 5.8 ± 0.9), and 3.6 ± 0.2 for F2, containing D4 microbiota (Shannon index 5.8 ± 0.9). On family level the microbiota composition within each fermentation reflected well its specific donor microbiota (Supplementary Table S4 and Supplementary Table S5) as previously observed in similar PolyFermS setup32. Within F1 microbiota of IR and untreated control reactor, Firmicutes (56%) and Bacteroidetes (26%) were the predominant phyla. In correspondence with the fecal D3 microbiota, F1 microbiota was characterized by high levels of bacterial families Bacteroidaceae (IR: 23 ± 5%; CR: 29 ± 6%) and Ruminococcaceae (IR: 18 ± 3%; CR: 11 ± 7%). Whereas F2 microbiota was dominated by Bacteroidetes (58%) over Firmicutes (36%) and characterized by high levels of Prevotellaceae (IR: 53 ± 6%; CR: 53 ± 4%) and Ruminococcaceae (IR: 19 ± 4%; CR: 18 ± 2%), also in correspondence with its donor fecal microbiota. Some bacterial families were detected at different abundances in vitro compared to the fecal sample. For example, in F1 increased abundances of Acidaminococcaceae (12 ± 1%; 0.3% in fecal sample) and Enterobacteriaceae (15 ± 4%; undetected in fecal sample) and in F2 Prevotellaceae (36 ± 6%; 17.2% in fecal sample) and unclassified Lactobacillales (4 ± 1%; undetectable in fecal sample) were detected (Supplementary Table S2 and Supplementary Table S5). A clear spatial separation and clustering of F1 and F2 microbiota was observed by Principle Coordinate Analysis (PCoA) on weighted UniFrac distance matrix on OTU level, indicating a distinct and stable microbial profile of the two donor microbiota in vitro (Fig. 1B).
Bacterial fermentation activity was monitored by SCFA analysis of fermentation effluents of PolyFermS reactors along the complete fermentation period. After the initial colonization-stabilization period of 12 and 15 days, total SCFA concentrations in IR effluents were stable with 153 (±9) mM for 20 days in F1 and 123 (±10) mM for 36 days in F2, respectively. Acetate was the predominant SCFA produced in both fermentations (F1: 55 ± 6 mM; F2: 69 ± 4 mM). Butyrate and propionate levels differed between both fermentations with F1 characterized by higher butyrate levels (F1: 38 ± 4 mM and F2: 29 ± 6 mM) and F2 characterized by higher propionate levels (F1: 21 ± 3 mM and F2: 42 ± 5 mM) (Fig. 1C). The metabolic profiles of IR over the whole fermentation periods of 36 and 42 days for F1 and F2, respectively, showed stable concentrations of the main SCFAs (Supplementary Figure S2).
Dietary fiber supplementation induces different metabolic and microbial responses in vitro depending on donor microbiota
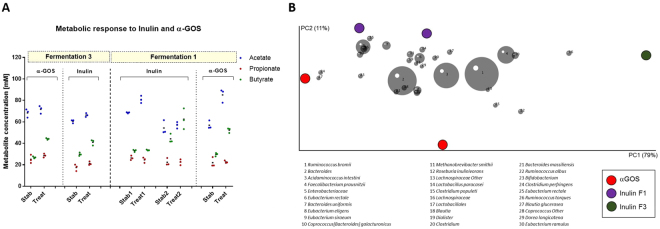
The effects of dietary fibers β-glucan, XOS, α-GOS and inulin on metabolic and microbial responses of stable PolyFermS microbiota of F1 and F2 were tested at a concentration of 4 g/L in the fermentation medium, mimicking an estimated daily intake of 9 g/day, for 7 days alternated by re-stabilization phases of 5 days, and two repetitions (noted I and II) were done for dietary fiber application within F1 and F2, except for α-GOS. Supplementation was performed through addition of sterile, non-heated fiber to the complex medium. Metabolic response of both microbiota was assessed by measuring SCFA production at the end of treatment (3 day sampling) and comparing to the production measured during the stabilization period before a fiber treatment is applied. Addition of dietary fibers yielded an overall increase in total SCFA production for both microbiota in F1 and F2 compared to stabilization, with mean increase ranging from 3 to 54 mM (F1) and 16 to 38 mM (F2), indicating fermentation of all supplemented fibers (Supplementary Figure S3). Acetate production was significantly enhanced for both microbiota upon supplementation with XOS and α-GOS, whereas during β-glucan treatment acetate levels remained stable. Butyrate and propionate productions were also increased by fiber supplementation but with microbiota-dependent response. Butyrate production increased (between 2 and 96%) in F1 (D3), while propionate production was enhanced (between 3 and 40%) in F2 (D4) microbiota upon β-glucan, XOS, α-GOS and inulin supplementation (Fig. 2A and B and Supplementary Table S6A). In addition, inulin also raised butyrate production in F2 (D4). Metabolic interactions within the PolyFermS microbiota upon fiber treatment were observed when inulin was supplemented. This is demonstrated by an increase in butyrate, when acetate levels remained stable or decreased (F1 treatment 2 and F2), but when acetate concentrations increased, butyrate levels remained stable (F1 treatment 1), suggesting cross-feeding of butyrate-producers on acetate.
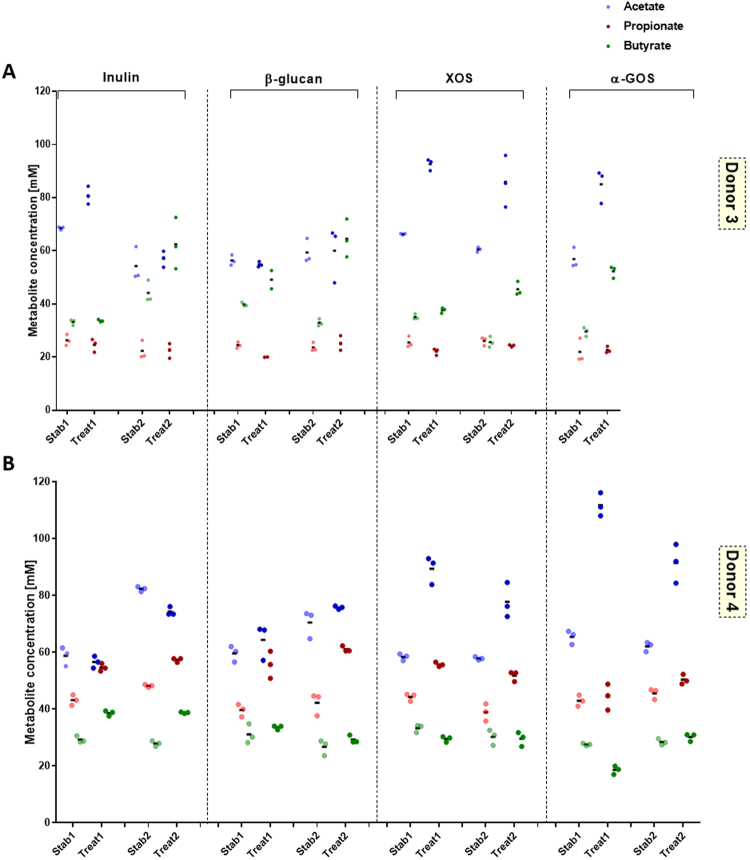
Figure 2.
Effect of dietary fiber supplementation on fermentation metabolite concentration (A and B) and microbiota of fermentation 1 (F1) (donor 3) and fermentation 2 (F2) (donor 4). Mean (black horizontal line) from 3 consecutive measurements of acetate, propionate and butyrate concentrations (mM) in the respective reactor at end of stabilization and treatment phase for F1 (D3 microbiota) (A) and F2 (D4 microbiota) (B). Two replicates are shown for each dietary fiber treatment.
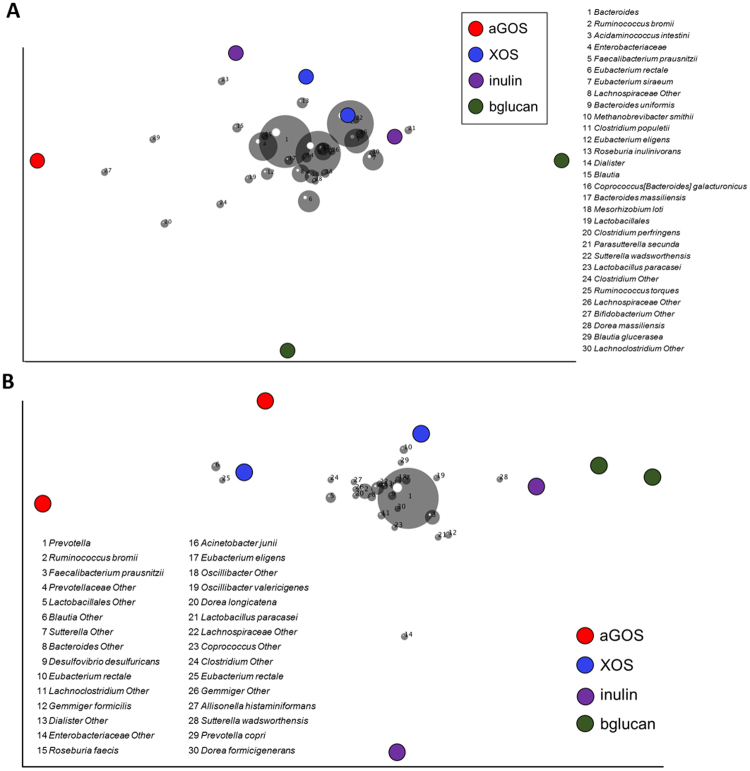
Interestingly we did not observe a consistent change of a single OTU upon fiber supplementation, confirming the current hypothesis of high functional redundancy within the gut microbiota and suggesting a multi-strain response causing observed changes in SCFA production. Principle coordinate analysis (PCoA) on weighted UniFrac distances of the fiber-supplemented PolyFermS microbiota allowed us to identify taxa that stratify the microbiota of F1 and F2 in relation to the dietary fiber supplementation and their abundances (Fig. 3A and B). Due to the different microbiota of F1 (Bacteroidaceae-Ruminococcaceae dominated) and F2 (Prevotellaceae-Ruminococcaceae dominated), the response on OTU level upon a dietary fiber was in some cases different. For example, α-GOS supplementation resulted in consistent higher levels compared to stabilization period of Lachnospiraceae in F2 microbiota during both treatment periods (I: 6 to 12% and II: 5 to 10%; Supplementary Table S7B), also reflected by a Blautia (6) and Eubacterium rectale (25) OTU in the PCoA. Whereas in F1 microbiota Lachnospiraceae levels decreased upon α-GOS supplementation (18 to 13%, Supplementary Table S7A). In PCoA, β-glucan supplemented F1 microbiota was determined by Eubacteriaceae OTUs (E. siraeum (7) and E. rectale (6)), in accordance with higher Eubacteriaceae levels compared to stabilization periods (I: 10 to 23% and II: 7 to 20%; Supplementary Table S7A). On the other hand, β-glucan supplemented F2 microbiota was separated in the PCoA by a Sutterella wadsworthensis OTU (28). An increased relative abundance of Prevotellaceae in both repetitions of β-glucan supplementation was also observed (I: 26 to 61% and II: 54 to 58%; Supplementary Table S7B) compared to the previous stabilization period. Interestingly, inulin showed consistent changes in relative abundance upon supplementation, independent of the donor microbiota. After inulin supplementation, the relative abundance of Ruminococcaceae increased in both fermentations (F1: 22 to 28% and F2: I: 13 to 19% and II: 16 to 22%; Supplementary Table S7B). No consistent changes in relative abundance of main bacterial families could be detected upon XOS supplementation in both microbiota (Supplementary Table 7). However, in the PCoA the XOS supplemented microbiota of F2 were both determined by an E. rectale OTU (10 and 25; Fig. 3B), which indicates a higher abundance of E. rectale in XOS supplemented F2 microbiota.
Figure 3.
Effect of dietary fiber supplementation on microbiota (A and B) of fermentation 1 (F1; donor 3) and fermentation 2 (F2; donor 4). Principle components analysis (PCA) biplot showing variation among the PolyFermS microbiota of F1 (B) and F2 (D) after supplementation with dietary fibers XOS, a GOS, b glucan and inulin. Variables included in the PCA were relative abundance of OTUs (>0,05%) and OTUs are represented as numbers.
Reproducible metabolic response to dietary fiber supplementation in PolyFermS model with same microbiota
Advances in research are based on the reproducibility of previously published data or investigated results and findings35. In this study, we performed a third fermentation with D3 microbiota 15 months after F1 with treatments inulin and α-GOS. In order to assess the microbial composition similarities or eventual differences after one year, the relative abundances (V4 region of 16S rRNA gene) of the different phyla and families were compared (Supplementary Table S8). At phylum level, fecal microbiota of donor D3 at both time points used to inoculate F1 and F3 was dominated by Firmicutes (F1 D3: 51% and F3 D3: 50%) and Bacteroidetes (F1 D3: 25% and F3 D3: 31%). The key bacterial families defining D3 microbiota remained stable with dominance of Bacteroidaceae (F1 D3: 17% and F3 D3: 22%) and Ruminococcaceae (F1 D3: 21% and F3 D3: 16%) and presence of Verrucomicrobiaceae (F1 D3: 6% and F3 D3: 4%) and Methanobacteriaceae (F1 D3: 9% and F3 D3: 6%).
In order to assess reproducibility and stability of the D3 microbiota in our PolyFermS model, the fermentation effluent microbiota of the non-treated reactors were compared to fecal donor and F1 and F2 microbiota. PCoA of UniFrac distances (Fig. 4A and B) showed F1 and F3 microbiota (D3) were more similar to each other on OTU composition (unweighted) and abundance (weighted) compared to F2 microbiota (D4), demonstrating the reproducible conservation of donor microbiota profile in vitro. The abundance of the dominant bacterial families was also comparable between F3 and F1 microbiota in inoculum reactors with Bacteroidaceae (F3: 29%; F1: 26%) and Ruminococcaceae (F3: 19%; F1: 19%) as predominant families (Supplementary Table S8B).Total SCFA production was also comparable between F3 and F1 with 129 ± 7 mM and 123 ± 10 mM, respectively (Fig. 4C). There was a slight difference in SCFA profile in the IR effluents, with lower butyrate levels in F3 (22%) compared to F1 (31%) but comparable acetate (F3: 48%; F1: 45%) and propionate (F3: 20%; F1: 17%) concentrations (Fig. 4C).
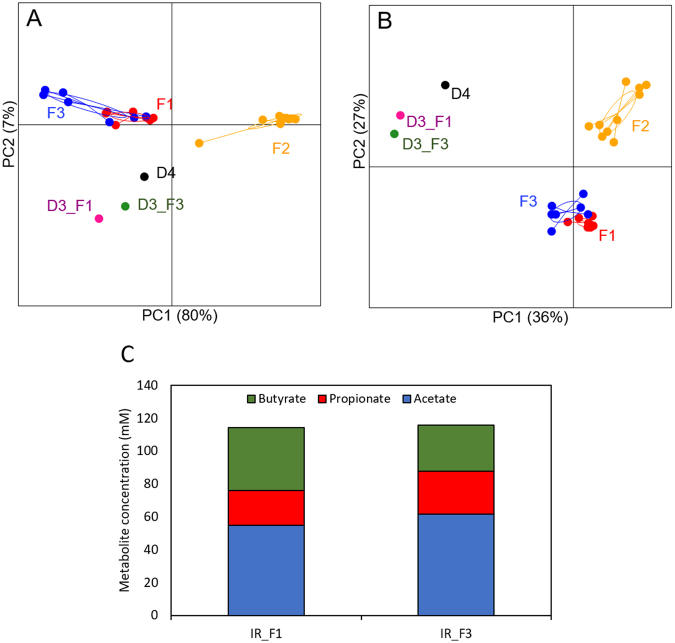
Figure 4.
Reproducability over time of PolyFermS model inoculated with same healthy fecal donor (D3); F3 was operated 15 months after F1. PCoA plots of weighted (A) and unweighted (B) were performed based on the UniFrac distance matrix generated from sequencing V4 region of 16S rRNA genes in samples from donor’s feces and fermentation effluents of F1 (D3), F2 (D4) and F3 (D3). Each circle represents a sample from feces and effluent samples from F1 (D3) (red), F2 (D4) (orange) and F3 (D3) (purple). Bar-plot representation of the mean acetate (blue), propionate (red) and butyrate (green) concentrations (mM) (C) of PolyFermS effluent samples of inoculum reactor (IR) during stable operation phase and connection with experimental reactors (IR_F1: day 1–day 36; IR_F3: day 1–day 23); D3, donor 3; F1, fermentation 1; F2, fermentation 2; D4, donor 4; F3, fermentation 3.
After reaching stable SCFA profile in reactors during stabilization, supplementation with inulin or α-GOS resulted in significant increases of total SCFAs (Fig. 5A; Supplementary Table S6B). Similar to F1, addition of α-GOS (from 28 ± 2 to 44 ± 1 mM, during stabilization and treatment, respectively) or inulin (from 30 ± 2 to 41 ± 3 mM) enhanced butyrate production, while inulin stimulated acetate production (from 61 ± 2 to 66 ± 2 mM). α-GOS supplementation resulted in increased levels of Ruminococcaceae (from 24 to 31% during stabilization and treatment, respectively) and Bacteroidaceae (from 22 to 26%), (Supplementary Table S9). Simultaneously a decrease in Eubacteriaceae (17 to 10%) was observed. The relative abundance of Ruminococcaceae was increased by inulin supplementation (from 41 to 48%), while abundance of the Eubacteriaceae decreased (from 15 to 9%).
Figure 5.
Repetition of α-GOS and inulin supplementation to D3 microbiota in PolyFermS resulted in comparable higher butyrate productions and stimulates specific OTUs (PCA). (A) Mean (black horizontal line) from 3 consecutive measurements of acetate, propionate and butyrate concentrations (mM) in the respective reactor at end of stabilization and treatment phase for fermentation 1(F1) and fermentation 3 (F3) (donor 3).Two replicates are shown for each dietary fiber treatment. (B) Principle components analysis (PCA) biplot showing variation in the PolyFermS microbiota of F1 and F3 after supplementation with dietary fibers a GOS and inulin. Variables included in the PCA were relative abundance of OTUs (>0,05%) and OTUs are represented as numbers.
Discussion
In this study the effect of supplementation of four different dietary fibers on human gut microbiota was investigated at the levels of metabolic and bacterial composition using a continuous in vitro fermentation system, modeling adult proximal colon conditions. By using two distinct fecal microbiota composition we could demonstrate a consistent and donor-dependent butyrogenic or propionigenic response towards the fiber treatments.
Both on metabolic and phylogenetic levels we successfully maintained two distinct colon microbial communities in the PolyFermS reactors reflecting the corresponding fecal microbiota donor. Both fecal microbiota were dominated by Firmicutes and Bacteroidetes species and differed mainly on family level within the Bacteroidetes phylum with higher levels of Bacteroidaceae species in D3, low in D4, and high levels of Prevotellaceae in D4, low in D3. Several other reports observed that individuals with high levels of Prevotella have low levels of Bacteroides and vice versa34,36,37, suggesting niche competition within the human gut microbiota. Abundance of Bacteroides has been associated with animal/fat-rich diets36,37, while high Prevotella abundance has been associated with plant-rich and vegetarian diets36,38. Difference in abundance of Bacteroidetes phylum between the two donor microbiota was preserved and even enhanced in our PolyFermS model for proximal colon microbiota, despite identical chime-simulating fermentation medium composition. The PolyFermS abundance levels fall within the range reported in a recently published large cohort of 1106 fecal microbiota of Western European individuals with Bacteroides: 0.1–72% and Prevotella: 0–56%39. Stable SCFA profiles were obtained in the untreated control reactors of F1 and F2, reflecting maintenance of functional stability of bacterial community during continuous operation of 36 and 43 days, respectively. The Prevotellaceae-dominated microbiota (F2) was characterized by high propionate production and can be explained by propionate –producing capacity of Prevotella species40. It was earlier reported that higher fecal propionate levels are associated with Prevotella species41,42. In another cohort it was shown that individuals with more than 20% Prevotella have higher levels of methylmalonyl-CoA mutase, a key enzyme involved in propionate production, in their fecal metaproteome43. The high butyrate production in F1 (Bacteroidaceae-dominated) microbiota is likely due butyrate-producing Lachnospiraceae and Ruminococcaceae species, which were the second and third dominating families within F1. Bacteroidaceae species do not produce butyrate within the human gut40,44, but may contribute to the butyrate pool by their acetate production, used as co-substrate during butyryl-CoA-transferase route in gut bacteria45.
Our data showed that all dietary fibers supplemented in the PolyFermS microbiota increased SCFA production, displaying fermentability of all tested substrates by both microbiota. Overall, F1 microbiota responded to all dietary fibers by increased butyrate production, whereas F2 microbiota showed increased propionate production. This specific response was consistent among the different treatments and the three fermentations. Similar inter-individual differences in butyrogenic or propionigenic response were observed in static batch experiments with fecal microbiota supplemented with wheat bran particles or inulin46, and with FOS and two arabinoxylan variants47. Comparable to our observations, Chen et al.47 showed that Prevotella-dominated microbiota responded by higher propionate production upon fiber supplementation in vitro. Both in F1 and F2, XOS and αGOS resulted in strong increase in acetate levels, which is produced by almost all heterotrophic gut bacteria48. Both dietary fibers were short-chain types, which makes them easier fermentable49 and it was shown in vitro that various intestinal bacteria can use GOS and XOS50. This broad-range utilization may explain the different effects measured on microbiota composition upon XOS supplementation. Metabolic cross-feeding between acetate- and butyrate- producers resulting in higher butyrate levels was observed with inulin in both PolyFermS microbiota, and repetitions. Inulin can be degraded by different Bifidobacterium spp., Lactobacillus spp.51 and some butyrate-producing Roseburia spp.52, F. prausnitzii and Eubacterium rectale53. Bifidobacteria and lactobacilli produce acetate and lactate, which can in turn be utilized by butyrate-producing bacteria. During these cross-feeding interactions on inulin-type fructans both commensalism (cross-feeding on acetate and lactate) and substrate competition occurs between both bacterial groups in co-culture experiments54. We observed a consistent increase in Ruminococcaceae, but no increase of Actinobacteriaceae (bifidobacteria), which suggests that in our set-up the inulin-degrading Ruminococcaceae (e. g. F. prausnitzii) produced butyrate while consuming the available acetate in the mixed microbial environment. Indeed, increased Ruminococcaceae levels upon inulin treatment was linked with an increase in a F. prausnitzii OTU, which is in accordance with in vitro55 and human observations56,57. Β-glucan supplementation resulted in increased butyrate and Eubacteriaceae levels in F1 microbiota, and increased propionate and Prevotellaceae levels in F2. Both changes in microbial composition explain the change in metabolic profile as Eubacteriaceae species produce butyrate and acetate, which becomes available for cross-feeding interactions, while Prevotellaceae produce propionate40. The increase in Bacteroides-Prevotella group and propionate production was also observed in vitro with oat β-glucan58. Prevotella can better ferment complex polysaccharides from the diet than Bacteroides59, which may explain their competitive advantage upon β-glucan supplementation.
Overall, we did not detect consistent or systematic changes in the microbiota composition upon dietary fiber supplementation. It appears that the microbiota modulation by dietary fibers occurs at species level as demonstrated by Chung et al.55 in a continuous in vitro fermentation with three different fecal microbiota and dietary fibers as sole carbohydrate source. Due to the higher inter-individual variation at species level within the human gut microbiota, it can be expected that a single fiber will not induce a strong specific modification of the microbiota at species level in different individuals. Indeed, human intervention studies with dietary fibers showed marked inter-individual microbiota changes, which were depended on the individuals’ dominant microbiota composition9.
Compared with in vivo and human studies, there are limitations of the in vitro approach, since the models do not replicate all the conditions that occur in the colon, resulting in enriching and diminishing of bacterial populations. However, our data correspond well with dietary fiber fermentation in vivo and allowed insight into the complex cross-feeding mechanisms. In particular, we managed to show that dietary fibers induce dynamic responses depending on an individual’s specific microbiota. Main strength of in vitro fermentation models for prebiotic research is that one can follow the in situ SCFA production upon treatment, whereas in human intervention studies fecal SCFA concentrations are only a proxy for colonic fermentation and mainly a result of absorption of the SCFA in the intestine and lack thereof. Our approach of repeated dietary fiber supplementation to a stable and active gut microbiota in a continuous fermentation model allowed elucidating direct and indirect metabolic and compositional shifts that can occur in the human gut using long-term supplementation. This is in contrast to often used static batch incubations with fecal microbiota and dietary fibers, which reflect a direct effect of a fiber on the growth and activity of fast substrate utilizers and often neglects the indirect effects or long term shifts that might have the most profound effect on microbiome composition.
To summarize, our study showed that two distinct fecal microbial consortia maintained in vitro in the PolyFermS continuous intestinal fermentation model inoculated with immobilized adult fecal microbiota responded differently to dietary fiber supplementation on metabolic and compositional level. Irrespective to the dietary fiber supplemented. The Bacteroidaceae-Ruminococcaceae dominated microbiota produced more butyrate while the Prevotellaceae-Ruminococcaceae dominated microbiota produced more propionate. No fiber-specific change on phylogenetic level was observed, but changes in abundance of specific families or species level OTUs within a microbiota together with cross-feeding interactions between the different functional groups could explain the observed changes in SCFA profiles. Our data suggest that the metabolic profile of SCFA may be the most suitable and robust read-out to characterize the microbiota-modulating effect of a fiber and emphasize on the importance to understand inter-individual responses to a prebiotic treatment for mechanistic understanding and human application.
Material and Methods
Fecal bacteria immobilization
Fecal samples were donated by two healthy individuals (male, age 33 and 32), who did not receive antibiotic or probiotic supplementation for at least 3 months before donation. Fecal samples were collected in a sterile 50 mL Falcon tube in an airtight container together with one Anaerogen sachet (Oxoid) to obtain anaerobic conditions until transfer into an anaerobic chamber (10% CO2, 5% H2 and 85% N2) within 3 h (Coy Laboratories, Ann Arbor, MI, USA). Fecal bacteria were immobilized in 1–2 mm gel beads consisting of gellan gum (2.5%, w/v), xanthan (0.25%, w/v) and sodium citrate (0.2%, w/v) under anaerobic conditions as previously described in detail30,32. Sixty mL of freshly produced fecal beads were transferred in the IR bioreactor containing 140 mL of nutritive medium. For bead colonization, three consecutive fed-batch fermentations were carried out by replacing 100 mL fresh nutritive medium every 8–12 h. Bacteria, growing close to the bead surface are continuously released into the growth medium due to active cell growth in the high-biomass-density peripheral layer29,60.
Nutritive medium
The nutritive medium was based on the composition described by Macfarlane et al.61 for simulation of the chyme in the adult human colon. It included (g L−1 of distilled water): pectin (citrus) (2), xylan (oat spelts) (2), arabinogalactan (larch) (2), guar gum (1), inulin (1), soluble potato starch (5), mucine (4), casein acid hydrolysate (3), peptone water (5), tryptone (5), yeast extract (4.5), L-cysteine HCl (0.8), bile salts (0.4), KH2PO4 (0.5), NaHCO3 (1.5), NaCl (4.5), KCl (4.5), MgSO4 anhydrated (0.61), CaCl2*2 H2O (0.1), MnCl2* 4 H2O (0.2), FeSO4* 7H20 (0.005), hemin (0.05) and Tween 80 (1 mL). Prior sterilization (20 min, 120 °C), the pH of the medium was adjusted to 5.7. One mL of a filter-sterilized (0.2 μm pore-size) vitamin solution62 was added to the sterilized and cooled down medium. All components of the fermentation medium were purchased from Sigma-Aldrich Chemie (Buchs, Switzerland), except for peptone water (Oxoid AG, Pratteln, Switzerland), inulin (Orafti®, RPN Food-technology AG, Sursee, Switzerland), bile salts (Oxoid AG), tryptone (Becton Dickinson AG, Allschwill, Switzerland), yeast extract (Merck, Darmstadt, Germany), KH2PO4 (VWR International AG), NaHCO3 (Fluka, Buchs, Switzerland), NaCl (VWR international AG, Dietikon, Switzerland), KCl (Fluka, Buchs, Switzerland) and KH2PO4 (VWR International AG).
Four different dietary fibers (inulin-type fructan, β-glucan, XOS, α-GOS) were investigated (Supplementary Table S1) and supplemented to sterile nutritive medium at a concentration of 4 g L−1, calculated for an estimated daily intake of 9 g L−1, accounting for the reactor volume of 0.2 L compared to 0.75 L for the proximal colon volume, and a chime medium supply of 0.6 L medium per day, giving a mean retention time of 8 h. Complete hydration of dietary fibers was allowed for 24 h under high speed stirring at 4 °C, as presented below.
Simulation of proximal colon microbiota and treatment with dietary fibers
Experimental set-up
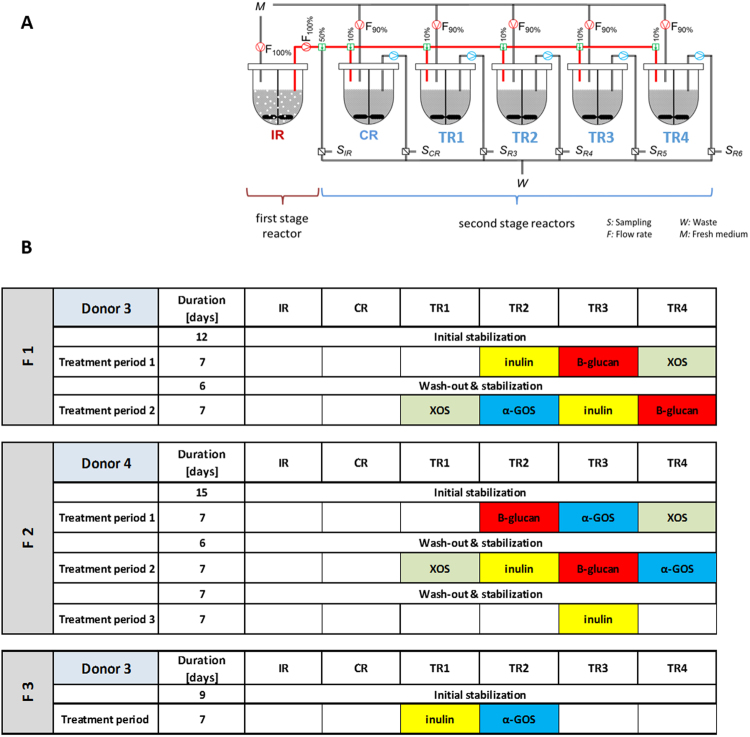
The set-up of the PolyFermS model was adapted for the aim of the study and is schematized in Fig. 6A. The continuous fermentations were carried out for up to 42 days in a two-stage design with a total of six bioreactors (Sixfors, Infors, Bottmingen, Switzerland). Each reactor was operated with conditions selected to mimic the adult proximal colon (pH 5.7, stirring at 120 rpm, 37 °C, and mean retention time of 8 h). Anaerobiosis was maintained through continuous headspace flushing with CO2, and a constant pH of 5.7 was maintained by addition of 2.5 M NaOH. IR had an operation volume of 200 mL and was inoculated with 60 mL of fecal beads and connected via a peristaltic pump (Reglo, Ismatec, Glattbrugg, Switzerland) to all second stage reactors, one control reactor (CR) and four treatment reactors (TR1-4) operated in parallel. All reactors were operated at a working volume of 200 mL. Fresh sterile nutritive medium was continuously supplied to IR at a flow rate of 25 mL/h, and second stage reactors were inoculated with 5% (v/v) (1.25 mL/h) IR effluent and supplied with 95% (v/v) (23.75 mL/h) fresh fermentation medium.
Figure 6.
Experimental set-up (A) and time schedule (B) of the continuous fermentation model with inoculum reactor (IR) and control (CR) and treatment (TR) reactors. CR and TR’s were fed with effluent from IR and with nutritive medium during stabilization and washout periods. TRs were fed with effluent from IR and supplemented fermentation medium (4 g/L dietary fiber) during treatment periods.
Experimental procedure
An initial colonization and stabilization phase of up to 15 days, prior experimentation was done (Fig. 6B). Each treatment was performed for 7 days for reaching a stable state, monitored by metabolite analysis and base consumption. The treatment periods were alternated with re-stabilization phases of 5–7 days, aiming to washout effects of the previous applied treatment and re-establishment of a microbiota composition similar to that in IR. The DFs (Supplementary Table S1) were added to the medium (4 g/L) and connected to assigned treatment reactor.
Effluent samples were taken daily and separated into bacterial pellet (10 min of centrifugation at 14.000 g at 4 °C) and supernatant, and stored at −20 °C until further analysis. Stability of the reactor microbial communities was monitored by daily measurements of main fermentation metabolites concentrations in sample supernatant. Standard observed variations in the PolyFermS fermentation metabolites are normally lower than 10% and used to define functional microbial stability, before starting and analyzing samples of a treatment period.
Microbial metabolite analysis
High performance liquid chromatography (HPLC) analysis (Thermo Fisher Scientific Inc. Accela, Wohlen, Switzerland) was performed to determine the concentrations of SCFAs (acetate, butyrate and propionate), branched-chain fatty acids (BCFAs) (isobutyrate, valerate and isovalerate) and intermediate metabolites (lactate and formate) produced by the microbiota in the reactor effluents. Analyses were performed with a Hitachi LaChrome device (Merck, Dietikon, Switzerland) using a Cation-H refill cartridge (30 × 4.6 mm) connected to an Aminex® HPX-87H (300 × 7.8 mm) column. Due to biofilm formation, supernatants of IR and TR reactors were diluted in ultrapure water and filtered through a 0.22 or 0.45 μm nylon membrane (Infochroma AG, Zug, Switzerland) into glass vials and sealed with crimp-caps. Around 40 μL of the sample were injected into the HPLC with a flow rate of 0.4 mL min−1 and H2SO4 as an eluent.
Microbial community analysis
Genomic DNA extraction
The genomic DNA was extracted from 200 mg feces and pellet of 2 mL PolyFermS effluent using the FastDNA® SPIN Kit for Soil (MP Biomedicals, Illkirch, France). Total DNA concentration (ng/μL) and purity was determined by spectrophotometry using Nanodrop (Nanodrop ND 1000 Spectrophotometer, Thermo Scientific, Wilmington, USA).
Microbiota profiling with 16S rRNA gene amplicon sequencing
The bacterial composition of the fecal and PolyFermS samples was determined using tag-encoded 16S rRNA gene Miseq-based (Illumina, CA, USA) high throughput sequencing. DNA samples of the last day of each experimental period of each PolyFermS reactor were selected for assessing the bacterial composition and its stability in the PolyFermS model and the shifts after prebiotic treatments. The V4 region of the 16S rRNA gene was amplified with modified primers 515 F (TATGGTAATTGTGTGNCAGCMGCCGCGGTAA) and 806 R (AGTCAGTCAGCCGGACTACHVGGGTWTCTAAT). Library preparation and sequencing was performed by StarSEQ (Mainz, Germany) using for sequencing one MiSeq cell and the V2 2 × 250 bp paired end Next Tera chemistry supplemented with 20% of PhiX.
The raw data set containing pair-ended reads with corresponding quality scores were merged using settings as previously mentioned63. The minimum length of merged reads was 200 bp. Quantitative Insight Into Microbial Ecology (QIIME) open source software package (1.7.0, 1.8.0 and 1.9.0) was used for subsequent analysis steps64. Purging the dataset from chimeric reads and constructing de novo Operational Taxonomic Units (OUT) was conducted using the UPARSE pipeline65. The custom human intestinal 16S rRNA database (HITdb) was used as a reference database66. Alpha and beta diversity analysis was performed as previously described using iterative subsampling (36,000 reads/sample)67.
Statistical analysis
All statistical analysis for HPLC were carried out using the SigmaPlot 13.0 version, San Jose, California, USA. HPLC Data are expressed as means ± Standard deviations (SD) of three days at the end of the stabilization and treatment period of each fermentation. HPLC data were compared between stabilization and treatment phase using the nonparametric Shapiro-Wilk test. P-values < 0.05 were considered significant. Brown-Forsythe test was used to determine differences between stabilization and treatment periods.
Electronic supplementary material
Acknowledgements
We thank Dr. Marco Meola for preparation of enterotype plot and assistance during 16S data analysis, Alfonso Die for technical assistance during SCFA analysis and people from Genetic Diversity Center at ETH Zurich for support during 16S amplicon sequencing preparation. This study was funded by DSM Nutritional Products Ltd. AGe and TW were supported by a research grant of Swiss National Science Foundation (project number: 35150, Bern, Switzerland).
Author Contributions
S.A.P., T.d.W., R.E.S. and C.L. conceived the experiment, S.A.P., L.B. and T.d.W. conducted the experiments. A.Gr.analysed 16S rRNA sequence data and L.K. prepared Figures 1B, 3A, 3B, 4A, 4B and 5B. T.d.W., A.G., S.A.P. and C.L. interpreted results. S.A.P. and A.G. wrote the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-22438-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. PNAS. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 5.Walker AW, Lawley TD. Therapeutic modulation of intestinal dysbiosis. Pharmacological research: the official journal of the Italian Pharmacological Society. 2013;69:75–86. doi: 10.1016/j.phrs.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lampe JW, Navarro SL, Hullar MA, Shojaie A. Inter-individual differences in response to dietary intervention: integrating omics platforms towards personalised dietary recommendations. The Proceedings of the Nutrition Society. 2013;72:207–218. doi: 10.1017/S0029665113000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Louis P, Scott KP, Duncan SH, Flint HJ. Understanding the effects of diet on bacterial metabolism in the large intestine. Journal of applied microbiology. 2007;102:1197–1208. doi: 10.1111/j.1365-2672.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- 9.Walker AW, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. The ISME journal. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korpela K, et al. Gut microbiota signatures predict host and microbiota responses to dietary interventions in obese individuals. PloS one. 2014;9:e90702. doi: 10.1371/journal.pone.0090702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harmon JP, Moran NA, Ives AR. Species response to environmental change: impacts of food web interactions and evolution. Science. 2009;323:1347–1350. doi: 10.1126/science.1167396. [DOI] [PubMed] [Google Scholar]
- 12.Bindels LB, Delzenne NM, Cani PD, Walter J. Towards a more comprehensive concept for prebiotics. Nature reviews. Gastroenterology & hepatology. 2015;12:303–310. doi: 10.1038/nrgastro.2015.47. [DOI] [PubMed] [Google Scholar]
- 13.Cummings JH, Bingham SA, Heaton KW, Eastwood MA. Fecal weight, colon cancer risk, and dietary intake of nonstarch polysaccharides (dietary fiber) Gastroenterology. 1992;103:1783–1789. doi: 10.1016/0016-5085(92)91435-7. [DOI] [PubMed] [Google Scholar]
- 14.Duncan SH, et al. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. International journal of systematic and evolutionary microbiology. 2006;56:2437–2441. doi: 10.1099/ijs.0.64098-0. [DOI] [PubMed] [Google Scholar]
- 15.Laparra JM, Sanz Y. Interactions of gut microbiota with functional food components and nutraceuticals. Pharmacological research: the official journal of the Italian Pharmacological Society. 2010;61:219–225. doi: 10.1016/j.phrs.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nature reviews. Gastroenterology & hepatology. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 17.Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker AW, Duncan SH, McWilliam Leitch EC, Child MW, Flint HJ. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Applied and environmental microbiology. 2005;71:3692–3700. doi: 10.1128/AEM.71.7.3692-3700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duncan SH, Louis P, Thomson JM, Flint HJ. The role of pH in determining the species composition of the human colonic microbiota. Environmental microbiology. 2009;11:2112–2122. doi: 10.1111/j.1462-2920.2009.01931.x. [DOI] [PubMed] [Google Scholar]
- 20.Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiological reviews. 1990;70:567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- 21.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut microbes. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberfroid MB. Inulin-type fructans: functional food ingredients. The Journal of nutrition. 2007;137:2493S–2502S. doi: 10.1093/jn/137.11.2493S. [DOI] [PubMed] [Google Scholar]
- 24.Juskiewicz J, Zdunczyk Z, Frejnagel S. Caecal parameters of rats fed diets supplemented with inulin in exchange for sucrose. Archives of animal nutrition. 2007;61:201–210. doi: 10.1080/17450390701297735. [DOI] [PubMed] [Google Scholar]
- 25.Grootaert C, et al. Comparison of prebiotic effects of arabinoxylan oligosaccharides and inulin in a simulator of the human intestinal microbial ecosystem. FEMS microbiology ecology. 2009;69:231–242. doi: 10.1111/j.1574-6941.2009.00712.x. [DOI] [PubMed] [Google Scholar]
- 26.Van den Abbeele P, et al. Arabinoxylans and inulin differentially modulate the mucosal and luminal gut microbiota and mucin-degradation in humanized rats. Environmental microbiology. 2011;13:2667–2680. doi: 10.1111/j.1462-2920.2011.02533.x. [DOI] [PubMed] [Google Scholar]
- 27.Rossi M, et al. Fermentation of fructooligosaccharides and inulin by bifidobacteria: a comparative study of pure and fecal cultures. Applied and environmental microbiology. 2005;71:6150–6158. doi: 10.1128/AEM.71.10.6150-6158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson GR, et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nature reviews. Gastroenterology & hepatology. 2017;14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 29.Payne AN, Zihler A, Chassard C, Lacroix C. Advances and perspectives in in vitro human gut fermentation modeling. Trends in biotechnology. 2012;30:17–25. doi: 10.1016/j.tibtech.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 30.Tanner SA, et al. In vitro continuous fermentation model (PolyFermS) of the swine proximal colon for simultaneous testing on the same gut microbiota. PloS one. 2014;9:e94123. doi: 10.1371/journal.pone.0094123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dostal A, Fehlbaum S, Chassard C, Zimmermann MB, Lacroix C. Low iron availability in continuous in vitro colonic fermentations induces strong dysbiosis of the child gut microbial consortium and a decrease in main metabolites. FEMS microbiology ecology. 2013;83:161–175. doi: 10.1111/j.1574-6941.2012.01461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zihler Berner A, et al. Novel Polyfermentor intestinal model (PolyFermS) for controlled ecological studies: validation and effect of pH. PloS one. 2013;8:e77772. doi: 10.1371/journal.pone.0077772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fehlbaum S, et al. Design and Investigation of PolyFermS In Vitro Continuous Fermentation Models Inoculated with Immobilized Fecal Microbiota Mimicking the Elderly Colon. PloS one. 2015;10:e0142793. doi: 10.1371/journal.pone.0142793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freedman LP, Cockburn IM, Simcoe TS. The Economics of Reproducibility in Preclinical Research. PLoS biology. 2015;13:e1002165. doi: 10.1371/journal.pbio.1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koren O, et al. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS computational biology. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez I, et al. The gut microbiota of rural papua new guineans: composition, diversity patterns, and ecological processes. Cell reports. 2015;11:527–538. doi: 10.1016/j.celrep.2015.03.049. [DOI] [PubMed] [Google Scholar]
- 39.Falony G, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 40.Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environmental microbiology. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 41.Salonen A, de Vos WM. Impact of diet on human intestinal microbiota and health. Annual review of food science and technology. 2014;5:239–262. doi: 10.1146/annurev-food-030212-182554. [DOI] [PubMed] [Google Scholar]
- 42.De Filippis F, et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. 2016;65:1812–1821. doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 43.Kolmeder CA, et al. Faecal Metaproteomic Analysis Reveals a Personalized and Stable Functional Microbiome and Limited Effects of a Probiotic Intervention in Adults. PloS one. 2016;11:e0153294. doi: 10.1371/journal.pone.0153294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanca A, et al. Potential and active functions in the gut microbiota of a healthy human cohort. Microbiome. 2017;5:79. doi: 10.1186/s40168-017-0293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Louis P, et al. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. Journal of bacteriology. 2004;186:2099–2106. doi: 10.1128/JB.186.7.2099-2106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Paepe K, Kerckhof FM, Verspreet J, Courtin CM. & Van de Wiele, T. Inter-individual differences determine the outcome of wheat bran colonization by the human gut microbiome. Environmental microbiology. 2017;19:3251–3267. doi: 10.1111/1462-2920.13819. [DOI] [PubMed] [Google Scholar]
- 47.Chen T, et al. Fiber-utilizing capacity varies in Prevotella- versus Bacteroides-dominated gut microbiota. Scientific reports. 2017;7:2594. doi: 10.1038/s41598-017-02995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luc, F. G. D. V. Ecological Interactions of Bacteria in the Human Gut. Springer Science + Business Media, LLC, 639–680 (2009).
- 49.Sanchez JI, et al. Arabinoxylan-oligosaccharides (AXOS) affect the protein/carbohydrate fermentation balance and microbial population dynamics of the Simulator of Human Intestinal Microbial Ecosystem. Microbial biotechnology. 2009;2:101–113. doi: 10.1111/j.1751-7915.2008.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott KP, Martin JC, Duncan SH, Flint HJ. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS microbiology ecology. 2014;87:30–40. doi: 10.1111/1574-6941.12186. [DOI] [PubMed] [Google Scholar]
- 51.Riviere A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the HumanGut. Frontiers in microbiology. 2016;7:979. doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Falony G, et al. In vitro kinetics of prebiotic inulin-type fructan fermentation by butyrate-producing colon bacteria: implementation of online gas chromatography for quantitative analysis of carbon dioxide and hydrogen gas production. Applied and environmental microbiology. 2009;75:5884–5892. doi: 10.1128/AEM.00876-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Vuyst L, Moens F, Selak M, Riviere A, Leroy F. Summer Meeting 2013: growth and physiology of bifidobacteria. Journal of applied microbiology. 2014;116:477–491. doi: 10.1111/jam.12415. [DOI] [PubMed] [Google Scholar]
- 54.Moens F, Weckx S, De Vuyst L. Bifidobacterial inulin-type fructan degradation capacity determines cross-feeding interactions between bifidobacteria and Faecalibacterium prausnitzii. International journal of food microbiology. 2016;231:76–85. doi: 10.1016/j.ijfoodmicro.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 55.Chung WS, et al. Modulation of the human gut microbiota by dietary fibres occurs at the species level. BMC biology. 2016;14:3. doi: 10.1186/s12915-015-0224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramirez-Farias C, et al. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. The British journal of nutrition. 2009;101:541–550. doi: 10.1017/S0007114508019880. [DOI] [PubMed] [Google Scholar]
- 57.Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environmental microbiology. 2010;12:304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- 58.Hughes SA, Shewry PR, Gibson GR, McCleary BV, Rastall RA. In vitro fermentation of oat and barley derived beta-glucans by human faecal microbiota. FEMS microbiology ecology. 2008;64:482–493. doi: 10.1111/j.1574-6941.2008.00478.x. [DOI] [PubMed] [Google Scholar]
- 59.Rampelli S, et al. Metagenome Sequencing of the Hadza Hunter-Gatherer Gut Microbiota. Current biology: CB. 2015;25:1682–1693. doi: 10.1016/j.cub.2015.04.055. [DOI] [PubMed] [Google Scholar]
- 60.Cinquin C, Le Blay G, Fliss I, Lacroix C. Immobilization of infant fecal microbiota and utilization in an in vitro colonic fermentation model. Microbial ecology. 2004;48:128–138. doi: 10.1007/s00248-003-2022-7. [DOI] [PubMed] [Google Scholar]
- 61.Macfarlane GT, Macfarlane S, Gibson GR. Validation of a Three-Stage Compound Continuous Culture System for Investigating the Effect of Retention Time on the Ecology and Metabolism of Bacteria in the Human Colon. Microbial ecology. 1998;35:180–187. doi: 10.1007/s002489900072. [DOI] [PubMed] [Google Scholar]
- 62.Michel C, et al. In vitro prebiotic effects of Acacia gums onto the human intestinal microbiota depends on both botanical origin and environmental pH. Anaerobe. 1998;4:257–266. doi: 10.1006/anae.1998.0178. [DOI] [PubMed] [Google Scholar]
- 63.Zachariassen LF, et al. Sensitivity to oxazolone induced dermatitis is transferable with gut microbiota in mice. Scientific reports. 2017;7:44385. doi: 10.1038/srep44385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 66.Ritari J, Salojarvi J, Lahti L, de Vos WM. Improved taxonomic assignment of human intestinal 16S rRNA sequences by a dedicated reference database. BMC genomics. 2015;16:1056. doi: 10.1186/s12864-015-2265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krych L, Hansen CH, Hansen AK, van den Berg FW, Nielsen DS. Quantitatively different, yet qualitatively alike: a meta-analysis of the mouse core gut microbiome with a view towards the human gut microbiome. PloS one. 2013;8:e62578. doi: 10.1371/journal.pone.0062578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.