Abstract
We present here a highly efficient sensor for bacteria that provides an olfactory output, allowing detection without the use of instrumentation, and with a modality that does not require visual identification. The sensor platform uses nanoparticles to reversibly complex and inhibits lipase. These complexes are disrupted in the presence of bacteria, restoring enzyme activity and generating scent from odorless pro-fragrance substrate molecules. This system provides rapid (15 min) sensing and very high sensitivity (102 cfu/mL) detection of bacteria using the human sense of smell as an output.
Keywords: nanoparticles, human olfaction, enzymes, self-assembly, bacteria
Graphical Abstract
The human olfactory system has evolved to detect extremely low concentrations of volatile organic compounds present in complex environments.1 Humans can discriminate more than 1 trillion olfactory stimuli, several orders of magnitude greater than their capability in visual discrimination. 2 This sensitivity and versatility makes olfaction a promising platform for biotechnological applications,3 however there have been few examples of the application of translation of sensor responses to olfactory outputs. 4–7
Nanotechnology provides new opportunities to redefine the bounds of human perception.8 There have been a wide variety of examples where the intrinsic properties of nanomaterials have been used to generate visual output, 9, 10 with additional examples of nanomaterials modulating other colorimetric processes.11–13 Engineered nanomaterials have also been shown to influence the behavior of fragrance molecules.14 In a recent study, Weder et al. demonstrated cellulose nanocrystals functionalized with pro-fragrance molecules that could be used to control the production of volatile compounds.4 These covalently bound complexes remain odorless until functional groups are cleaved in response to specific external stimuli, generating pungent aroma molecules.5 Taken together, we hypothesized that pro-fragrances in combination with surface-engineered nanomaterials could provide reactive constructs to transduce molecular interactions into outputs that could be ‘read out’ through our sense of smell, providing a useful sensor modality for detection of bacteria that provides a potential strategy for combatting the threat of bacterial drinking water contamination that contributes to over 1.5 million deaths worldwide a year.15,16
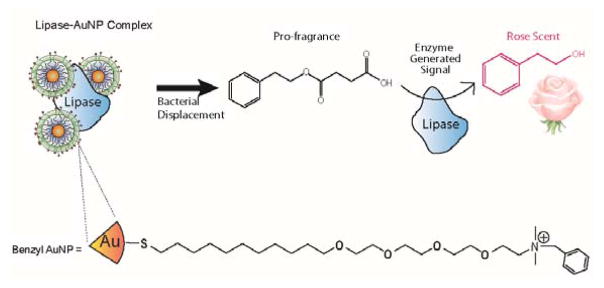
We use a supramolecular-based approach to generate an effective smell-based sensor platform for bacteria. The system is comprised of three tunable components: 1) surface functionalized nanoparticles, 2) pro-fragrance molecules, and 3) enzymes to cleave the pro-fragrances to generate the olfactory output (Figure 1). In this sensor, the surface moieties of the nanoparticles behave as both selective recognition elements for analytes present in solution and to reversibly inhibit the complexed enzymes.17 The pro-fragrance molecules18 provide a ‘turn-on’ response for the sensor system, going from odorless to strongly odiferous upon cleavage by the enzyme. Finally, the enzyme provides a strategy for amplifying the output, generating multiple fragrance molecules per recognition event.17 Bringing these components together provides a sensitive sensor system for bacteria, allowing human subjects to rapidly detect bacteria in solution at levels as low as 102 cfu/mL, a relevant limit of detection for overall bacterial load in drinking water, and consistent with other recently published sensor systems.19,20,21,22
Figure 1.
Schematic representation of sensor elements used in this study. Cationic AuNPs bind with the anionic enzyme inhibiting the catalysis of the pro-fragrance into scent. Bacteria present in solution compete for the AuNP surface and displace the enzyme inducing the production of the rose fragrance.
Results and Discussion
Our sensor design uses nanoparticles to both recognize the bacteria and to inhibit the fragrance-generating enzyme. We chose AuNPs possessing ligands with terminal benzyl headgroups, as these nanoparticles have been shown to interact strongly with the anionic cell surface of bacteria. 23,24 We used the robust and industrially used Candida Rugosa lipase as the enzymatic amplifier, 25 relying on the negative charge of the protein to provide electrostatic complementarity with the cationic nanoparticle, and hence inhibiting catalysis.11,17,26,27 Given the ability of human olfaction to discern an enormous variety of scents, we had a wide range of pro-fragrance options to choose from. We ultimately chose the succinic acid ester of phenylethyl alcohol (SAEPE) as our substrate/pro-fragrance, due to the low odor threshold of phenylethyl alcohol,28 coupled with the orthogonality of the pleasant rose scent with odors commonly found in contaminated drinking water.
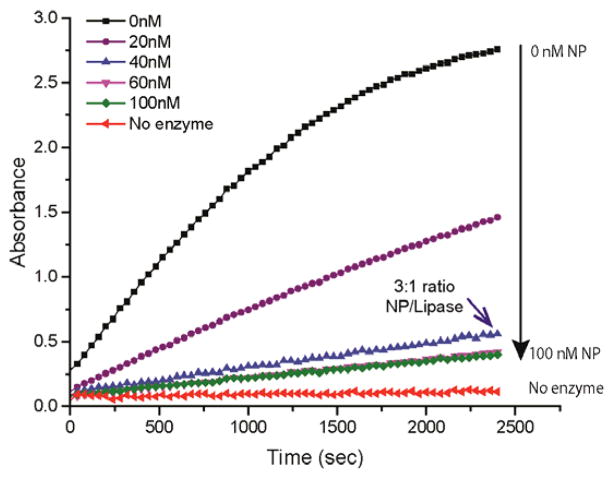
We initially performed a colorimetric assay to optimize the AuNP:lipase ratio required for inhibition. These studies were performed using p-nitrophenylbutyrate (pNPB) in sodium phosphate buffer solution (5 mM, pH 7.4). As shown in Figure 2, an approximately 3:1 AuNP to lipase ratio provided essentially complete inhibition of the lipase. This AuNP:lipase ratio was used to generate the nanozyme complex for all further studies. This colorimetric assay was able to detect both Gram positive and negative bacteria, including: E. coli, B. subtilis, M. luteus, and P. aeruginosa (Figure S1), indicating the generality of the enzyme activation process.
Figure 2.
Lipase inhibition assay in the presence of benzyl AuNP. Lipase (15 nM) was incubated with a series of benzyl AuNP concentrations before adding the colorimetric substrate p-NPB (0.6 mM).
We next turned to bacterial sensing using the enzyme platform, beginning with an instrument-based analytical strategy. These studies used E. coli as a non-pathogenic “safe” bacteria strain to minimize health concerns in both the instrumental and human studies. 29 Solutions of the sensor elements were incubated for 30 minutes prior to the addition of the pro-fragrance. We then used headspace gas chromatography to quantify the production of scent generated by our bacterial sensor.30 The concentration of the volatile product present in the headspace of the sample vial was quantified according to an external calibration curve (see Supporting Information). As shown in Figure 3, the uninhibited lipase cleaves significantly more pro-fragrance than the nanoparticle-enzyme complex and controls. Significantly, no signal was observed using the substrate alone and bacteria, indicating that the bacteria do not hydrolyze the pro-fragrance in the timeframe studied. As expected, the sensor system generated measurable and distinctly different signals in the presence of 104 and 106 cfu/mL of E. coli.
Figure 3.
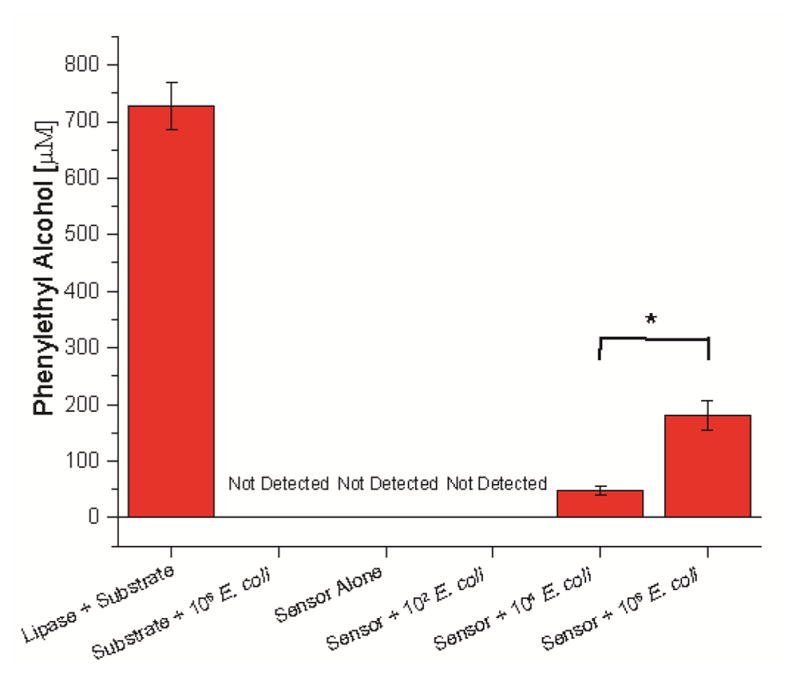
Headspace gas chromatography analysis of sensor response to increasing concentrations of bacteria. Samples were prepared in triplicate. Error bars represent standard deviations of the measurements. *= p< 0.05, ***=p<0.001.
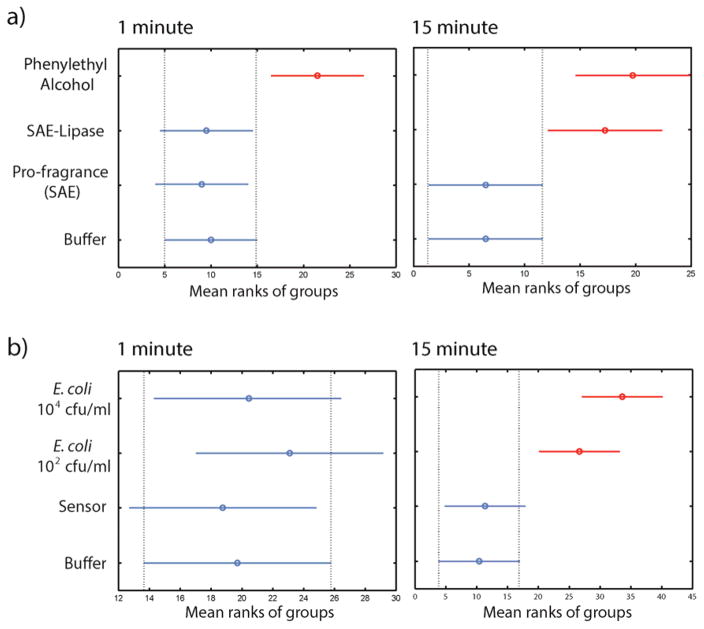
Having established the generation of fragrance output, we next determined the ability of humans to serve as “detectors”. Ten volunteers were asked to smell glass vials at two time points: 1 minute as a control, and 15 minutes for sensing, with the interval chosen to ensure olfactory clearance.31 They ranked the samples in order from least/no smell (1) to strongest smell (5), and the raw ranking order data were analyzed using the Kruskal-Wallace H-test. Initial studies focused on the sensitivity of the scent response. As expected, participants were able to detect the phenylethyl alcohol at both time points (Figure 4a). In contrast, the enzyme-substrate pair was not detected at 1 minute, but readily discerned at 15 minutes. In our sensing studies, no significant difference in response was observed across conditions at 1 minute under any conditions; however after 15 minutes participants were successfully able to detect E. coli concentrations at both 102 and 104 cfu/mL (Figure 4b) with high significance relative to the controls. Interestingly, 102 cfu/mL of E. coli did not produce a detectable signal using gas chromatography, demonstrating that human olfaction was more sensitive than the chromatographic method.
Figure 4.
Human olfactory detection studies. (A) Lipase activity test in the presence of the pro-fragrance SAEPE was carried out with six participants. SAEPE only and 5 mM Phosphate buffer were used as the negative control. The hydrolyzed form of SAEPE was used as the positive control (strong standard). Hydrolyzed SAEPE and SAEPE in the presence of uninhibited lipase are significantly different from the negative controls SAEPE alone (p<0.01 and p<0.01, respectively) after 15 minutes. (B) With ten participants, olfactory detection of E. coli at 102 and 104 cfu/mL were compared to the controls of just buffer and sensor only after 15 minutes. The olfactory signals from the vials which contained 102 and 104 cfu/mL of E. coli are significantly different from the signal from the sensor-only vial (p<0.001 and p<0.0001, respectively).
Conclusions
In summary, we report here the development of a supramolecular-based sensor that uses the human olfactory system to read out the response. This sensor was able to detect bacteria with high sensitivity. These studies demonstrate that by controlling the behavior of responsive nanomaterials at the molecular level, we can alter how human beings observe their surroundings in a manner that is otherwise impossible. We believe this responsive strategy can be broadly applied to other surface functionalized nanoparticles and enzymes to provide sensing of a wide variety of analytes, with the availability of an almost limitless number of aroma profiles providing versatility unavailable with other transduction strategies.
Materials and Methods
All reagents/materials were purchased from Fisher Scientific and used as received. Benzyl functionalized AuNPs were synthesized according to previous reports.32
Bacteria Growth Conditions
Bacteria were cultured in LB medium at 37 °C and 275 rpm until stationary phase. The cultures were then harvested by centrifugation and washed with 0.85 % sodium chloride solution for three times. Concentrations of resuspended bacterial solution were determined by optical density measured at 600 nm. 5 mM sodium phosphate buffer was used to make dilutions of bacterial solutions.
Plate Reader Assay
Lipase inhibition assay was done at 25 °C with the final concentrations in Costar clear 96 well plate of 15 nM lipase, 0.6 mM pNPB, and 20, 40, 60, 80, 100 nM benzyl AuNP. Lipase and benzyl AuNP were first incubated for 30 minutes in 96 well plate to insure their interaction reaches equilibrium, then 10 μL of substrate p-NPB was added into the well. The activity of lipase was monitored every 30 seconds for a total of 40 minutes time frame at the absorbance of 405 nm.
Human Trial Assays
Olfactory detection of lipase activity
Four different solutions were made in 20 mL glass vials with a final volume of 1 mL each. The volume of 1 mL was chosen to maintain the easy-to-use format of the sensor for eventual on-site detection use. 5 mM sodium phosphate buffer and 4 mM SAEPE were used as the negative controls and the rose scent (2-Phenylethyl alcohol) was used as the positive control, a strong standard. The activity of lipase was assessed by incubating 100 nM of lipase with 4 mM of SAEPE for 20 minutes. The participants were asked to smell these samples and rank them in the order from 1 to 5 with 1 has the lightest smell and 5 has the strongest smell.
Olfactory detection of E. coli
The same procedure was followed as above for buffer and sensor samples. For the E. coli-containing vials, 100 nM lipase was incubated with 300 nM benzyl AuNP for 30 minutes, and then 10 μL of E. coli was added into each vial so that the final concentrations of E. coli in each vial are 102 and 104 cfu/mL.
Gas Chromatography Head-Space Analysis
Headspace phenylethyl alcohol was measured using a gas chromatography (model GC-17A, Shimadzu Co., Tokyo, Japan) equipped with a solid-phase microextraction (SPME) auto injector (model AOC-5000, Shimadzu Co., Tokyo, Japan). Samples (1 mL) in 20 mL glass vials capped with aluminum caps with polytetrafluoroethylene (PTFE)/silicone septa. Samples were prepared using 500 nM lipase, 1.5 μM benzyl AuNP, and 4 mM of SAEPE. A 50/3 μm divinylbenzene (DVB)/carboxen/polydimethylsiloxane (PDMS) stable flex (SPME) fiber (Supelco Co., Bellefonte, PA) was then inserted into the vial headspace for 2 min to absorb volatiles. The fiber was transferred to the GC injector port (250 °C) for 3 min. The injection port was operated in split mode, and the split ratio was set at 20:1. Volatiles were separated on a fused-silica capillary Equity-1 Supelco column (30 × 0.25 mm inner diameter × 25 μm) coated with 100% PDMS at an initial oven temperature of 70 °C to final temperature of 220 °C over 10 min (step rate 15 °C/min). A flame ionization detector was used at a temperature of 250 °C. Phenylethyl alcohol concentrations were determined from peak areas using a standard curve made from dilutions of phenylethyl alcohol in 5 mM sodium phosphate buffer. Each measurement was performed in triplicate and results were expressed as mean values ± standard deviation.
Kruskal-Wallace H-test
Kruskal-Wallis test is a non-parametric version of one-way ANOVA which is applied when the assumption of normal (Gaussian) distribution is not met. This test can compare the medians of multiple samples to determine if they come from the same population or not. This methodology uses ranks of the data to compare the test statistics. To do so, the results from all groups are pooled and arranged in rank order from smallest to largest. The numeric index of this ordering is then used to evaluate the null hypothesis (sample are coming from the same distribution) using chi-square statistics. MATLAB software (MATLAB and Statistics Toolbox Release 2012b, The MathWorks, Inc., Natick, Massachusetts, United States) was used to perform Kruskal-Wallis test.33,34
Supplementary Material
Acknowledgments
This research was supported by Firmenich SA, the NIH (GM077173), the National Academies of Science, Pakistan-U.S. Science and Technology Cooperation Program, and the Manning Fund at the University of Massachusetts. The authors thank Prof. Julian McClements, Prof. Eric Decker, and Jean Alamed, from the Department of Food Science at the University of Massachusetts for the use and assistance using the GC-Headspace instrumentation. VR thanks Michael Famulok for the thoughtful suggestion leading to the genesis of this project.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
Additional experiments including (1) colorimetric assay for bacterial detection, (2) NMR of profragrance, (3) GC calibration curve, and (4) serial dilutions of the rose fragrance.
References
- 1.Sela L, Sobel N. Human Olfaction: a Constant State of Change-Blindness. Exp Brain Res. 2010;205:13–29. doi: 10.1007/s00221-010-2348-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushdid C, Magnasco MO, Vosshall LB, Keller A. Humans Can Discriminate More Than 1 Trillion Olfactory Stimuli. Science. 2014;343:1370–1372. doi: 10.1126/science.1249168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hellwig M, Henle T. Baking, Ageing, Diabetes: A Short History of the Maillard Reaction. Angew Chemie Int Ed. 2014;53:10316–10329. doi: 10.1002/anie.201308808. [DOI] [PubMed] [Google Scholar]
- 4.Kuhnt T, Herrmann A, Benczédi D, Foster EJ, Weder C. Functionalized Cellulose Nanocrystals as Nanocarriers for Sustained Fragrance Release. Polym Chem. 2015;6:6553–6562. [Google Scholar]
- 5.Kuhnt T, Herrmann A, Benczédi D, Weder C, Foster EJ. Controlled Fragrance Release from Galactose-Based pro-Fragrances. RSC Ad. 2014;4:50882–50890. [Google Scholar]
- 6.Xu YQ, Zhang ZY, Ali MM, Sauder J, Deng XD, Giang K, Aguirre SD, Pelton R, Li YF, Filipe CDM. Turning Tryptophanase into Odor-Generating Biosensors. Angew Chem Int Ed. 2014;53:2620–2622. doi: 10.1002/anie.201309684. [DOI] [PubMed] [Google Scholar]
- 7.Mohapatra H, Phillips ST. Using Smell To Triage Samples in Point-of-Care Assays. Angew Chem Int Ed. 2012;51:11145–11148. doi: 10.1002/anie.201207008. [DOI] [PubMed] [Google Scholar]
- 8.Whitesides GM. Reinventing Chemistry. Angew Chemie Int Ed. 2015;54:3196–3209. doi: 10.1002/anie.201410884. [DOI] [PubMed] [Google Scholar]
- 9.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective Colorimetric Detection of Polynucleotides based on the Distance-Dependent Optical Properties of Gold Nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 10.de la Rica R, Stevens MM. Plasmonic ELISA for the Ultrasensitive Detection of Disease Biomarkers with the Naked Eye. Nat Nanotechnol. 2012;7:821–824. doi: 10.1038/nnano.2012.186. [DOI] [PubMed] [Google Scholar]
- 11.Miranda OR, Chen HT, You CC, Mortenson DE, Yang XC, Bunz UHF, Rotello VM. Enzyme-Amplified Array Sensing of Proteins in Solution and in Biofluids. J Am Chem Soc. 2010;132:5285–5289. doi: 10.1021/ja1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang ZW, Le NDB, Gupta A, Rotello VM. Cell Surface-based Sensing with Metallic Nanoparticles. Chem Soc Rev. 2015;44:4264–4274. doi: 10.1039/c4cs00387j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miranda OR, Li XN, Garcia-Gonzalez L, Zhu ZJ, Yan B, Bunz UHF, Rotello VM. Colorimetric Bacteria Sensing Using a Supramolecular Enzyme-Nanoparticle Biosensor. J Am Chem Soc. 2011;133:9650–9653. doi: 10.1021/ja2021729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan B, Landis RF, Jerri Ha, Normand V, Benczédi D, Ouali L, Rotello VM. Hybrid Organic-Inorganic Colloidal Composite “Sponges” via Internal Crosslinking. Small. 2015;11:1302–1309. doi: 10.1002/smll.201401753. [DOI] [PubMed] [Google Scholar]
- 15.WHO, UNICEF. Progress on Drinking Water and Sanitation: 2014 Update. Geneva, Switzerland: WHO Press; 2014. [Google Scholar]
- 16.WHO. Guidelines for Drinking-Water Quality. 4. Geneva, Switzerland: WHO Press; 2011. [Google Scholar]
- 17.Miranda OR, Li XN, Garcia-Gonzalez L, Zhu ZJ, Yan B, Bunz UHF, Rotello VM. Colorimetric Bacteria Sensing Using a Supramolecular Enzyme-Nanoparticle Biosensor. J Am Chem Soc. 2011;133:9650–9653. doi: 10.1021/ja2021729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrmann A. Controlled Release of Volatiles under Mild Reaction Conditions: From Nature to Everyday Products. Angew Chem Int Ed Engl. 2007;46:5836–5863. doi: 10.1002/anie.200700264. [DOI] [PubMed] [Google Scholar]
- 19.Tripathi SM, Bock WJ, Mikulic P, Chinnappan R, Ng A, Tolba M, Zourob M. Long period grating based biosensor for the detection of Escherichia coli bacteria. Biosens Bioelectron. 2012;35(1):308–312. doi: 10.1016/j.bios.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Knoll W, Dostalek J. Bacterial Pathogen Surface Plasmon Resonance Biosensor Advanced by Long Range Surface Plasmons and Magnetic Nanoparticle Assays. Anal Chem. 2012;84(19):8345–8350. doi: 10.1021/ac301904x. [DOI] [PubMed] [Google Scholar]
- 21.Chang JB, Mao S, Zhang Y, Cui SM, Zhou GH, Wu XG, Yang CH, Chen JH. Ultrasonic-assisted self-assembly of monolayer graphene oxide for rapid detection of Escherichia coli bacteria. Nanoscale. 2013;5(9):3620–3626. doi: 10.1039/c3nr00141e. [DOI] [PubMed] [Google Scholar]
- 22.Bartram J, Cotruvo J, Exner M, Fricker C, Glasmacher A. Heterotrophic Plate Count Measurement in Drinking Water Safety Management - Report of an Expert Meeting Geneva, 24–25 April 2002. Int J Food Microbiol. 2004;92(3):241–247. doi: 10.1016/j.ijfoodmicro.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Robinson SM, Gupta A, Saha K, Jiang Z, Moyano DF, Sahar A, Riley MA, Rotello VM. Functional Gold Nanoparticles as Potent Antimicrobial Agents against Multi-Drug-Resistant Bacteria. ACS Nano. 2014;8:10682–10686. doi: 10.1021/nn5042625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller KP, Wang L, Benicewicz BC, Decho AW. Inorganic Nanoparticles Engineered to Attack Bacteria. Chem Soc Rev. 2015;44:7787–7807. doi: 10.1039/c5cs00041f. [DOI] [PubMed] [Google Scholar]
- 25.Jeong Y, Duncan B, Park MH, Kim C, Rotello VM. Reusable Biocatalytic Crosslinked Microparticles Self-Assembled from Enzyme-Nanoparticle Complexes. Chem Commun (Camb) 2011;47:12077–12079. doi: 10.1039/c1cc14448k. [DOI] [PubMed] [Google Scholar]
- 26.Rana S, Le NDB, Mout R, Saha K, Tonga GY, Bain RES, Miranda OR, Rotello CM, Rotello VM. A multichannel nanosensor for instantaneous readout of cancer drug mechanisms. Nat Nanotechnol. 2015;10:65–69. doi: 10.1038/nnano.2014.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rana S, Le NDB, Mout R, Duncan B, Elci SG, Saha K, Rotello VM. A Multichannel Biosensor for Rapid Determination of Cell Surface Glycomic Signatures. Acs Cent Sci. 2015;1:191–197. doi: 10.1021/acscentsci.5b00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsukatani T, Miwa T, Furukawa M, Costanzo RM. Detection Thresholds for Phenyl Ethyl Alcohol Using Serial Dilutions in Different Solvents. Chem Senses. 2003;28:25–32. doi: 10.1093/chemse/28.1.25. [DOI] [PubMed] [Google Scholar]
- 29.Yang W, Zerbe H, Petzl W, Brunner RM, Guenther J, Draing C, von Aulocke S, Schuberth HJ, Seyfert HM. Bovine TLR2 and TLR4 Properly Transduce Signals from Staphylococcus aureus and E-coli, but S-aureus Fails to both Activate NF-kappa B in Mammary Epithelial Cells and to Quickly Induce TNF Alpha and Interleukin-8 (CXCL8) Expression in the Udder. Mol Immunol. 2008;45:1385–1397. doi: 10.1016/j.molimm.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 30.Poole CF. Gas Chromatography. Waltham: Elsevier; 2012. Print. [Google Scholar]
- 31.Philpott CM, Wolstenholme CR, Goodenough PC, Clark a, Murty GE. Olfactory Clearance: What Time Is Needed in Clinical Practice? J Laryngol Otol. 2008;122:912–917. doi: 10.1017/S0022215107000977. [DOI] [PubMed] [Google Scholar]
- 32.Tonga GY, Jeong Y, Duncan B, Mizuhara T, Mout R, Das R, Kim ST, Yeh YC, Yan B, Hou S, et al. Supramolecular Regulation of Bioorthogonal Catalysis in Cells Using Nanoparticle-Embedded Transition Metal Catalysts. Nat Chem. 2015;7:597–603. doi: 10.1038/nchem.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonald John H. Handbook of Biological Statistics. Baltimore, MD: Sparky House Publishing; 2009. Print. [Google Scholar]
- 34.Miller JC, Miller JN. Statistics for Analytical Chemistry. Chichester: E. Horwood; 1984. Print. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.