Summary
Cellular senescence is a damage response aimed to orchestrate tissue repair. We have recently reported that cellular senescence, through the paracrine release of interleukin‐6 (IL6) and other soluble factors, strongly favors cellular reprogramming by Oct4, Sox2, Klf4, and c‐Myc (OSKM) in nonsenescent cells. Indeed, activation of OSKM in mouse tissues triggers senescence in some cells and reprogramming in other cells, both processes occurring concomitantly and in close proximity. In this system, Ink4a/Arf‐null tissues cannot undergo senescence, fail to produce IL6, and cannot reprogram efficiently; whereas p53‐null tissues undergo extensive damage and senescence, produce high levels of IL6, and reprogram efficiently. Here, we have further explored the genetic determinants of in vivo reprogramming. We report that Ink4a, but not Arf, is necessary for OSKM‐induced senescence and, thereby, for the paracrine stimulation of reprogramming. However, in the absence of p53, IL6 production and reprogramming become independent of Ink4a, as revealed by the analysis of Ink4a/Arf/p53 deficient mice. In the case of the cell cycle inhibitor p21, its protein levels are highly elevated upon OSKM activation in a p53‐independent manner, and we show that p21‐null tissues present increased levels of senescence, IL6, and reprogramming. We also report that Il6‐mutant tissues are impaired in undergoing reprogramming, thus reinforcing the critical role of IL6 in reprogramming. Finally, young female mice present lower efficiency of in vivo reprogramming compared to male mice, and this gender difference disappears with aging, both observations being consistent with the known anti‐inflammatory effect of estrogens. The current findings regarding the interplay between senescence and reprogramming may conceivably apply to other contexts of tissue damage.
Keywords: interleukin‐6, p16Ink4a, plasticity, pluripotency, reprogramming, SASP, senescence
1. INTRODUCTION
Senescence is a cellular response to damage characterized by a stable cell cycle arrest and by the secretion of cytokines and other soluble factors with pleiotropic functions, collectively known as senescence‐associated secretory phenotype or SASP (Freund, Orjalo, Desprez & Campisi, 2010; Muñoz‐Espín & Serrano, 2014). The primary role of senescence is thought to be the orchestration of tissue remodeling and repair. This has been demonstrated in a variety of settings, including tissue repair in the skin and liver (Demaria et al., 2014; Krizhanovsky et al., 2008), embryonic development (Davaapil, Brockes & Yun, 2017; Muñoz‐Espín et al., 2013; Storer et al., 2013), placenta formation (Chuprin et al., 2013), as well as cancer regression (Ventura et al., 2007; Xue et al., 2007). In general, senescent cells are efficiently cleared as part of a successful tissue repair process. However, upon severe or chronic damage, senescence‐orchestrated tissue repair may fail and senescent cells may accumulate, contributing to disease and aging (Muñoz‐Espín & Serrano, 2014).
The power of cellular senescence in inducing tissue remodelling has been further extended to processes of cellular reprogramming in vivo. The transgenic expression of the four transcription factors abbreviated as OSKM (Oct4, Sox2, Klf4, and c‐Myc) (Takahashi & Yamanaka, 2006) in adult mice induces dedifferentiation and cellular reprogramming within multiple tissues (Abad et al., 2013; Ohnishi et al., 2014). However, in addition to reprogramming, the activation of OSKM also results in cellular damage and senescence, both in vitro (Banito et al., 2009) and in vivo (Chiche et al., 2017; Mosteiro et al., 2016). Therefore, OSKM induces two opposite cellular fates, namely senescence and reprogramming, that coexist in vivo in separate, but proximal, subsets of cells (Chiche et al., 2017; Mosteiro et al., 2016). Importantly, it has been demonstrated that senescence plays an active role in facilitating in vivo reprogramming through the paracrine action of the SASP, being interleukin‐6 (IL6) a critical mediator (Chiche et al., 2017; Mosteiro et al., 2016). Of note, IL6 plays an important role also during in vitro reprogramming (Brady et al., 2013; Mosteiro et al., 2016). Moreover, the concept that senescence promotes cellular plasticity has been further extended to the activation of somatic stem/progenitor cells. In particular, the SASP can confer somatic stem/progenitor features onto proximal epithelial cells in several tissues (Ritschka et al., 2017).
The tumor suppressor genes p53, p21, Ink4a, and Arf act as cell‐autonomous barriers for cellular reprogramming (Banito et al., 2009; Kawamura et al., 2009; Li et al., 2009; Marion et al., 2009; Utikal et al., 2009; Zhao et al., 2008). These barriers are conceivably activated by cellular damages associated to reprogramming, most notably replication stress (Marion et al., 2009), which result in proliferation arrest and, consequently, inhibit reprogramming (Hanna et al., 2009). At the same time, p53 and the genetic locus Ink4a/Arf also affect reprogramming, although in opposite directions, through cell extrinsic mechanisms (Mosteiro et al., 2016). In the absence of p53, the induction of OSKM leads to exacerbated damage and senescence in tissues, which results in high levels of IL6 that further enhance reprogramming (Mosteiro et al., 2016). The Ink4a/Arf locus plays a complex role in reprogramming: it promotes reprogramming through the paracrine influence of senescence (Mosteiro et al., 2016), and, at the same time, it is a cell‐autonomous barrier for reprogramming (Li et al., 2009). In vivo, the absence of Ink4a/Arf severely impairs OSKM‐senescence, IL6 levels are modestly increased, and reprogramming is very inefficient (Mosteiro et al., 2016). Therefore, the positive cell‐autonomous impact of Ink4a/Arf deficiency is completely obscured in vivo by the absence of senescence and IL6 secretion (Mosteiro et al., 2016). The emerging picture is that tissue damage and senescence provide a tissue microenvironment that is critical for OSKM reprogramming in vivo.
In this report, we dissect genetically the role of Ink4a, Arf, p53, p21, and Il6 on in vivo reprogramming.
2. RESULTS
2.1. Ink4a is required for OSKM‐induced senescence and reprogramming
The Ink4a/Arf genetic locus encodes two tumor suppressor genes, namely p16Ink4a and p19Arf, acting on separate pathways: p16Ink4a is an inhibitor of the CDK46/cyclin D kinases and, therefore, functions as a positive regulator of the Retinoblastoma family of proteins; p19Arf is an inhibitor of the ubiquitin ligase MDM2, thereby, stabilizing p53 (Gil & Peters, 2006). Previously, we have reported that the activation of an inducible OSKM transgene (abbreviated as i4F for inducible four factors) in mice simultaneously lacking Ink4a and Arf fails to trigger senescence and to produce high levels of IL6, and consequently, reprogramming is severely compromised (Mosteiro et al., 2016). To strengthen the role of IL6 during in vivo reprogramming, we tested whether reprogramming can be rescued in i4F;Ink4a/Arf‐null mice by the exogenous administration of recombinant IL6 (rIL6). Of note, we have previously observed that rIL6 administration elevates in vivo reprogramming in wild‐type i4F mice (Mosteiro et al., 2016). We focused on the pancreas because it is the tissue that shows the fastest and more widespread senescence, dedifferentiation, and reprogramming (Mosteiro et al., 2016). We observed that i4F;Ink4a/Arf‐null mice treated with rIL6 tend to present a higher degree of reprogramming, assessed by the extent of pancreatic dysplasia, upon 7 days of doxycycline treatment (0.2 mg/ml) compared to i4F; Ink4a/Arf‐null controls (Figure S1).
To genetically dissect the individual contribution of Ink4a and Arf during in vivo reprogramming, we generated reprogrammable mouse lines with null alleles for either Ink4a (Krimpenfort, Quon, Mooi, Loonstra & Berns, 2001) or Arf (Kamijo et al., 1997). After a short treatment with doxycycline, we quantified the following four readouts: the presence of senescent cells (by staining for senescence‐associated β‐galactosidase or SAβG), the mRNA levels of several factors characteristically expressed by senescent cells (collectively known as the SASP), the extent of dysplasia (which we use as a surrogate for cell dedifferentiation and partial reprogramming), and the occurrence of advanced or full reprogramming (by staining for the pluripotency marker NANOG).
We compared i4F;Ink4a‐null and i4F mice of mixed genetic background, but derived from the same parents and therefore with the same mixed genetic background. After 10 days of doxycycline treatment (0.2 mg/ml), we observed that the degree of pancreatic dysplasia (Figure 1a) and reprogramming (Figure 1b) was strongly decreased in i4F;Ink4a‐null mice compared to their i4F counterparts. In parallel to this, i4F;Ink4a‐null pancreas presented limited senescence after OSKM activation (Figure 1b), and little expression of senescence‐associated genes like Arf, p53, and p21 as well as Il6 and other factors involved in the SASP, such as Tnf, Il1a, Il1b, Il1rn, Mmp3, and Pai1 (Figure 1c). Of note, p21 mRNA was not induced in i4F mice (Figure 1c), nor in any of the genetic combinations studied in this work (see later). In relation to this, we have previously reported that upon in vivo OSKM activation, the protein levels of p21 are highly induced in a p53‐independent manner (Mosteiro et al., 2016). These observations suggest that during in vivo reprogramming p21 protein levels are regulated post‐transcriptionally, possibly through one or some of the multiple post‐transcriptional mechanisms that are known to regulate p21 (Scoumanne, Cho, Zhang & Chen, 2011; Warfel & El‐Deiry, 2013; Zhang & Chen, 2008).
Figure 1.
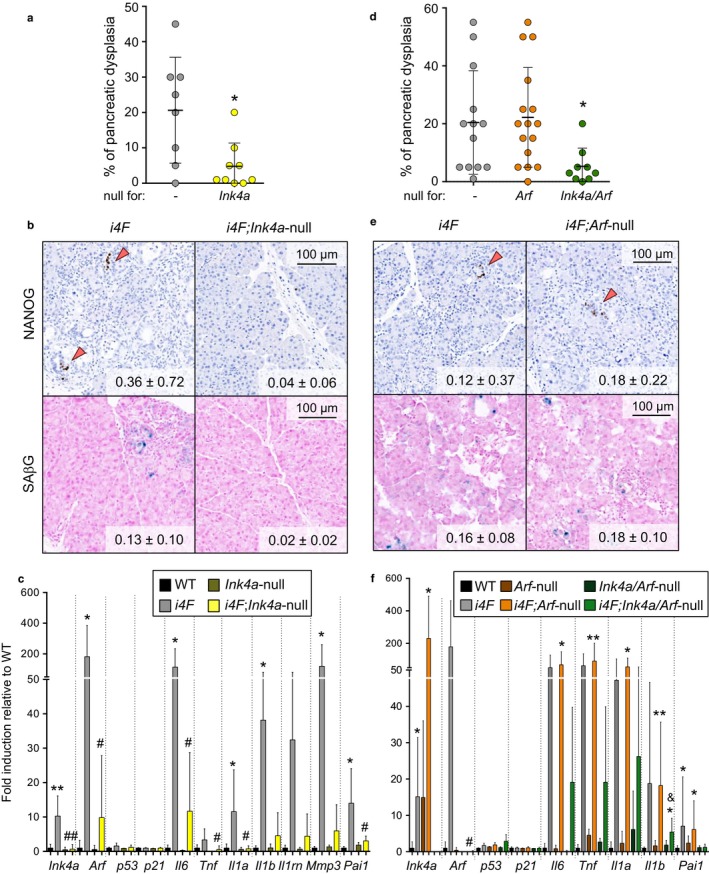
Ink4a is required, and Arf is dispensable for in vivo OSKM‐induced senescence and reprogramming. a. Percentage of dysplasia in the pancreas of i4F and i4F;Ink4a‐null mice treated with 0.2 mg/ml doxycycline for 10 days and analyzed at the end of the treatment. Values correspond to the percentage (%) of pancreatic area affected (evaluated blindly). b. NANOG immunohistochemistry and SAβG staining in the pancreas of the same mice as in panel a. Images are representative for at least six mice (n ≥ 6) in NANOG and three mice (n ≥ 3) in SAβG. Values correspond to ‰ of NANOG+ cells and % of SAβG area quantified with an automated software. c. mRNA levels of the indicated genes in the pancreas of the same mice as in panel a. The number of mice tested is: n = 5 in WT and Ink4a‐null; n = 8 in i4F; n = 10 in i4F;Ink4a‐null. d. Percentage of dysplasia in the pancreas of i4F, i4F;Arf‐null, and i4F;Ink4a/Arf‐null mice treated with 0.2 mg/ml doxycycline for 7 days and analyzed at the end of the treatment. Values correspond to the percentage (%) of pancreatic area affected (evaluated blindly). e. NANOG immunohistochemistry and SAβG staining in the pancreas of the same mice as in panel d. Images are representative for at least 13 mice (n ≥ 13) in NANOG and three mice (n ≥ 3) in SAβG. Values correspond to ‰ of NANOG+ cells and % of SAβG area quantified with an automated software. f. mRNA levels of the indicated genes in the pancreas of the same mice as in panel d. The number of mice tested is: n = 3 in WT, Arf‐null and Ink4a/Arf‐null; n = 10 in i4F; n = 12 in i4F;Arf‐null and n = 6 in i4F;Ink4a/Arf‐null. All the mice tested were males of 8–10 weeks of age. All values are expressed as average ± SD Statistical significance compared to WT controls was assessed using the unpaired two‐tailed Student's t test with Welch's correction: p < .05, *; p < .01, **. Comparisons of each genotype to i4F controls are indicated in the same manner but using the symbol “#.” Comparisons between i4F;Arf‐null and i4F;Ink4a/Arf‐null are indicated in the same manner but using the symbol “&”
Next, we evaluated the extent of in situ reprogramming in the pancreas of i4F;Arf‐null mice. We compared in parallel the behavior of i4F, i4F;Arf‐null, and i4F;Ink4a/Arf‐null mice, all in a pure C57BL6 genetic background. Interestingly, i4F;Arf‐null mice presented similar levels of pancreatic dysplasia compared to i4F mice after 7 days of doxycycline treatment (0.2 mg/ml), while i4F;Ink4a/Arf‐null pancreatic tissue was almost unaffected (Figure 1d). We also observed that the amounts of NANOG+ cells as well as senescent cells in the pancreas of i4F;Arf‐null and i4F mice were comparable (Figure 1e). Finally, we confirmed that the expression of SASP factors in i4F;Arf‐null pancreas was increased to similar levels as those observed in i4F pancreas upon OSKM activation (Figure 1f). All together, these results demonstrate that Ink4a is required for OSKM‐induced senescence and secretion of IL6, while Arf plays a minimal role in these processes. This is in agreement with our previous data showing that Il6 and Tnf are induced in mouse embryonic fibroblasts (MEFs) upon radiation‐induced senescence and this is abolished when either Ink4a or Ink4a/Arf are knocked‐down, but not when Arf is knocked‐down (Mosteiro et al., 2016).
2.2. Absence of p21 increases OSKM‐induced senescence and reprogramming
The cell cycle inhibitor p21CIP1 participates in senescence (Muñoz‐Espín & Serrano, 2014; Sharpless & Sherr, 2015), and it is a cell‐autonomous barrier for in vitro reprogramming (Banito et al., 2009; Kawamura et al., 2009). The protein levels of p21 are elevated in tissues expressing OSKM and this occurs independently of p53 (Mosteiro et al., 2016) and, possibly, through post‐transcriptional mechanisms (see above Figure 1c). To investigate the role of p21 during in vivo reprogramming, we generated reprogrammable mice deficient for p21 (Brugarolas et al., 1995). We first evaluated the reprogramming efficiency in the pancreas of i4F;p21‐null mice compared to i4F controls, all of pure C57BL6 genetic background, after 7 days of doxycycline treatment (0.2 mg/ml). Interestingly, in the case of i4F;p21‐null pancreas, we observed that the degree of pancreatic dysplasia was significantly higher than in i4F pancreas (Figure 2a). We also observed an increased accumulation of reprogrammed (NANOG+) and senescent (SAβG+) cells in the pancreas of i4F;p21‐null mice compared to i4F controls (Figure 2b). In agreement, the levels of SASP factors, such as Il6, Tnf, and Pai1, presented the same tendency, that is, i4F<i4F;p21‐null (Figure 2c). Therefore, p21 is a negative regulator of OSKM‐induced senescence and reprogramming in vivo.
Figure 2.
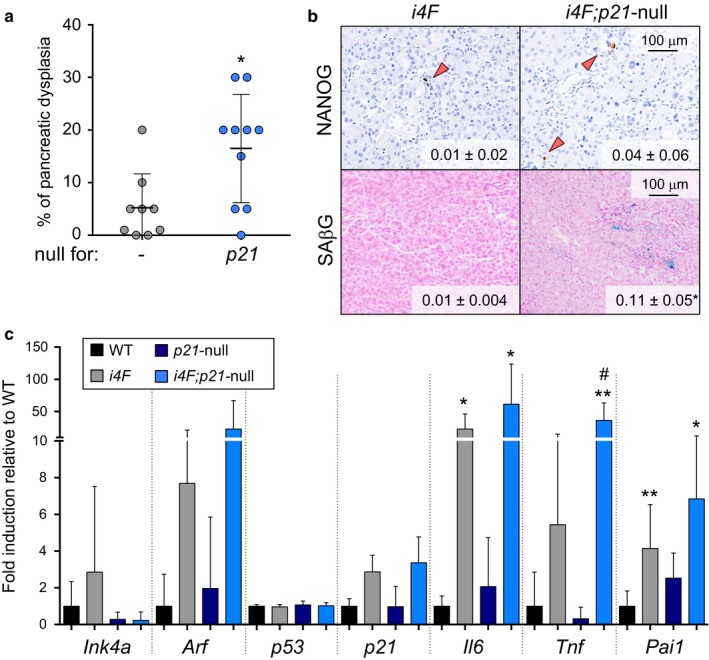
p21 limits in vivo OSKM‐induced senescence and reprogramming. a. Percentage of dysplasia in the pancreas of i4F and i4F; p21‐null mice treated with 0.2 mg/ml doxycycline for 7 days and analyzed at the end of the treatment. Values correspond to the percentage (%) of pancreatic area affected (evaluated blindly). b. NANOG immunohistochemistry and SAβG staining in the pancreas of the same mice as in panel a. Images are representative for at least 9 mice (n ≥ 9) in NANOG and 3 mice (n ≥ 3) in SAβG. Values correspond to ‰ of NANOG+ cells and % of SAβG area quantified with an automated software. c. mRNA levels of the indicated genes in the pancreas of the same mice as in panel a. The number of mice tested is: n = 6 in WT; n = 8 in p21‐null; n = 9 in i4F; and n = 10 in i4F;p21‐null. All the mice tested were females of 12–16 weeks of age. All values are expressed as average ± SD Statistical significance compared to WT controls was assessed using the unpaired two‐tailed Student's t test with Welch's correction: p < .05, *; p < .01, **. Comparisons between i4F and i4F;p21‐null are indicated in the same manner but using the symbol “#”
2.3. Absence of p53 renders IL6 production and reprogramming independent of Ink4a
We have previously reported that p53‐null mice present massive levels of senescence and reprogramming upon OSKM activation, and both processes are partially inhibited by in vivo treatment with anti‐IL6 antibodies (Mosteiro et al., 2016). Given the essential role of Ink4a in senescence and IL6 secretion, we investigated its importance in a p53‐deficient context and, for this, we generated p53/Ink4a/Arf mice. Interestingly, we observed that, after 7 days of doxycycline treatment (0.2 mg/ml), i4F;p53‐null;Ink4a/Arf‐null mice had a similar degree of dysplasia as i4F;p53‐null mice, all being in a pure C57BL6 genetic background (Figure 3a). Similarly, the amount of reprogramming assessed by the presence of NANOG+ cells in the pancreas was comparable between i4F;p53‐null and i4F;p53‐null;Ink4a/Arf‐null mice (Figure 3b). We next evaluated the accumulation of senescent cells and observed that it was decreased i4F;p53‐null;Ink4a/Arf‐null pancreas compared to i4F;p53‐null pancreas (Figure 3b), reinforcing the importance of Ink4a in the induction of OSKM‐driven senescence (Mosteiro et al., 2016). However, no significant differences were detected regarding the upregulation of the SASP between i4F;p53‐null and i4F;p53‐null;Ink4a/Arf‐null mice (Figure S2). This indicates that in a p53‐null context, the SASP becomes independent of Ink4a/Arf. In this regard, it is known that p53 is a negative regulator of the SASP and its deficiency results in a massive upregulation of the SASP (Coppé et al., 2008). Our current observations with i4F;p53‐null;Ink4a/Arf‐null mice also suggest that the SASP, rather than the extent of senescence, is the key factor responsible for a high reprogramming efficiency. All together, we conclude that the absence of p53 has a dominant effect over the absence of Ink4a with regard to cytokine secretion and this is key in determining the efficiency of reprogramming.
Figure 3.
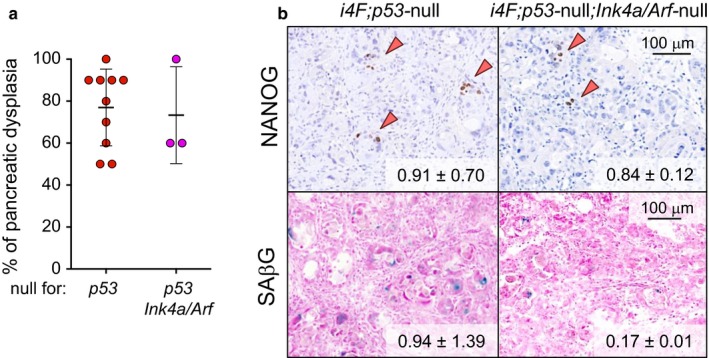
Deletion of p53 is dominant over the deletion of Ink4a/Arf during in vivo OSKM‐induced senescence and reprogramming. a. Percentage of dysplasia in the pancreas of i4F;p53‐null and i4F;p53‐null;Ink4a/Arf‐null mice treated with 0.2 mg/ml doxycycline for 7 days and analyzed at the end of the treatment. Values correspond to the percentage (%) of pancreatic area affected (evaluated blindly). b. NANOG immunohistochemistry and SAβG staining in the pancreas of the same mice as in panel a. Images are representative for at least four mice (n ≥ 4) except in i4F; p53‐null; Ink4a/Arf‐null (n = 2). Values correspond to ‰ of NANOG+ cells and % of SAβG area quantified with an automated software. All the mice tested are males of 10–14 weeks of age
2.4. Il6 mutation impairs in vivo reprogramming
Damaged and senescent cells promote the reprogramming of their neighboring cells through secreted factors, being IL6 one of the key mediators of this cross talk (Mosteiro et al., 2016). Moreover, IL6 plays an important role during in vitro reprogramming (Brady et al., 2013) and treatment with anti‐IL6 antibodies decreases reprogramming both in vitro (Mosteiro et al., 2016) and in vivo (Chiche et al., 2017; Mosteiro et al., 2016), while exogenous administration of rIL6 enhances in vivo reprogramming (Mosteiro et al., 2016) (see also Figure S1). To further strengthen the role of IL6 during in vivo reprogramming, we generated reprogrammable mouse lines (i4F) combined with mutant alleles of Il6 (Kopf et al., 1994). These groups of mice were of mixed genetic background but were derived from the same parents, thereby sharing a comparable genetic background. We observed that after 7 days of doxycycline treatment (0.2 mg/ml), the pancreatic dysplasia in i4F;Il6‐mutant mice was much reduced compared to their i4F counterparts (Figure 4a). In this genetic background and at the time point analyzed, the number of NANOG+ cells was too low to assess differences between genotypes (Figure 4b), but we observed a suggestive reduction in the levels of senescent cells in the pancreas (Figure 4b) together with a residual induction of Ink4a, Arf, and several SASP factors, such as Tnf, Il1a, Il1b, and Pai1 (Figure 4c). This is in agreement with the previously reported role of IL6 in the induction of paracrine senescence (Kuilman et al., 2008). All together, these results reinforce the critical role of IL6 for in vivo reprogramming.
Figure 4.
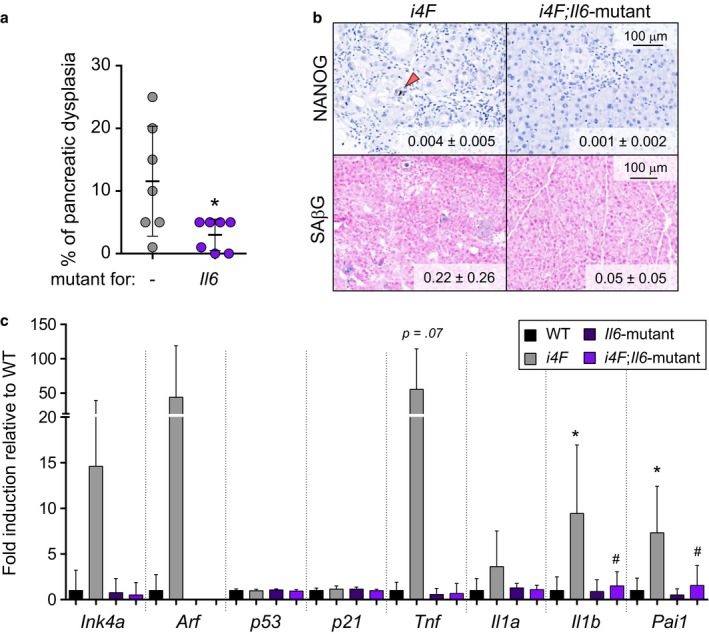
Deletion of Il6 impairs in vivo OSKM‐induced senescence and reprogramming. a. Percentage of dysplasia in the pancreas of i4F and i4F;Il6‐mutant mice treated with 0.2 mg/ml doxycycline for 7 days and analyzed at the end of the treatment. Values correspond to the percentage (%) of pancreatic area affected (evaluated blindly). b. NANOG immunohistochemistry and SAβG staining in the pancreas of the same mice as in panel a. Images are representative for at least four mice (n ≥ 4) in NANOG and at least three mice (n ≥ 3) in SAβG. Values correspond to ‰ of NANOG+ cells and % of SAβG area quantified with an automated software. c. mRNA levels of the indicated genes in the pancreas of the same mice as in panel a. The number of mice tested is n = 5 in WT and Il6‐mutant and n = 7 in i4F and i4F;Il6‐null. All the mice tested were males of 9–18 weeks of age. All values are expressed as average ± SD Statistical significance compared to WT controls was assessed using the unpaired two‐tailed Student's t test with Welch's correction: p < .05, *. Comparisons of each genotype to i4F controls are indicated in the same manner but using the symbol “#”
2.5. In vivo reprogramming is decreased in female mice
All the above analyses were performed using groups of young mice of the same sex, usually males, with the only exception of the experiment shown in Figure 2 that was performed with females. We noted that, upon OSKM induction, control female i4F mice presented reduced levels of senescence, dysplasia and reprogramming compared to control i4F males. To substantiate this quantitatively, we compiled our current data in young (8–12 weeks of age) i4F males from experiments in Figures 1a,d, and 4a and compared them with i4F females of the same age from the experiment in Figure 2a. Interestingly, this comparison revealed that young female mice are indeed less prone to undergo reprogramming compared to young male mice (Figure 5a). It is well established that the hormone‐bound form of the estrogen receptor inhibits NκB (Kalaitzidis & Gilmore, 2017; Naugler et al., 2007). NFκB is a major inducer of the SASP and IL6 and, indeed, pharmacological inhibition of NF‐κB reduces OSKM‐induced senescence and reprogramming in vivo (Mosteiro et al., 2016). Therefore, the observed reduced reprogramming of female mice could reflect an attenuated production of IL6 in response to OSKM. To support this, we evaluated the levels of Il6 mRNA in the pancreas of male and female mice. Interestingly, compared to males, females presented a much lower upregulation of Il6 upon OSKM activation (Figure 5b).
Figure 5.

Female mice present reduced in vivo reprogramming. a. Percentage of dysplasia in the pancreas of i4F mice from young male (n = 28, from Figures 1a,d, and 4a), young female (n = 9, from Figure 2a), old male (n = 7) and old female (n = 5), treated with 0.2 mg/ml doxycycline for 7 days, and analyzed at the end of the treatment. Young male and female mice were 8–12 weeks of age; old male and female mice were ≥54 weeks of age. Values correspond to the percentage (%) of pancreatic area affected (evaluated blindly). b. mRNA levels of the indicated genes in the pancreas of the indicated mice, treated with 0.2 mg/ml doxycycline for 7 days, and analyzed at the end of the treatment. The number of mice tested is n = 8 in male WT; n = 9 in female WT; n = 8 in male i4F and n = 9 in female i4F. All values are expressed as average ± SD Statistical significance was assessed using the unpaired two‐tailed Student's t test with Welch's correction: p < .001, ***. Some nonsignificant (n.s.) comparisons are also indicated
During aging, there is a general increase in systemic inflammation and IL6 levels, both in males and females (Cevenini, Monti & Franceschi, 2013; Maggio, Guralnik, Longo & Ferrucci, 2006) and, accordingly, we observed that aged mice undergo in vivo reprogramming more efficiently than young mice (Mosteiro et al., 2016). Aging also results in lower levels of estrogens in female mice (Nelson et al., 1981). Based on this, we hypothesized that aged female mice should have an in vivo reprogramming efficiency similar to the one in aged male mice. To address this, we compared the degree of pancreatic dysplasia in a group of old females and males (≥54 weeks of age). Interestingly, the gender differences observed in young mice disappeared in old mice (Figure 5a).
In summary, the gender‐specific difference observed in reprogramming and its attenuation with aging is consistent with the concept that the levels of IL6 strongly influence in vivo reprogramming and suggest that estrogens negatively regulate in vivo reprogramming.
3. DISCUSSION
The activation of OSKM in vivo triggers not only the reprogramming of a small population of cells, but also induces damage in many other cells (Chiche et al., 2017; Mosteiro et al., 2016). We have previously identified a positive causal connection between damaged cells and reprogramming in tissues upon in vivo OSKM activation (Mosteiro et al., 2016). In this report, we have further explored the in vivo interplay between senescence and reprogramming. To summarize our findings and to facilitate comparison between the different genotypes, we have compiled the key data from Figures 1, 2, 3, 4 into unified panels (Figure 6a–c). For this, we have pooled the groups of control i4F male mice in Figures 1a,d, and 4a, thereby increasing the statistical potency of the comparisons.
Figure 6.

Genetic dissection of OSKM‐induced reprogramming and senescence. a. Compilation of senescence induction (SAβG+ cells) by OSKM in the pancreas of the indicated mice from Figures 1, 2, 3, 4. N values are: i4F, n = 10; i4F;Il6‐mutant, n = 3; i4F;Ink4a/Arf‐null, n = 3; i4F;Ink4a‐null, n = 3; i4F;Arf‐null, n = 5; i4F;p53‐null, n = 4; i4F;p53‐null;Ink4a/Arf‐null, n = 2; female i4F, n = 3; female i4F;p21‐null, n = 4. b. Compilation of dysplasia induction by OSKM in the pancreas of the indicated mice from Figures 1, 2, 3, 4. Compilation of data from Figures 1, 2, 3, 4. N values are: i4F, n = 28; i4F; Il6‐mutant, n = 7; i4F;Ink4a/Arf‐null, n = 9; i4F;Ink4a‐null, n = 9; i4F; Arf‐null, n = 16; i4F;p53‐null, n = 10; i4F;p53‐null;Ink4a/Arf‐null, n = 3; female i4F, n = 9; female i4F;p21‐null, n = 10. c. Compilation of reprogramming induction (NANOG+ cells) by OSKM in the pancreas of the indicated mice from Figures 1, 2, 3, 4. Compilation of data from Figures 1, 2, 3, 4. N values are: i4F, n = 23; i4F; Il6‐mutant, n = 4; i4F;Ink4a/Arf‐null, n = 9; i4F;Ink4a‐null, n = 9; i4F;Arf‐null, n = 15; i4F;p53‐null, n = 5; i4F;p53‐null;Ink4a/Arf‐null, n = 2; female i4F, n = 9; female i4F;p21‐null, n = 10. d. Correlation between the ‰ NANOG+ cells (assessed by IHC) and the mRNA levels of Il6 (relative to WT mice and assessed by qRT–PCR). Compilation of all the paired values obtained in this study (n = 78). Statistical significance was evaluated by two‐tailed Student's t test (p < .0001) and Pearson correlation (r = .60). In a–c, all values are expressed as average ± SEM Statistical significance was assessed using the unpaired two‐tailed Student's t test with Welch's correction: p < .05, *; p < .01, **; p < .001, ***; p < .0001, ****
Importantly, we have confirmed genetically the key role of IL6 in the cross talk between OSKM‐induced senescence and OSKM‐induced reprogramming. We have observed that tissues mutant for IL6 present a reduction in senescence (Figure 6a), which is in agreement with the known role of IL6 in reinforcing senescence in a paracrine and autocrine manner (Kuilman et al., 2008). Consequently, the SASP is also reduced, and this is accompanied by reduced pancreatic dysplasia (Figure 6b) and reprogramming (Figure 6c). In line with this, we have also observed that young female mice present lower levels of OSKM‐induced senescence, pancreatic dysplasia, and reprogramming compared to male mice (Figure 6a–c). This gender difference can be explained by the known negative regulation of NFκB activity and IL6 production by estrogens (Kalaitzidis & Gilmore, 2017; Naugler et al., 2007). We have previously reported that aged mice are more prone to undergo reprogramming than young ones (Mosteiro et al., 2016) and we proposed that this may reflect the higher levels of systemic inflammation and IL6 characteristic of aged mice (Cevenini et al., 2013; Maggio et al., 2006). Estrogen levels are reduced during mouse female aging (Nelson et al., 1981), and indeed, we have also observed that reprogramming was similarly high in aged female and male mice.
To further reinforce the relationship between IL6 and reprogramming, we have represented in the same graph all the paired values of Il6 mRNA and NANOG+ cells that we have accumulated in this study (n = 78 mice with paired data). Importantly, despite the variability of the data, a modest but significant positive association can be detected between the amount of NANOG+ cells and the expression of Il6 in the pancreas (Figure 6d). IL6 has been implicated in several models of cell dedifferentiation and tissue regeneration (see, e.g., Dubois‐Pot‐Schneider et al., 2014; Gallucci et al., 2000; Lin, Kondo, Ishida, Takayasu & Mukaida, 2003; Schmidt‐Arras & Rose‐John, 2016). Our observations on the critical role of IL6 in the cross talk between damage and reprogramming suggest that the reprogrammable mouse model recapitulates aspects of tissue repair upon injury.
The secretion of IL6 by damaged cells requires a functional Ink4a/Arf locus (Mosteiro et al., 2016). Ink4a/Arf encodes two different tumor suppressors, p16Ink4a and p19Arf, which have separate promoters and reading frames, generating two nonhomologous proteins (Gil & Peters, 2006). p16Ink4a inhibits CDK4, 6/cycD kinases, maintaining the retinoblastoma family proteins in their nonproliferative unphosphorylated form. On the other hand, p19Arf induces cell cycle arrest through MDM2‐mediated stabilization of p53. Here, we show that Ink4a‐null mice fail to activate an efficient senescent response upon induction of OSKM (Figure 6a), which results in lower levels of IL6, reduced pancreatic dysplasia (Figure 6b) and lower levels of reprogramming (Figure 6c). In contrast, deletion of Arf does not affect neither OSKM‐induced senescence nor in vivo reprogramming (Figure 6a–c). Therefore, Ink4a, but not Arf, appears to be critical for the interplay between senescence and reprogramming.
We have also demonstrated that OSKM‐induced senescence does not require p53 (Mosteiro et al., 2016). Indeed, tissues lacking p53 present a stronger damage response upon OSKM activation, leading to increased accumulation of senescent cells and increased reprogramming efficiency (Figure 6a–c). We wondered if Ink4a was also critical for senescence and reprogramming in the absence of p53. Triply deficient p53/Ink4a/Arf mice presented levels of senescence clearly lower than p53‐null mice, but higher than in Ink4a‐deficient mice (Figure 6a). Interestingly, however, p53/Ink4a/Arf triply deficient tissues behaved equal to p53‐null tissues regarding their high levels of IL6 and reprogramming (Figure 6b,c). Therefore, the absence of p53 has a dominant effect that renders IL6 production independent of Ink4a. In fact, it is known that p53 is a negative regulator of the SASP and its deficiency results in a massive upregulation of the SASP (Coppé et al., 2008).
To shed some light on other known regulators of senescence and reprogramming, we investigated the role of p21. Mice deficient for p21 present higher levels of OSKM‐induced senescence, SASP production, dysplasia, and reprogramming compared to control i4F mice (Figure 6a–c), indicating that it is a barrier for both OSKM‐induced senescence and reprogramming. Of interest, p21 protein levels are notably increased upon OSKM activation, both in wild‐type as well as in p53‐null tissues (Mosteiro et al., 2016), and our current data indicate that this is not due to an upregulation of p21 mRNA levels (Figures 1c,f, 4c and S1). These observations suggest that, upon in vivo OSKM activation, the levels of p21 protein are elevated through post‐transcriptional mechanisms that are known to play a role in p21 regulation (Scoumanne et al., 2011; Warfel & El‐Deiry, 2013; Zhang & Chen, 2008). In this context, it is worth mentioning that the upregulation of p21 during developmentally programmed senescence is also independent of p53 (Muñoz‐Espín et al., 2013; Storer et al., 2013).
All together, we have genetically dissected some of the key players in the cross talk between damage and reprogramming in vivo. These findings may have important implications for the understanding of tissue responses to damage.
4. EXPERIMENTAL PROCEDURES
4.1. Reprogrammable mice
To generate reprogrammable mice combined with null alleles for each the different tumor suppressor genes and IL6, we used the reprogrammable mouse line known as i4F‐B, which carries a ubiquitous doxycycline‐inducible OSKM transgene, abbreviated as i4F, and inserted into the Pparg gene (Abad et al., 2013). Reprogrammable i4F‐B mice were crossed with mutant mice for Ink4a/Arf, Ink4a (Krimpenfort et al., 2001), Arf (Kamijo et al., 1997), p53 (Jacks et al., 1994), p21 (Brugarolas et al., 1995), or Il6 (Kopf et al., 1994). Most of these strains were in a pure C57BL/6J.Ola.Hsd genetic background and were compared to i4F mice of the same genetic background. The exception was the i4F;Ink4a‐null and i4F;Il6‐mutant mice that were in a mixed genetic background enriched for C57BL/6J.Ola.Hsd and were compared to i4F mice derived from the same parental mice and therefore with similar mixed genetic background.
4.2. Animal procedures
Animal experimentation at the CNIO, Madrid, was performed according to protocols approved by the CNIO‐ISCIII Ethics Committee for Research and Animal Welfare (CEIyBA). In general, male mice of 10–13 weeks of age were treated with 0.2 mg/ml doxycycline in the drinking water (supplemented with 7.5% sucrose) for 7 days, except for the i4F;Ink4a‐null mice and the corresponding controls that were treated for 10 days. In the case of i4F;p21‐null mice and the corresponding i4F controls, we used female mice of 10–13 weeks of age. When indicated (Figure 5a), we used old mice of ≥54 weeks of age. Treatment with rIL6 (Abyntek Biopharma, S.L, #AI081) was concomitant to doxycycline treatment and was performed by intraperitoneal injection with 5 μg of rIL6 three times per week during 1 week.
4.3. Analysis of mRNA levels
Total RNA was isolated by acid guanidinium thiocyanate–phenol–chloroform extraction from frozen pancreas tissue. Up to 5 µg of total RNA was reverse transcribed into cDNA using iScript™ Advanced cDNA Synthesis Kit for RT–qPCR (Bio‐Rad #172‐5038). Quantitative real‐time PCR was performed using GoTaq® qPCR Master Mix (Promega #A6002) in an ABI PRISM 7700 thermocycler (Applied Biosystem). For input normalization, we used the housekeeping gene Actin.
| Forward primer | Reverse primer | |
|---|---|---|
| Actin | 5′‐GGCACCACACCTTCTACAATG‐3′ | 5′‐GTGGTGGTGAAGCTGTAGCC‐3′ |
| Arf | 5′‐GCCGCACCGGAGGCACCACACCATCCT‐3′ | 5′‐TTGAGCAGAAGAGCTGCTACGT‐3′ |
| Il1a | 5′‐AAGTCTCCAGGGCAGAGAGG‐3′ | 5′‐CTGATTCAGAGAGAGATGGTCAA‐3′ |
| Il1b | 5′‐AAAAGCCTCGTGCTGTCG‐3′ | 5′‐AGGCCACAGGTATTTTGTCG‐3′ |
| Il1rn | 5′‐TTGTGCCAAGTCTGGAGATG‐3′ | 5′‐GTTGTGCAGAGGAACCATCC‐3′ |
| Il6 | 5′‐GTTCTCTGGGAAATCGTGGA‐3′ | 5′‐GGTACTCCAGAAGACCAGAGGA‐3′ |
| Ink4a | 5′‐TACCCCGATTCAGGTGAT‐3′ | 5′‐TTGAGCAGAAGAGCTGCTACGT‐3′ |
| Mmp3 | 5′‐CGGGGAGAAGTCCTGTTTTT‐3′ | 5′‐GGAAGAGATGGCCAAAATGA‐3′ |
| Pai1 | 5′‐CCAACATCTTGGATGCTGAA‐3′ | 5′‐GCCAGGGTTGCACTAAACAT‐3′ |
| P53 | 5′‐CATCCTTTAACTCTAAGGCCTCATTC‐3′ | 5′‐AAGATCCGCGGGCGTAA‐3′ |
| P21 | 5′‐CCTTCTCGTGAGACGCTTAC‐3′ | 5′‐GTGGGTCTGACTCCAGCCC‐3′ |
| Tnf | 5′‐GCCTCTTCTCATTCCTGCTT‐3′ | 5′‐CTCCTCCACTTGGTGGTTTG‐3′ |
4.4. Immunohistochemistry
Tissue samples were fixed in 10% neutral‐buffered formalin (4% formaldehyde in solution), paraffin‐embedded, and cut in 3 µm sections, which were mounted in superfrost®plus slides and dried. For different staining methods, slides were deparaffinized in xylene and re‐hydrated through a series of graded ethanol until water. Serial sections were stained with hematoxylin and eosin. For immunohistochemistry, an automated immunostaining platform was used (Ventana discovery XT; Roche). Antigen retrieval was first performed with high pH buffer (CC1m; Roche), endogenous peroxidase was blocked, and slides were then incubated with NANOG antibody (Cell Signalling Technology, 8822). After the primary antibody, slides were incubated with the corresponding secondary antibodies and visualization systems (OmniRabbit, Ventana; Roche) conjugated with horseradish peroxidase (Chromomap, Ventana; Roche). Immunohistochemical reaction was developed using 3,30‐diaminobenzidine tetrahydrochloride (DAB) as a chromogen and nuclei were counterstained with hematoxylin. Finally, the slides were dehydrated, cleared, and mounted with a permanent mounting medium for microscopic evaluation. Whole digital slides were acquired with a slide scanner (Mirax Scan; Zeiss), and images captured with the Pannoramic Viewer Software (3DHISTECH). Image analysis and quantification were performed in a completely automated manner using with the AxioVision software package (Zeiss). For each staining, several slides were quantified per mouse (at least three mice per group).
4.5. SAβG staining of histological sections
SAβG staining was performed in tissue cryosections preserved in OCT freezing medium using the Senescence β‐Galactosidase Staining Kit (Cell Signaling, #9860). Briefly, tissue cryosections of 12 µm were fixed at room temperature 5 min with a solution containing 2% formaldehyde and 0.2% glutaraldehyde in PBS, washed three times with PBS, and incubated 48 hrs at 37°C with the staining solution containing X‐gal in N‐N‐dimethylformamide (pH 6.0). Sections were counterstained with nuclear fast red. Image analysis and quantification were performed in a completely automated manner as indicated above for the immunohistochemistry.
4.6. Statistical methods
Mice were allocated to their experimental groups according to their predetermined genotype, and therefore, there was no randomization. Investigators were not blinded to the experimental groups. Quantitative PCR data were obtained from independent biological replicates (n values correspond to the biological replicates, that is number of mice; technical replicates of the PCR were not considered in the n value). Data in Figures 1, 2, 3, 4, 5 are represented as average ± SD; data in Figure 6 are represented as average ± SEM. Statistical significance was assessed using Student´s t test (two‐tailed, unpaired) with Welch´s correction (Pearson correction in the case of Figure 6d).
CONFLICT OF INTEREST
Authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
L.M. performed most of the experiments and participated in the experimental design, data analysis, figure preparation, discussion, and writing; C.P. contributed to most of the experimental work; A. de M. supervised the histological stainings and quantifications, and performed the histopathological evaluations; M.S. contributed to the experimental design, discussion, and writing.
Supporting information
Mosteiro L, Pantoja C, de Martino A, Serrano M. Senescence promotes in vivo reprogramming through p16INK4a and IL‐6. Aging Cell. 2018;17:e12711 https://doi.org/10.1111/acel.12711
Funding information
L.M. was recipient of an FPU contract from the Spanish Ministry of Education (MECD). Work in the laboratory of M.S. was funded by the CNIO and by grants from the Spanish Ministry of Economy co‐funded by the European Regional Development Fund (RETOS project), the European Research Council (ERC Advanced Grant), the European Union (RISK‐IR project), and the Botin Foundation and Banco Santander (Santander Universities Global Division). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- Abad, M. , Mosteiro, L. , Pantoja, C. , Cañamero, M. , Rayon, T. , Ors, I. , … Serrano, M. (2013). Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature, 502, 340–345. [DOI] [PubMed] [Google Scholar]
- Banito, A. , Rashid, S. T. , Acosta, J. C. , Li, S. , Pereira, C. F. , Geti, I. , … Gil, J. (2009). Senescence impairs successful reprogramming to pluripotent stem cells. Genes & Development, 23, 2134–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady, J. J. , Li, M. , Suthram, S. , Jiang, H. , Wong, W. H. , & Blau, H. M. (2013). Early role for IL‐6 signalling during generation of induced pluripotent stem cells revealed by heterokaryon RNA‐Seq. Nature Cell Biology, 15, 1244–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas, J. , Chandrasekaran, C. , Gordon, J. I. , Beach, D. , Jacks, T. , & Hannon, G. J. (1995). Radiation‐induced cell cycle arrest compromised by p21 deficiency. Nature, 377, 552–557. [DOI] [PubMed] [Google Scholar]
- Cevenini, E. , Monti, D. , & Franceschi, C. (2013). Inflamm‐ageing. Current Opinion in Clinical Nutrition and Metabolic Care, 16, 14–20. [DOI] [PubMed] [Google Scholar]
- Chiche, A. , Le Roux, I. , von Joest, M. , Sakai, H. , Aguín, S. B. , Cazin, C. , … Li, H. (2017). Injury‐induced senescence enables in vivo reprogramming in skeletal muscle. Cell Stem Cell, 20, 407–414. [DOI] [PubMed] [Google Scholar]
- Chuprin, A. , Gal, H. , Biron‐Shental, T. , Biran, A. , Amiel, A. , Rozenblatt, S. , & Krizhanovsky, V. (2013). Cell fusion induced by ERVWE1 or measles virus causes cellular senescence. Genes & Development, 27, 2356–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppé, J.‐P. , Patil, C. K. , Rodier, F. , Sun, Y. , Muñoz, D. P. , Goldstein, J. , … Campisi, J. (2008). Senescence‐associated secretory phenotypes reveal cell‐nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biology, 6, e301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davaapil, H. , Brockes, J. P. , & Yun, M. H. (2017). Conserved and novel functions of programmed cellular senescence during vertebrate development. Development, 144, 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaria, M. , Ohtani, N. , Youssef, S. A. , Rodier, F. , Toussaint, W. , Mitchell, J. R. , … Campisi, J. (2014). An essential role for senescent cells in optimal wound healing through secretion of PDGF‐AA. Developmental Cell, 31, 722–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois‐Pot‐Schneider, H. , Fekir, K. , Coulouarn, C. , Glaise, D. , Aninat, C. , Jarnouen, K. , … Corlu, A. (2014). Inflammatory cytokines promote the retrodifferentiation of tumor‐derived hepatocyte‐like cells to progenitor cells. Hepatology, 60, 2077–2090. [DOI] [PubMed] [Google Scholar]
- Freund, A. , Orjalo, A. V. , Desprez, P. Y. , & Campisi, J. (2010). Inflammatory networks during cellular senescence: Causes and consequences. Trends in Molecular Medicine, 16, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallucci, R. M. , Simeonova, P. P. , Matheson, J. M. , Kommineni, C. , Guriel, J. L. , Sugawara, T. , & Luster, M. I. (2000). Impaired cutaneous wound healing in interleukin‐6‐deficient and immunosuppressed mice. FASEB Journal, 14, 2525–2531. [DOI] [PubMed] [Google Scholar]
- Gil, J. , & Peters, G. (2006). Regulation of the INK4b–ARF–INK4a tumour suppressor locus: All for one or one for all. Nature Reviews Molecular Cell Biology, 7, 667–677. [DOI] [PubMed] [Google Scholar]
- Hanna, J. , Saha, K. , Pando, B. , van Zon, J. , Lengner, C. J. , Creyghton, M. P. , … Jaenisch, R. (2009). Direct cell reprogramming is a stochastic process amenable to acceleration. Nature, 462, 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks, T. , Remington, L. , Williams, B. O. , Schmitt, E. M. , Halachmi, S. , Bronson, R. T. , & Weinberg, R. A. (1994). Tumor spectrum analysis in p53‐mutant mice. Current Biology, 4, 1–7. [DOI] [PubMed] [Google Scholar]
- Kalaitzidis, D. , & Gilmore, T. D. (2017). Transcription factor cross‐talk: The estrogen receptor and NF‐κB. Trends in Endocrinology and Metabolism, 16, 46–52. [DOI] [PubMed] [Google Scholar]
- Kamijo, T. , Zindy, F. , Roussel, M. F. , Quelle, D. E. , Downing, J. R. , Ashmun, R. A. , … Sherr, C. J. (1997). Tumor Suppression at the Mouse INK4a Locus Mediated by the Alternative Reading Frame Product p19 ARF. Cell, 91, 649–659. [DOI] [PubMed] [Google Scholar]
- Kawamura, T. , Suzuki, J. , Wang, Y. V. , Menendez, S. , Morera, L. B. , Raya, A. , … Izpisúa Belmonte, J. C. (2009). Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature, 460, 1140–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf, M. , Baumann, H. , Freer, G. , Freudenberg, M. , Lamers, M. , Kishimoto, T. , … Köhler, G. (1994). Impaired immune and acute‐phase responses in interleukin‐6‐deficient mice. Nature, 368, 339–342. [DOI] [PubMed] [Google Scholar]
- Krimpenfort, P. , Quon, K. C. , Mooi, W. J. , Loonstra, A. , & Berns, A. (2001). Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature, 413, 83–86. [DOI] [PubMed] [Google Scholar]
- Krizhanovsky, V. , Yon, M. , Dickins, R. A. , Hearn, S. , Simon, J. , Miething, C. , … Lowe, S. W. (2008). Senescence of activated stellate cells limits liver fibrosis. Cell, 134, 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman, T. , Michaloglou, C. , Vredeveld, L. C. W. , Douma, S. , van Doorn, R. , Desmet, C. J. , … Peeper, D. S. (2008). Oncogene‐induced senescence relayed by an interleukin‐dependent inflammatory network. Cell, 133, 1019–1031. [DOI] [PubMed] [Google Scholar]
- Li, H. , Collado, M. , Villasante, A. , Strati, K. , Ortega, S. , Cañamero, M. , … Serrano, M. (2009). The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature, 460, 1136–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Z.‐Q. , Kondo, T. , Ishida, Y. , Takayasu, T. , & Mukaida, N. (2003). Essential involvement of IL‐6 in the skin wound‐healing process as evidenced by delayed wound healing in IL‐6‐deficient mice. Journal of Leukocyte Biology, 73, 713–721. [DOI] [PubMed] [Google Scholar]
- Maggio, M. , Guralnik, J. M. , Longo, D. L. , & Ferrucci, L. (2006). Interleukin‐6 in aging and chronic disease: A magnificent pathway. Journals of Gerontology. Series A, Biological Sciences and Medical Sciences, 61, 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion, R. M. , Strati, K. , Li, H. , Murga, M. , Blanco, R. , Ortega, S. , … Blasco, M. A. (2009). A p53‐mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature, 460, 1149–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosteiro, L. , Pantoja, C. , Alcazar, N. , Marión, R. M. , Chondronasiou, D. , Rovira, M. , … Serrano, M. (2016). Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science, 354, aaf4445. [DOI] [PubMed] [Google Scholar]
- Muñoz‐Espín, D. , Cañamero, M. , Maraver, A. , Gómez‐López, G. , Contreras, J. , Murillo‐Cuesta, S. , … Serrano, M. (2013). Programmed cell senescence during mammalian embryonic development. Cell, 155, 1104–1118. [DOI] [PubMed] [Google Scholar]
- Muñoz‐Espín, D. , & Serrano, M. (2014). Cellular senescence: From physiology to pathology. Nature Reviews Molecular Cell Biology, 15, 482–496. [DOI] [PubMed] [Google Scholar]
- Naugler, W. E. , Sakurai, T. , Kim, S. , Maeda, S. , Kim, K. , Elsharkawy, A. M. , & Karin, M. (2007). Gender disparity in liver cancer due to sex differences in MyD88‐dependent IL‐6 production. Science, 317, 121–124. [DOI] [PubMed] [Google Scholar]
- Nelson, J. F. , Felicio, L. S. , Osterburg, H. H. , & Finch, C. E. (1981). Altered profiles of estradiol and progesterone associated with prolonged estrous cycles and persistent vaginal cornification in aging C57BL/6J mice. Biol Reprod. 24(4), 784–794. [DOI] [PubMed] [Google Scholar]
- Ohnishi, K. , Semi, K. , Yamamoto, T. , Shimizu, M. , Tanaka, A. , Mitsunaga, K. , … Yamada, Y. (2014). Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell, 156, 663–677. [DOI] [PubMed] [Google Scholar]
- Ritschka, B. , Storer, M. , Mas, A. , Heinzmann, F. , Ortells, M. C. , Morton, J. P. , … Keyes, W. M. (2017). The senescence‐associated secretory phenotype induces cellular plasticity and tissue regeneration. Genes & Development, 31, 172–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt‐Arras, D. , & Rose‐John, S. (2016). IL‐6 pathway in the liver: From physiopathology to therapy. Journal of Hepatology, 64, 1403–1415. [DOI] [PubMed] [Google Scholar]
- Scoumanne, A. , Cho, S. J. , Zhang, J. , & Chen, X. (2011). The cyclin‐dependent kinase inhibitor p21 is regulated by RNA‐binding protein PCBP4 via mRNA stability. Nucleic Acids Research, 39, 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless, N. E. , & Sherr, C. J. (2015). Forging a signature of in vivo senescence. Nature Reviews Cancer, 15, 397–408. [DOI] [PubMed] [Google Scholar]
- Storer, M. , Mas, A. , Robert‐Moreno, A. , Pecoraro, M. , Ortells, M. C. , Di Giacomo, V. , … Keyes, W. M. (2013). Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell, 155, 1119–1130. [DOI] [PubMed] [Google Scholar]
- Takahashi, K. , & Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Utikal, J. , Polo, J. M. , Stadtfeld, M. , Maherali, N. , Kulalert, W. , Walsh, R. M. , … Hochedlinger, K. (2009). Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature, 460, 1145–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura, A. , Kirsch, D. G. , McLaughlin, M. E. , Tuveson, D. A. , Grimm, J. , Lintault, L. , … Jacks, T. (2007). Restoration of p53 function leads to tumour regression in vivo. Nature, 445, 661–665. [DOI] [PubMed] [Google Scholar]
- Warfel, N. , & El‐Deiry, W. (2013). p21WAF1 and tumourigenesis: 20 years after. Current Opinion in Oncology, 25, 52–58. [DOI] [PubMed] [Google Scholar]
- Xue, W. , Zender, L. , Miething, C. , Dickins, R. A. , Hernando, E. , Krizhanovsky, V. , … Lowe, S. W. (2007). Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature, 445, 656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , & Chen, X. (2008). Posttranscriptional Regulation of p53 and Its Targets by RNA‐Binding Proteins. Current Molecular Medicine, 8, 845–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Yin, X. , Qin, H. , Zhu, F. , Liu, H. , Yang, W. , … Deng, H. (2008). Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell, 3, 475–479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


