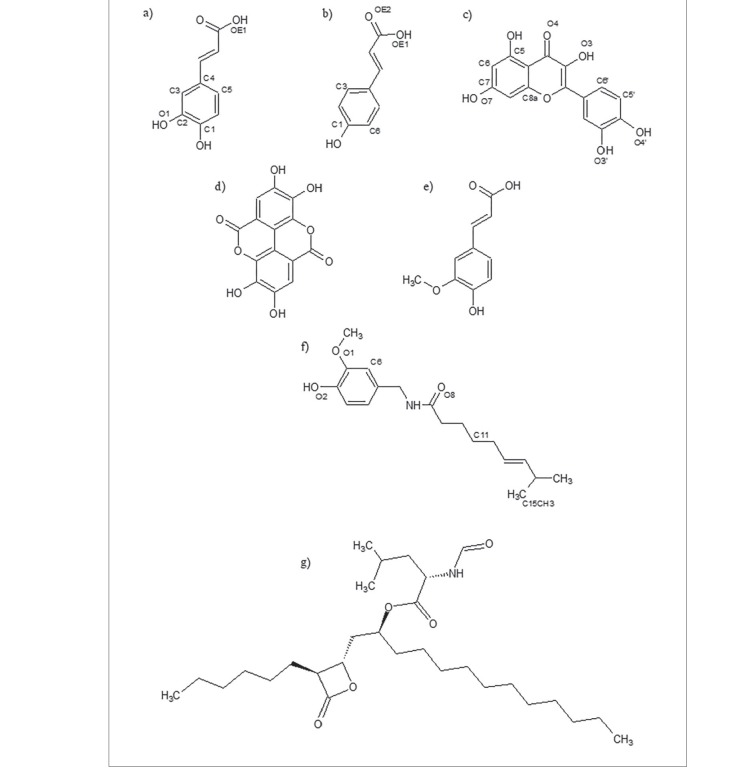
Summary
The inhibitory activity and binding characteristics of caffeic acid, p-coumaric acid, quercetin and capsaicin, four phenolic compounds found in hot pepper, against porcine pancreatic lipase activity were studied and compared to hot pepper extract. Quercetin was the strongest inhibitor (IC50=(6.1±2.4) µM), followed by p-coumaric acid ((170.2±20.6) µM) and caffeic acid ((401.5±32.1) µM), while capsaicin and a hot pepper extract had very low inhibitory activity. All polyphenolic compounds showed a mixed-type inhibition. Fluorescence spectroscopy studies showed that polyphenolic compounds had the ability to quench the intrinsic fluorescence of pancreatic lipase by a static mechanism. The sequence of Stern-Volmer constant was quercetin, followed by caffeic and p-coumaric acids. Molecular docking studies showed that caffeic acid, quercetin and p-coumaric acid bound near the active site, while capsaicin bound far away from the active site. Hydrogen bonds and π-stacking hydrophobic interactions are the main pancreatic lipase-polyphenolic compound interactions observed.
Key words: polyphenolic compounds, pancreatic lipase, enzymatic inhibition, molecular docking, anti-obesity effect
Introduction
Obesity is a global public health concern that has been described as a risk factor for diseases such as cardiovascular, hypertension, type II diabetes, and some forms of cancer (1). The seriousness of this concern is evidenced by the warning issued by the World Health Organization that 2.8 million people die every year because of obesity-related diseases (2). Several strategies have been developed to overcome this problem, among them the use of drugs and surgeries, or most frequently changes in alimentary habits. These changes include not only diets low in caloric intake, but also inclusion of foods rich in bioactive compounds that exert specific beneficial effects, such as polyphenolic compounds. Several mechanisms may explain the anti-obesity actions of polyphenolic compounds, including the interaction and inhibition of digestive enzymes like pancreatic lipase (EC 3.1.1.3) (3, 4), an enzyme responsible for the hydrolysis of 50-70% of ingested lipids (5).
Spices are examples of bioactive-rich foods that have been identified for their beneficial effects on human health (6). Pancreatic lipase inhibition has been reported by different polyphenolic compound-rich extracts from herbs and spices such as tea (Camellia sinensis) (7), anise myrtle (Syzygium anisatum) (8), chinese liquorice (Glycyrrhiza uralensis) (9) and muscadine grapes (Vitis rotundifolia) (10). Inhibition of pancreatic lipase by individual polyphenolic compounds has also been reported, mainly as inhibition percentage and inhibition saturation kinetics, showing IC50 values (concentration of compound/sample required to inhibit enzyme activity by 50%) (9, 11). Among polyphenolic compounds, quercetin has been reported as one of the best pancreatic lipase inhibitors, while phenolic acids have shown low inhibitory activity (1, 10). Sergent et al. (1) obtained IC50 values for quercetin and orlistat of (21.5±9.4) and (32.0±8.5) µM, respectively. Polyphenolic compounds are mostly found in food as glycosylated derivatives; however, the relevance of studying pure polyphenolic compounds in their form of aglycones comes from the fact that these compounds interact with the digestive enzymes on the brush border cells of the small intestine mainly as aglycones (12). Polyphenolic compounds extracted from hot peppers are recognized for their antioxidant activity (13), and anti-obesity properties, related to an increase in energy expenditure (14); however, no information about its possible inhibitory activity against pancreatic lipase has been reported. According to Chen and Kang (15), jalapeño hot peppers contain several phenolic acids such as p-coumaric and caffeic acids, and flavonoids like quercetin and luteolin. Hot peppers also contain capsaicin, a compound unique to the Capsicum species, and responsible for their pungency. Capsaicin is known for its anti-inflammatory properties (15), but barely studied for its ability to inhibit digestive enzymes. Interaction of quercetin with pancreatic lipase has been reported, but there are discrepancies on the reported results. Both competitive (10) and non-competitive (16) inhibition modes have been reported. In the case of phenolic acids such as caffeic acid, p-coumaric acid and capsaicin, as far as we know there are no reports describing the mechanism of their inhibition of pancreatic lipase.
The aim of the present study is to determine the inhibition mechanisms of some polyphenolic compounds (caffeic acid, p-coumaric acid and quercetin) present in jalapeño hot peppers towards pancreatic lipase by evaluating their IC50 values, kinetic parameters and intrinsic fluorescence, and by performing docking analysis to predict their interactions with residues of pancreatic lipase binding site. The inhibitory effect of a dry smoked jalapeño pepper polyphenolic extract was also analyzed.
Materials and Methods
Materials
Porcine pancreatic lipase type II, p-nitrophenyl laureate (pNPL), caffeic acid, p-coumaric acid, capsaicin, quercetin, Tris, bovine serum albumin (BSA) and Triton X-100 were purchased from Sigma-Aldrich (St. Louis, MO, USA). Methanol and ethanol were purchased from J.T.Baker (Center Valley, PA, USA). Orlistat capsules from Redustat® (Liomont® Laboratories, Toluca, Mexico) were purchased in a local drugstore.
Extraction of polyphenolic compounds from dry smoked jalapeño pepper (chipotle)
Extraction of polyphenolic compounds was carried out according to Moreno-Escamilla et al. (13), the phenolic content was determined by the Folin-Ciocalteu’s method, and results were expressed in µg of gallic acid equivalents (GAE) per g of extract.
Pancreatic lipase activity assay
Commercial pancreatic lipase was prepared according to McDougall et al. (17) with some modifications. Pancreatic lipase was dissolved in 20 mM Tris buffer (pH=7.2) at the concentration of 2.0 mg/mL, centrifuged at 1411´g in Eppendorf 5810R centrifuge (Eppendorf AG, Hamburg, Germany) for 20 min, and the supernatant was filtered in a polyvinylidene fluoride (PVDF) membrane filter (0.22 µm; Durapore®, Hercules, CA, USA). Purity of the lipase and co-lipase was estimated by SDS-PAGE analysis (93% purity; EDAS 290, Kodak®, New York, NY, USA). Protein concentration was determined by Bradford method (18), reading the absorbance of cationic form of the dye Coomassie Blue G250 at 595 nm, and results were expressed in mg/mL using bovine serum albumin (BSA; 1 mg/mL) as standard. Pancreatic lipase substrate (pNPL, 0.08% mass per volume) was dissolved in 5 mM sodium acetate solution (pH=5.0, 1% Triton X-100). Final pNPL concentrations were 11.7-124.2 μM. Caffeic acid, p-coumaric acid and capsaicin were dissolved separately at three different concentrations (25, 50 and 75 μg/mL) in methanol. Due to its lower solubility, quercetin was dissolved in ethanol at the same concentrations. An Orlistat solution was prepared at 1.2 mg/mL in methanol (19). Stability of the compounds in the solvent was tested for the time of assay by measuring their UV-Vis spectra (Agilent 8453 HP UV-Vis spectrometer G1103A; Agilent Technologies, Santa Clara, CA, USA).
The control assay contained 500 μL of Tris 200 mM working buffer solution (pH=8.2), 260 μL of distilled water, 140 μL of pancreatic lipase solution and 40 μL of substrate solution. For the inhibition studies, 60 μL of each polyphenolic compound (or extract) and 200 μL of distilled water were used. The substrate was added to start the reaction, then the absorbance was recorded at 400 nm during 1000 s at 37 °C. All samples were assayed in triplicate.
The inhibition percentage was calculated according to Dalar and Konczak (20) from the measured absorbances of control and sample (endpoint):
where Acb is the absorbance of the control blank, Ac is the absorbance of the control, Asb is the absorbance of the sample blank and As is the absorbance of the sample. Blanks were prepared without the pancreatic lipase solution and the substrate solution, respectively. IC50 was calculated from a substrate concentration versus inhibition percentage plot.
Accurate Ki values of polyphenolic compounds were determined by calculating the apparent maximum reaction rate (vmax) and Michaelis-Menten constant (Km) by both linear (Lineweaver-Burk) and non-linear (Michaelis- -Menten) analyses First, apparent vmax and Km values were obtained from the least-square regression lines by plotting the reciprocal of the substrate concentration [S] against the reciprocal of the enzyme reaction velocity as in the work by Alvarez-Parrilla et al. (21):
These values were used as initial values for the determination of the parameters by non-linear analysis using Sigma Plot v. 13.0 (22), by fitting the value of the experimental initial reaction rate (v0) as a function of the initial inhibitor concentration to the Michaelis-Menten equation (Eq. 3). The v0 values were obtained from a linear curve fitting (slopes) of a time-course plot of the enzymatic reaction:
Ki and Ki’ (dissociation constant of free enzyme and enzyme-substrate complex, respectively) values for a mixed- -type inhibition were obtained from the following equations (23):
 |
where apparent Km’ and vmax’ are the values in the presence of the inhibitor, and [I] is the inhibitor concentration. These calculations were done for each inhibitor concentration.
Polyphenolic compound-pancreatic lipase interaction measured by fluorescence spectra
Different volumes of caffeic acid, p-coumaric acid, quercetin and capsaicin (or extract) at 0.0125 M were added into a 10-mL flask (final concentrations 0.025-0.25·10-4 M). A volume of 1.0 mL of the pancreatic lipase solution (10-4 M) was added, and the solution was diluted to 10 mL with the working buffer and incubated at 37 °C for 1 h in darkness. Emission fluorescence spectra were measured in the 300-550 nm interval, at a fixed excitation wavelength of 280 nm in a RF 5301-PC spectrofluorometer (Shimadzu, Columbia, MD, USA). Even though the intrinsic fluorescence of pancreatic lipase is attributed to three amino acid residues, phenylalanine (Phe), tyrosine (Tyr), and tryptophan (Trp), the fluorescence of Phe and Tyr residues at this wavelength is negligible, so intrinsic fluorescence of the enzyme is mainly due to Trp (24). The polyphenolic compounds do not exhibit Förster resonance energy transfer (FRET) at that excitation wavelength (25). The Stern-Volmer equation was used to determine the presence of a quenching mechanism, by plotting F0/F against [Q]:
where F0 and F are the fluorescence intensities at the maximum λem in the absence and presence of an interacting molecule (polyphenol), respectively, and [Q] is the molar concentration of the interacting molecule. The kq is the bimolecular quenching constant, τ0 is the fluorescence lifetime in the absence of the quencher, and Ksv is the Stern-Volmer quenching constant (26). A τ0 of 1.59 ns was used for pancreatic lipase, according to previously published results (27).
Molecular docking
The crystal structure of pancreatic lipase (Protein Data Bank (PDB) (28) code 1ETH (29)) was used as template for the generation of pancreatic ligand complex models. Ligand structures: caffeic acid, p-coumaric acid and quercetin were obtained from crystallographic complexes with different proteins (PDB codes: 4EYQ (30), 1D7E (31) and 1E8W (32)). Capsaicin structure was generated and minimized using PyMOL (33). The docking analyses were performed with AutoDock Vina using the interphase installed in USCF Chimera (34) run with the default parameters and a search volume of approximately the same size as pancreatic lipase. The crystal structure of pancreatic lipase was considered rigid, and polyphenolic compounds and capsaicin structures were considered flexible during docking. The possible interactions of the best-scored solution of the models for each ligand were analyzed.
Statistical analysis of the enzyme kinetics and the binding experiments
All the experimental assays were run in triplicates. Results are expressed as mean values±standard deviation. Analysis of variance and Fisher’s least significant difference analysis were performed by SPSS v. 20 software (35) for the determination of statistical differences between treatments.
Results and Discussion
Pancreatic lipase activity
The effect of four compounds: caffeic acid, p-coumaric acid, quercetin and capsaicin (Fig. 1), commonly found in hot pepper, and the hot pepper extract, on the activity of pancreatic lipase were monitored at saturation conditions, using 62.1 μM pNPL. Enzyme inhibition percentage increased in a hyperbolic trend, as the concentration of the polyphenolic compounds increased. p-Coumaric acid and capsaicin appear to have weak inhibition power, since lower inhibition was observed even at higher concentrations (data not shown). The hot pepper extract showed no pancreatic lipase inhibition (36). To compare the inhibitory capacity of the analyzed compounds, IC50 values were determined for caffeic acid, p-coumaric acid, quercetin and orlistat (Table 1). Caffeic acid had the highest IC50, followed by p-coumaric acid and finally quercetin, which showed an IC50 statistically similar to that of orlistat. In agreement with these results, You et al. (10) observed that quercetin had lower IC50 (276.8 µM) than ellagic acid (342.4 µM). Both quercetin and ellagic acid have the same molecular mass of 302.2 g/mol, respectively; however, their molecular structures are different (Figs. 1c and d). Quercetin, which is a flavonoid, has two aromatic rings in its structure and one heterocyclic ring in a linear structure, while ellagic acid possesses two aromatic rings and two heterocyclic rings, presenting a more rigid structure. This emphasizes the relevance of the structure of flavonoid in comparison with other polyphenolic structures (37). For example, the differences between ellagic acid and quercetin might be associated with their different inhibitory activities against pancreatic lipase (17). Differences between phenolic acids and flavonoids such as quercetin may be responsible for our obtained different inhibitory capacities. Similar to our results, where phenolic acids had higher IC50 values than quercetin, Sergent et al. (1) observed that quercetin IC50 ((21.5±9.4) µM) was lower than that of ferulic acid ((123.9±13.4) µM; Fig. 1e) and not significantly different from Orlistat ((32.0±8.5) µM), detected using 4-methylumbelliferone as substrate.
Fig. 1.
The chemical structures of polyphenolic compounds: a) caffeic acid, b) p-coumaric acid, c) quercetin, d) ellagic acid, and e) ferulic acid; and the compounds: f) capsaicin, and g) orlistat. Elements (C, H and O) of caffeic acid, p-coumaric acid, quercetin and capsaicin for the molecular docking analysis are indicated
Table 1. Pancreatic lipase apparent catalytic parameters (vmax, Km, Ki and Ki´) and IC50 of the hydrolysis of pNPL in the presence of different polyphenolic compounds, capsaicin and orlistat.
| Compound | c/µM | vmax/(µM/s) | Km/µM | Ki/µM | Ki´/µM | IC50/µM |
|---|---|---|---|---|---|---|
| pNPL (control) | 11.7-124.2 | (0.11±0.03)a | (1.96±0.20)ab | n.d. | n.d. | n.d. |
| Caffeic acid | 300.21 | (0.10±0.02)a | (0.30±0.06)d | (370.2±18.4)a | (388.2±30.2)a | (401.5±32.1)a |
| p-Coumaric acid | 275.00 | (0.10±0.01)a | (0.95±0.10)c | (205.7±44.1)b | (290.7±39.7)b | (170.2±20.6)b |
| Quercetin | 14.53 | (0.10±0.02)a | (2.15±0.40)a | (12.0±4.4)d | (17.9±6.9)d | (6.1±2.4)c |
| Capsaicin | 40.47 | (0.10±0.02)a | (1.80±0.19)b | (66.0±14.3)c | (50.7±13.0)c | n.d. |
| Orlistat | 9.68 | (0.09±0.03)a | (0.30±0.05)d | (16.9±4.8)d | (19.0±2.8)d | (4.0±1.0)c |
The data are presented as mean value±standard deviation of triplicate analyses. Different letters in the same row indicate statistically significant values (Fisher’s least significant difference analysis, p≤0.05) with respect to control, or between treatments for dissociation constant of free enzyme and enzyme-substrate complex (Ki and Ki´). Km=Michaelis-Menten constant, n.d.=not determined
In contrast, no significant effect of the extract on pancreatic lipase activity was observed. Similar results have been reported for black chokeberry (Aronia melanocarpa L.) extracts (38). However, other studies have shown high to moderate pancreatic lipase inhibition by plant extracts such as cumin (Cuminum cyminum) (39), green tea (Camellia sinensis) (40), muscadine grape (Vitis rotundifolia) (10), and other edible plants, for example Beijing grass (Murdannia loriformis) (41). Here, the lack of pancreatic lipase-inhibitory effect could be explained by the difference in the content of bioactive compounds compared to other extracts with high inhibitory activity, or by the synergistic/antagonistic effects among bioactive compounds in the extracts (38). According to Chen and Kang (15), hot pepper extracts contain high mass fractions (per dry mass) of p-coumaric acid ((4.56±0.11) mg/g) and capsaicin ((9.05±0.05) mg/g), both of them with low pancreatic lipase-inhibitory activity, which may explain the low effect of the extract. Another phenolic acid mainly found in hot peppers is caffeic acid ((1.32±0.08) mg/g), while quercetin is the major flavonoid ((8.86±1.03) mg/g). The observed IC50 values of p-coumaric acid, caffeic acid and quercetin were compared to their content found by Chen and Kang (15). We found that only quercetin had a higher content than its IC50 value (almost 5-fold), whereas phenolic acids had lower content than their IC50 values (i.e. almost 6-fold for p-coumaric acid). In this way, the absence of the inhibitory effect of the hot pepper extract may be explained by the low content of capsaicin and phenolic acids (lower than their IC50 values).
The effects of caffeic acid, p-coumaric acid, quercetin and capsaicin on the enzyme kinetics of pancreatic lipase using pNPL as substrate were evaluated, and results are shown in Fig. 2, where the initial velocity v0, shown as absorbance change (λ=400 nm) in arbitrary units per second, versus substrate concentration (µM) was plotted. Even though all compounds had a significant effect (p<0.05) on v0, compared to control, quercetin seemed to be the best inhibitor. To characterize the inhibitory pattern, both linear and non-linear analyses of inhibition curves of each compound and the extract were performed. Linearized plots indicated a mixed-type inhibition, in agreement with other authors (5, 10, 26), and suggested two inhibition components, competitive and non-competitive. Next, non-linear regression fit of data to Eqs. 3–5 was performed to obtain the catalytic parameters of pancreatic lipase in the presence and absence of polyphenolic compounds (Table 1). Significant differences in apparent Km values with respect to control were observed among all compounds. The apparent vmax values showed no significant differences with respect to control. Similar apparent vmax and smaller apparent Km values of quercetin than of control support the idea that a mixed-type inhibition regulates the process (42). Phenolic acids possessed significantly lower apparent Km values than control. Quercetin had an apparent Km higher than control, and capsaicin did not exhibit statistically significant differences compared to control. According to Zhang et al. (43), a higher molecular mass structure might be beneficial for decreasing enzymatic activity (higher apparent Km value), which can be attributed for example to the increase of hydroxyl group in the structure of quercetin with respect to phenolic acids. Narita and Inouye (42) observed a mixed- -type inhibition when using p-coumaric acid as inhibitor of pancreatic α-amylase activity and explained that lower apparent Km values in the presence of inhibitors meant that the binding affinity of the substrate and the inhibitor for their respective sites increases when both are present (p-coumaric acid and substrate).
Fig. 2.
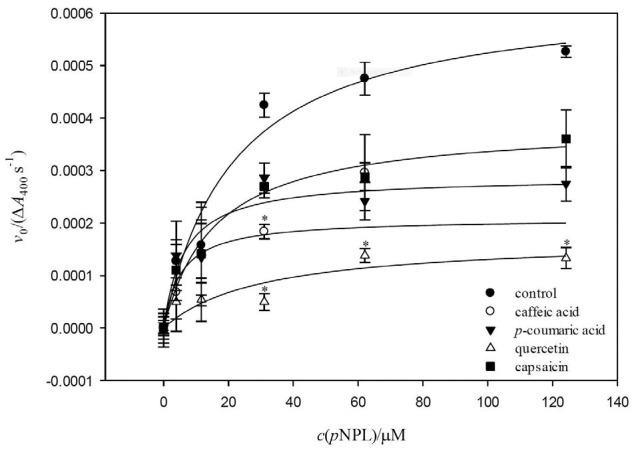
Effect of caffeic acid, p-coumaric acid, quercetin and capsaicin on the hydrolysis of p-nitrophenyl laureate (pNPL) catalyzed by pancreatic lipase. Symbols represent experimental initial reaction rate (v0 (expressed as ΔA400 nm/s))±standard deviation. The lines are Michaelis-Menten curves fitted to experimental data. Asterisk represents statistical difference between treatments (Fisher’s least significant difference analysis, p≤0.05)
Lower Ki than Ki´ values of the polyphenolic compounds (caffeic acid, p-coumaric acid and quercetin) were observed. These lower Ki´ values indicate that the inhibitor has more affinity for the enzyme-substrate complex than for the free enzyme in a mixed inhibition mechanism (42). The competitive component of mixed inhibition prevailed in the systems. All tested compounds except capsaicin had higher Ki´ values than their respective Ki values, indicating that they bind tighter to the free enzyme than to the substrate-bound form of pancreatic lipase. These results are in agreement with the competitive inhibition of pancreatic lipase activity by polyphenolic extract observed by You et al. (10). Our results may suggest that capsaicin might be binding to a different site in the enzyme, instead of the substrate binding site. In accordance with IC50 values, Ki and Ki´ values (Table 1) of quercetin were the lowest of all studied compounds. These results agree with those reported by You et al. (10). Liu (44) and Sergent et al. (1) also observed that phenolic acids were not as potent pancreatic lipase inhibitors as quercetin. Quercetin inhibitory activity on other pancreatic enzymes such as α-amylase has also been studied (45).
Binding of polyphenolic compounds to pancreatic lipase by fluorescence spectroscopy analysis
The fluorescence intensity and quenching of Trp residues from pancreatic lipase upon the incubation with polyphenolic compounds was monitored, and data were analyzed using Eq. 6. There are seven Trp residues in the pancreatic lipase that may be responsible for the fluorescence emission of this protein (24). According to crystal structure (PDB code 1ETH (28)), five of them are located in the N-terminal domain, while the other two are in the C-terminal domain, and three Trp residues are located near the active site of the enzyme. Pancreatic lipase also possesses 25 Phe residues, and 16 Tyr residues that may account for fluorescence. Changes in the fluorescence intensity of pancreatic lipase are attributed to the interaction of ligands, which results in a change in the polarity of the environment surrounding Trp residues, but changes in the environment of Phe and Tyr residues may also have minor contribution to pancreatic lipase fluorescence.
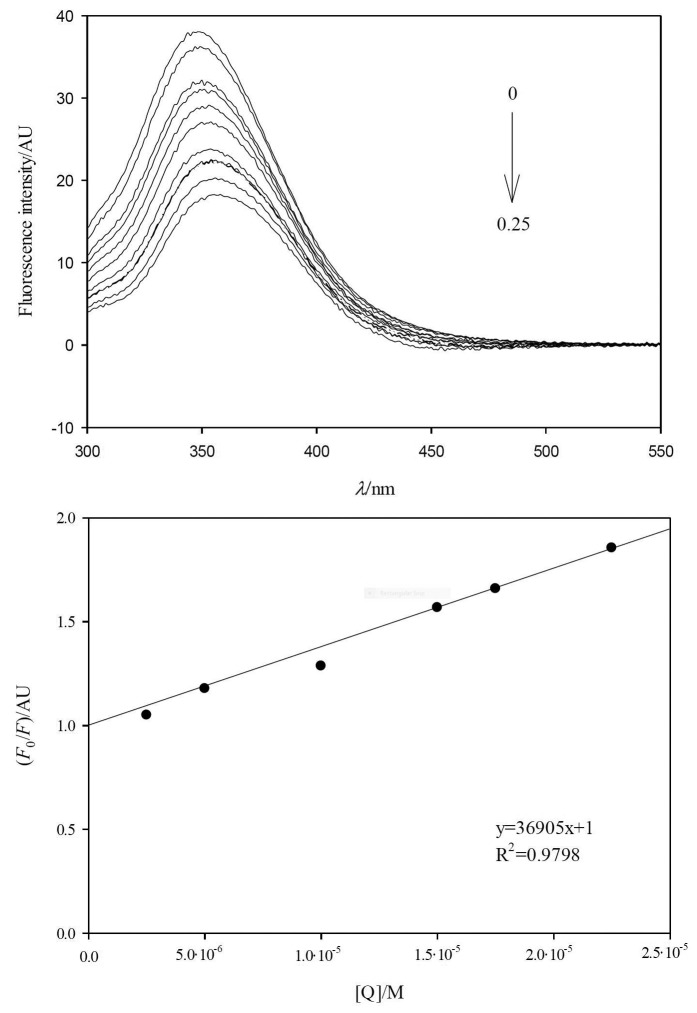
To evaluate the pancreatic lipase-polyphenolic compound interactions, series of emission spectra of pancreatic lipase at fixed concentration (2.0 mg/mL) were recorded upon the addition of different polyphenolic compound concentrations (2.5-25 μM). Fig. 3a shows that the addition of different concentrations of caffeic acid to pancreatic lipase decreased the fluorescence intensity. The same behaviour was observed of the rest of the polyphenolic compounds. A slight bathochromic shift of 12 nm was recorded in the presence of caffeic acid. The rest of the polyphenolic compounds showed similar bathochromic shifts (5-8 nm). This decrease in fluorescence intensity accompanied by a bathochromic shift in the maximum fluorescence would correspond to an increase in the polarity environment surrounding the Trp residues due to a conformational transition of the protein, which involves the exposure of Trp residues (46, 47). The small bathochromic displacement observed reflected that the environment of the Trp during the polyphenolic compounds-pancreatic lipase interaction did not change considerably (3, 26).
Fig. 3.
Effect of polyphenolic compounds on fluorescence spectra of pancreatic lipase: a) emission fluorescence spectra of pancreatic lipase (2.0 mg/mL) in the presence of different concentrations of caffeic acid (final concentrations: 0, 0.025, 0.05, 0.10, 0.125, 0.15, 0.175, 0.20, 0.225 and 0.25·10-4 M), λex=280 nm, and b) Stern-Volmer plot (Eq. 6) for the experimental data of pancreatic lipase fluorescence (λem=354 nm) at different caffeic acid concentrations as quencher [Q]. F0 and F=fluorescence intensities at the maximum of λem in the absence and presence of the polyphenolic compounds
Table 2 summarizes the Ksv and kq values of polyphenolic compounds. The linearity observed by plotting F0/F and [Q] (Fig. 3b) allowed to determine the existence of a quenching mechanism (Eq. 6). A linear Stern-Volmer plot is indicative of a single class of fluorophore equally accessible to the quencher (46). The sequence of calculated Ksv values for the polyphenolic compounds was quercetin, caffeic acid, p-coumaric acid and capsaicin. Our results agree with those of Li et al. (16), who studied the interaction of quercetin, isoquercetin and rutin with pancreatic lipase and observed that the sequence of Ksv values did not correspond to the inhibition sequence. Also, our Ksv value ((4.5±0.7)·10-4 M-1) for quercetin was similar to the value obtained by Li et al. (16) of 4.95·10-4 M-1. Contrarily to the calculated Ksv values observed by Gonçalves et al. (48) of pancreatic lipase and procyanidin fractions from grape seed, Li et al. (16) did not find a relation between the polyphenolic structures with higher molecular mass or higher number of hydroxyl groups, and higher Ksv values.
Table 2. Stern-Volmer and bimolecular quenching constant values of the pancreatic lipase interaction with caffeic acid, p-coumaric acid, quercetin and capsaicin.
| Compound | Ksv/(10-4 M -1) | R2 | kq/(10-13 M-1∙s-1) | ∆F/AU |
|---|---|---|---|---|
| Caffeic acid | (3.7±1.2)ab | 0.9798 | (2.3±0.8)ab | (19.1±1.8)a |
| p-Coumaric acid | (2.0±0.6)ab | 0.9089 | (1.3±0.2)ab | (18.7±2.3)a |
| Quercetin | (4.5±0.5)a | 0.9669 | (2.8±0.1)a | (18.1±2.0)a |
| Capsaicin | (0.40±0.01)c | 0.9885 | (0.25±0.03)c | (6.6±2.2)b |
ΔF corresponds to the difference between registered fluorescence intensity of analyzed compounds (caffeic acid, p-coumaric acid, quercetin and capsaicin) at c=1 and 25 µM. The data are presented as mean value±standard deviation of triplicate analyses. Different letters in the same row indicate statistically significant values (Fisher’s least significant difference analysis, p≤0.05). Ksv=Stern-Volmer quenching constant, kq=bimolecular quenching constant, F=fluorescence intensity
The calculated kq values were higher than 2.0·10-10 M-1·s-1, the maximum value for a dynamic mechanism in aqueous medium (16, 26). This indicates that the quenching process is an intrinsic and static process, involving only Trp residues (5, 16, 26). Static quenching has been previously reported for the interaction of pancreatic lipase with flavonoids, e.g. quercetin (16). This type of quenching is characterized by the formation of a non-fluorescent complex between the enzyme and the ligand, in which the enzyme segment contains the Trp residue (46). Thus, the observed changes in the maximum emission intensity reflected the direct interactions of the analyzed compounds with the areas located near the Trp, and the induced conformational changes of the enzyme (49).
It seems that the flavonoid quercetin, which has more hydroxyl groups in its structure than the phenolic acids (caffeic and p-coumaric acids) (37), was more effective in quenching the intrinsic fluorescence of pancreatic lipase (higher Ksv and kq values), suggesting more affinity (50). Gonçalves et al. (48) observed that Ksv and kq increased as the molecular mass of three different procyanidin fractions increased, while the IC50 decreased (higher inhibitory capacity) with respect to pancreatic lipase activity. This behaviour can be explained considering that even though the main interactions between polyphenolic compounds and pancreatic lipase are recognized as non-covalent (5, 7, 16, 24), the polar interactions (determined by molecular docking) between quercetin and pancreatic lipase would explain the lowest IC50 calculated for quercetin. Gonçalves et al. (3) described pancreatic lipase-polyphenolic compound interactions as reversible, mainly hydrogen bonds and π-stacking interactions; and others only mentioned these interactions as weak (26). The other non-polyphenolic compound, capsaicin, had Ksv and kq values significantly lower than phenolic acids and quercetin (data not shown), which indicates that the observed inhibition may follow a different mechanism than polyphenolic compounds (16).
Molecular docking
To predict the possible binding sites of polyphenolic compounds with pancreatic lipase, docking studies were carried out with all tested compounds (Fig. 4, Table 3). All polyphenolic compounds had one binding site. Binding sites for caffeic acid, p-coumaric acid and quercetin were located close to the enzyme active site, serine153 (Ser153), aspartic acid176 (Asp176) and histidine263 (His263), as described by Kokotos (51), while capsaicin had a binding site far away from the enzyme active site. The same possible binding site was observed for orlistat (data not shown), in agreement with other authors who reported that Ser153 is involved in the binding of this molecule with the enzyme (51, 52). The interaction of these compounds with residues near the active site may explain the competitive component of the observed mixed-inhibition of the polyphenolic compounds. The inhibitory mechanism might be explained considering that the inhibitor interacts with the substrate-enzyme binding. In other words, the inhibitor would bind either to free enzyme or substrate-enzyme complex, but near the substrate binding site. The non-competitive component for polyphenolic compound mixed-type inhibition could be explained considering that the binding to the active site occurs in a form that only affects the catalytic steps and not the substrate binding.
Fig. 4.
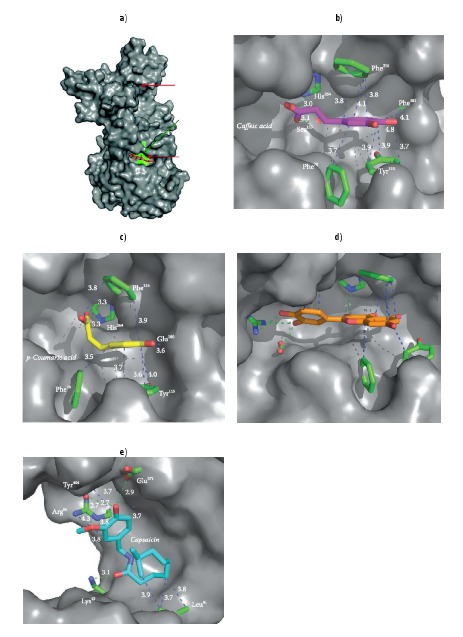
Crystal structure of pancreatic lipase (Protein Data Bank code 1ETH (28)) model of the binding sites for caffeic acid, p-coumaric acid, quercetin and capsaicin achieved by docking experiments: a) surface representation of the overall three-dimensional structure of pancreatic lipase (shown in dark grey colour at www.ftb.com.hr) indicating the active site and the potential binding sites for polyphenolic compounds (b-e). Specific interactions of: b) caffeic acid, c) p-coumaric acid, d) quercetin, and e) capsaicin. Proposed binding amino acids are pointed out (shown in green colour at www.ftb.com.hr). Hydrophobic bonds and polar interactions are drawn in dotted lines (shown as blue and green dotted lines, respectively, at www.ftb.com.hr)
Table 3. Binding of compounds to pancreatic lipase and details of interactions.
| Ligand atom | Residue atom | Residue number | Distance/Å | Type |
|---|---|---|---|---|
| Caffeic acid | ||||
| OE1 | NE | His264 | 3.0 | Polar |
| OE1 | OG | Ser153 | 3.1 | Polar |
| C3 | C2 | Phe78 | 3.7 | Hydrophobic |
| O1 | C1 | Tyr115 | 4.8 | Hydrophobic |
| O1 | C3 | 3.9 | Hydrophobic | |
| O1 | C4 | 3.9 | Hydrophobic | |
| C1 | C2 | Pro181 | 4.1 | Hydrophobic |
| C2 | C2 | Phe216 | 4.1 | Hydrophobic |
| C5 | C2 | 4.1 | Hydrophobic | |
| C4 | C1 | 3.8 | Hydrophobic | |
| p-Coumaric acid | ||||
| OE1 | NE | His264 | 3.3 | Polar |
| OE1 | NB | 3.3 | Polar | |
| C3 | C2 | Phe78 | 3.5 | Hydrophobic |
| C6 | C1 | Tyr115 | 3.6 | Hydrophobic |
| C1 | C5 | 4.0 | Hydrophobic | |
| C1 | C2 | Phe216 | 3.9 | Hydrophobic |
| OE2 | C5 | 3.8 | Hydrophobic | |
| Quercetin | ||||
| O3’ | OD1 | Asp80 | 3.0 | Polar |
| O3’ | NH2 | Arg257 | 3.1 | Polar |
| O4’ | NH1 | 3.1 | Polar | |
| C5’ | CH3 | Val26 | 3.8 | Hydrophobic |
| C6’ | CH3 | Ala261 | 3.6 | Hydrophobic |
| O3 | NE | His264 | 3.2 | Polar |
| O4 | C2 | Phe216 | 3.7 | Hydrophobic |
| C5 | C1 | 3.9 | Hydrophobic | |
| C6 | C2 | 3.9 | Hydrophobic | |
| C7 | C1 | 3.9 | Hydrophobic | |
| O7 | C3 | Tyr115 | 4.1 | Hydrophobic |
| C8a | C2 | Phe78 | 3.5 | Hydrophobic |
| Capsaicin | ||||
| O2 | EO1 | Glu371 | 2.9 | Polar |
| O2 | OG | Tyr404 | 2.7 | Polar |
| O1 | OG | 4.3 | Polar | |
| O1 | NH | Arg65 | 3.8 | Polar |
| C6 | NH1 | 3.8 | Hydrophobic | |
| O8 | NE | Lys42 | 3.1 | Polar |
| C11 | CE1 | Leu41 | 3.8 | Hydrophobic |
| C11 | CE2 | 3.7 | Hydrophobic | |
| C15CH3 | CE2 | 3.9 | Hydrophobic | |
His=histidine, Ser=serine, Phe=phenylalanine, Tyr=tyrosine, Pro=proline, Asp=aspartic acid, Arg=arginine, Val=valine, Ala=alanine, Glu=glutamic acid, Lys=lysine, Leu=leucine
The pancreatic lipase binding site for polyphenolic compounds was mainly formed by four pancreatic lipase amino acid residues: Phe78, Tyr115, His152 and Phe216 (Figs. 4b-e).Residues Tyr115, Ser153 and Proline181 (Pro181) were also involved in the caffeic acid-pancreatic lipase interaction (Fig. 4b). p-Coumaric acid also interacts with glutamic acid180 (Glu180) (Fig. 4c and Table 3; Fig. 1 for references). In the quercetin-pancreatic lipase complex, quercetin was found near Asp80, arginine257 (Arg257), valine260 (Val260) and alanine261 (Ala261) (Fig. 4d). In the capsaicin-pancreatic lipase complex, capsaicin interacted mainly with leucine41 (Leu41), lysine42 (Lys42), Arg65, Glu371 and Tyr404 (Fig. 4e). This different binding region for capsaicin, with respect to the polyphenolic compounds, could be attributed to the different structure of capsaicin (53), shown in Fig. 1. In this way, the non-competitive component of capsaicin could be in this site. Even though the polyphenolic compound binding sites were in the vicinity of the catalytic site, the small fluorescence shift may be attributed to the fact that no significant polarity changes were predicted around Trp residues.
He et al. (7) proposed that the binding forces between polyphenolic compounds and pancreatic lipase were mainly hydrophobic interactions. These hydrophobic and non-covalent interactions are mentioned in Table 3 and shown in Figs. 4b-e (blue doted lines; colour version available at www.ftb.com.hr). Among them, π-stacking interactions are formed between aromatic rings from polyphenolic compounds and pancreatic lipase aromatic amino acids, Phe and Tyr. Our results are in agreement with those reported by Kim et al. (54), which indicated that hydrophobic bonds (with residues like Phe) were the main interactions found between polyphenolic compounds and pancreatic lipase. Another hydrophobic bond predicted by the docking analysis was the hydrogen bond between the hydroxyl groups of polyphenolic compounds and polar groups of pancreatic lipase (5). Fewer polar interactions in Fig. 4 (shown as green dotted lines; see www.ftb.com.hr) were observed for the polyphenolic compound-pancreatic lipase complexes. Quercetin-pancreatic lipase complex presented more polar interactions compared to the rest of the polyphenolic compound-pancreatic lipase complexes.
The quercetin size and structural rigidity could be responsible for its binding characteristics (more polar interactions) and consequently its higher pancreatic lipase inhibitory potency. You et al. (10) reported a higher inhibitory potency (lower IC50) of quercetin than of ellagic acid (Fig. 1). Böhl et al. (55) described that quercetin (among other flavonoids) presented higher affinity towards proteins due to structural characteristics. Quercetin possesses characteristics such as catechol structure in B ring and double bond between C2 and C3 in C ring. Other factors such as number of aromatic rings and number of free hydroxyl groups present in their structure are associated with the ability of polyphenolic compounds to interact with proteins (37, 56).
∆F values calculated from fluorescence studies for polyphenolic compound-pancreatic lipase interactions could be associated with their binding with three amino acids close to Trp residues. According to docking results, caffeic acid, p-coumaric acid and quercetin bound with Phe78 and His264, found near Trp residues located at positions 86 and 252. In contrast, the capsaicin-pancreatic lipase complex was located in the vicinity of only one Trp residue (Trp30, which is close to Leu41 and Lys42). This suggested that polyphenolic compounds have a higher effect on protein conformation (Trp environment) than capsaicin and may explain the lower ΔF, Ksv and kq values obtained for the latter in the fluorescence studies.
Caffeic acid showed high π-stacking interactions with Phe78, Tyr115, Pro181 and Phe216, and two polar bonds with Ser153 and His264. A total of ten possible bonds between this polyphenolic compound and pancreatic lipase were observed. It seems that caffeic and p-coumaric acids showed an ability to form a complex with pancreatic lipase (no significant differences between Ksv and kq values) similar to that observed for quercetin, but through less polar interactions (2 against 4 or 5 observed for the other studied compounds). This higher number of hydrophobic interactions present in the phenolic acid-pancreatic lipase complex, instead of the more polar interactions present in quercetin-pancreatic lipase complex, could explain the lower inhibitory activity of the phenolic acids. Notwithstanding, further studies regarding the phenolic acid-pancreatic lipase binding are needed.
Conclusion
In summary, all the tested polyphenolic compounds showed mixed-type inhibition of pancreatic lipase, quercetin being the strongest inhibitor tested, with IC50 similar to that of orlistat. Phenolic acids showed intermediate inhibitory activity, while capsaicin and the jalapeño pepper extract showed the lowest inhibition. Binding studies showed that polyphenolic compounds might form a complex with pancreatic lipase throughout a static quenching mechanism. Molecular docking analysis showed that all compounds except capsaicin bound close to the active site of pancreatic lipase and that hydrogen bonds and π-stacking interactions were the main polyphenolic compound-pancreatic lipase interactions. This study confirmed stronger inhibitory activity of quercetin than of non-flavonoids, and pointed out that the reported anti-obesity effects of polyphenolic compounds or hot pepper extracts are probably unrelated to pancreatic lipase inhibition. Also, it showed that flavonoids and phenolic acids bind to similar sites in pancreatic lipase but subtle differences in binding may account for their different inhibitory potencies.
Acknowledgements
The authors are grateful for the financial support from CONACYT, Mexico (CB-2011-01-167932, CB-2011-01-167164 and Fronteras en la Ciencia 2015-563). A.I.M.G. and A.A.V.F. are very grateful for the scholarships from CONACYT and the support from UACJ.
References
- 1.Sergent T, Vanderstraeten J, Winand J, Beguin P, Schneider YJ. Phenolic compounds and plant extracts as potential natural anti-obesity substances. Food Chem. 2012;135:68–73. 10.1016/j.foodchem.2012.04.074 [DOI] [Google Scholar]
- 2.WHO Media Centre. Obesity and Overweight. Geneva, Switzerland: World Health Organization (WHO); 2014. Available from: http://www.who.int/mediacentre/factsheets/fs311/.
- 3.Gonçalves R, Mateus N, de Freitas V. Inhibition of α-amylase activity by condensed tannins. Food Chem. 2011;125:665–72. 10.1016/j.foodchem.2010.09.061 [DOI] [Google Scholar]
- 4.Vazquez-Flores AA, Wong-Paz JE, Lerma-Herrera MA, Martinez-Gonzalez AI, Olivas-Aguirre FJ, Aguilar CN, et al. Proanthocyanidins from the kernel and shell of pecan (Carya illinoinensis): Average degree of polymerization and effects on carbohydrate, lipid, and peptide hydrolysis in a simulated human digestive system. J Funct Foods. 2017;28:227–34. 10.1016/j.jff.2016.11.003 [DOI] [Google Scholar]
- 5.Wu X, He W, Zhang H, Li Y, Liu Z, He Z. Acteoside: A lipase inhibitor from the Chinese tea Ligustrum purpurascens kudingcha. Food Chem. 2014;142:306–10. 10.1016/j.foodchem.2013.07.071 [DOI] [PubMed] [Google Scholar]
- 6.Mercado-Mercado G, de la Rosa Carrillo L, Wall-Medrano A, López-Díaz JA, Álvarez-Parrilla E. Polyphenolic compounds and antioxidant capacity of tipically consumed spices in Mexico. Nutr Hosp. 2013;28:36–46 (in Spanish). 10.3305/nh.2013.28.1.6298 [DOI] [PubMed] [Google Scholar]
- 7.He Q, Lv Y, Yao K. Effects of tea polyphenols on the activities of α-amylase, pepsin, trypsin and lipase. Food Chem. 2007;101:1178–82. 10.1016/j.foodchem.2006.03.020 [DOI] [Google Scholar]
- 8.Sakulnarmrat K, Konczak I. Composition of native Australian herbs polyphenolic-rich fractions and in vitro inhibitory activities against key enzymes relevant to metabolic syndrome. Food Chem. 2012;134:1011–9. 10.1016/j.foodchem.2012.02.217 [DOI] [PubMed] [Google Scholar]
- 9.Won SR, Kim SK, Kim YM, Lee PH, Ryu JH, Kim JW, et al. A lipase inhibitor from the roots of Glycyrrhiza uralensis. Food Res Int. 2007;40:1046–50. 10.1016/j.foodres.2007.05.005 [DOI] [Google Scholar]
- 10.You Q, Chen F, Wang X, Jiang Y, Lin S. Anti-diabetic activities of phenolic compounds in muscadine against alpha-glucosidase and pancreatic lipase. Lebensm Wiss Technol. 2012;46:164–8. 10.1016/j.lwt.2011.10.011 [DOI] [Google Scholar]
- 11.Costamagna MS, Zampini IC, Alberto MR, Cuello S, Torres S, Pérez J, et al. Polyphenols rich fraction from Geoffroea decorticans fruits flour affects key enzymes involved in metabolic syndrome, oxidative stress and inflammatory process. Food Chem. 2016;190:392–402. 10.1016/j.foodchem.2015.05.068 [DOI] [PubMed] [Google Scholar]
- 12.Zhang B, Deng Z, Ramdath DD, Tang Y, Chen PX, Liu R, et al. Phenolic profiles of 20 Canadian lentil cultivars and their contribution to antioxidant activity and inhibitory effects on α-glucosidase and pancreatic lipase. Food Chem. 2015;172:862–72. 10.1016/j.foodchem.2014.09.144 [DOI] [PubMed] [Google Scholar]
- 13.Moreno-Escamilla JO, de la Rosa LA, López-Díaz JA, Rodrigo-García J, Núñez-Gastélum JA, Alvarez-Parrilla E. Effect of the smoking process and firewood type in the phytochemical content and antioxidant capacity of red jalapeño pepper during its transformation to chipotle pepper. Food Res Int. 2015;76:654–60. 10.1016/j.foodres.2015.07.031 [DOI] [PubMed] [Google Scholar]
- 14.Yang HJ, Kwon DY, Kim MJ, Kim DS, Kang S, Shin BK, et al. Red peppers with different pungencies and bioactive compounds differentially modulate energy and glucose metabolism in ovariectomized rats fed high fat diets. J Funct Foods. 2014;7:246–56. 10.1016/j.jff.2014.02.004 [DOI] [Google Scholar]
- 15.Chen L, Kang YH. Anti-inflammatory and antioxidant activities of red pepper (Capsicum annuum L.) stalk extracts: Comparison of pericarp and placenta extracts. J Funct Foods. 2013;5:1724–31. 10.1016/j.jff.2013.07.018 [DOI] [Google Scholar]
- 16.Li YQ, Yang P, Gas F, Zhang ZW, Wu B. Probing the interaction between 3 flavonoids and pancreatic lipase by methods of fluorescence spectroscopy and enzymatic kinetics. Eur Food Res Technol. 2011;233:63–9. 10.1007/s00217-011-1491-z [DOI] [Google Scholar]
- 17.McDougall GJ, Kulkarni NN, Stewart D. Berry polyphenols inhibit pancreatic lipase activity in vitro. Food Chem. 2009;115:193–9. 10.1016/j.foodchem.2008.11.093 [DOI] [Google Scholar]
- 18.Bradford MM. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 19.Mohammadi A, Haririan I, Rezanour N, Ghiasi L, Walker RB. A stability-indicating high performance liquid chromatographic assay for the determination of orlistat in capsules. J Chromatogr A. 2006;1116:153–7. 10.1016/j.chroma.2006.03.038 [DOI] [PubMed] [Google Scholar]
- 20.Dalar A, Konczak I. Phenolic contents, antioxidant capacities and inhibitory activities against key metabolic syndrome relevant enzymes of herbal teas from Eastern Anatolia. Ind Crops Prod. 2013;44:383–90. 10.1016/j.indcrop.2012.11.037 [DOI] [Google Scholar]
- 21.Alvarez-Parrilla E, de la Rosa LA, Rodrigo-Garcia J, Escobedo-Gonzalez R, Mercado-Mercado G, Moyers-Montoya E, et al. Dual effect of β-cyclodextrin (β-CD) on the inhibition of apple polyphenol oxidase by 4-hexylresorcinol (HR) and methyl jasmonate (MJ). Food Chem. 2007;101:1346–56. 10.1016/j.foodchem.2006.03.040 [DOI] [Google Scholar]
- 22.SigmaPlot v. 13.0, SYSTAT Sofware, Inc., San Jose, CA, USA; 2014. Available from: https://systatsoftware.com/products/sigmaplot/.
- 23.Tipton KF. Patterns of enzyme inhibition. In: Engel PC, editor. Enzymology Labfax. Oxford, UK: BIOS Scientific Publisher Limited; 1996. pp. 115-74. [Google Scholar]
- 24.Zhang J, Xiao L, Yang Y, Wang Z, Li G. Lignin binding to pancreatic lipase and its influence on enzymatic activity. Food Chem. 2014;149:99–106. 10.1016/j.foodchem.2013.10.067 [DOI] [PubMed] [Google Scholar]
- 25.Chandrakuntal K, Thomas NM, Pradeep GK, Laloraya M, Laloraya MM. Fluorescence resonance energy transfer between polyphenolic compounds and riboflavin indicates a possible accessory photoreceptor function for some polyphenolic compounds. Photochem Photobiol. 2006;82:1358–64. 10.1562/2005-08-09-RA-644 [DOI] [PubMed] [Google Scholar]
- 26.Wang S, Dong S, Zhang R, Shao H, Liu Y. Effects of proanthocyanidins on porcine pancreatic lipase: Conformation, activity, kinetics and thermodynamics. Process Biochem. 2014;49:237–43. 10.1016/j.procbio.2013.10.018 [DOI] [Google Scholar]
- 27.Ramos P, Coste T, Piémont E, Lessinger JM, Bousquet JA, Chapus C, et al. Time-resolved fluorescence allows selective monitoring of Trp30 environmental changes in the seven-Trp-containing human pancreatic lipase. Biochemistry. 2003;42:12488–96. 10.1021/bi034900e [DOI] [PubMed] [Google Scholar]
- 28.RCSB PDB (Protein Data Bank). Piscotaway, NJ, USA: Rutgers, The State University of New Jersey and La Jolla, CA, USA: San Diego Supercomputer Center (SDSC) and Skaggs School of Pharmacy and Pharmaceutical Sciences. Available from: https://www.rcsb.org/pdb/explore/explore.do?structureId=1eth.
- 29.Hermoso J, Pignol D, Kerferlec B, Crenon I, Chapus C, Fontecilla-Camps JC. Lipase activation by nonionic detergents: The crystal structure of the porcine lipase-colipase-tetraethylene glycol monooctyl ether. J Biol Chem. 1996;271:18007–16. 10.1074/jbc.271.30.18007 [DOI] [PubMed] [Google Scholar]
- 30.Tan K, Chang C, Cuff M, Osipiuk J, Landorf E, Mack JC, et al. Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-coumaric acid and related aromatic acids. Proteins. 2013;81:1709–26. 10.1002/prot.24305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Aalten DV, Crielaard W, Hellingwerf KJ, Joshua-Tor L. Conformational substates in different crystal forms of the photoactive yellow protein - Correlation with theoretical and experimental flexibility. Protein Sci. 2000;9:64–72. 10.1110/ps.9.1.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, et al. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol Cell. 2000;6:909–19. 10.1016/S1097-2765(05)00089-4 [DOI] [PubMed] [Google Scholar]
- 33.PyMOL molecular graphics system, New York, NY, USA: Schrödinger; 2014. Available from: https://www.pymol.org/.
- 34.UCSF. Chimera resource for biocomputing visualization, and informatics (RBVI), San Francisco, CA, USA: National Institutes of Health; 2014. Available from: https://www.cgl.ucsf.edu/chimera/.
- 35.SPSS v. 20; Armonk, NY, USA: IBM; 2014. Available from: https://www.ibm.com/analytics/mx/es/technology/spss/.
- 36.Yang X, Kong F. Effects of tea polyphenols and different teas on pancreatic α-amylase activity in vitro. Lebensm Wiss Technol. 2016;66:232–8. 10.1016/j.lwt.2015.10.035 [DOI] [Google Scholar]
- 37.Martinez-Gonzalez AI, Díaz-Sánchez ÁG, de la Rosa LA, Vargas-Requena CL, Bustos-Jaimes I, Alvarez-Parrilla E. Polyphenolic compounds and digestive enzymes: in vitro non-covalent interactions. Molecules. 2017;22:669. 10.3390/molecules22040669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Worsztynowicz P, Napierała M, Białas W, Grajek W, Olkowicz M. Pancreatic α-amylase and lipase inhibitory activity of polyphenolic compounds present in the extract of black chokeberry (Aronia melanocarpa L.). Process Biochem. 2014;49:1457–63. 10.1016/j.procbio.2014.06.002 [DOI] [Google Scholar]
- 39.Muthamma Milan KS, Dholakia H, Tiku PK, Vishveshwaraiah P. Enhancement of digestive enzymatic activity by cumin (Cuminum cyminum L.) and role of spent cumin as a bionutrient. Food Chem. 2008;110:678–83. 10.1016/j.foodchem.2008.02.062 [DOI] [Google Scholar]
- 40.Gondoin A, Grussu D, Stewart D, McDougall GJ. White and green tea polyphenols inhibit pancreatic lipase in vitro. Food Res Int. 2010;43:1537–44. 10.1016/j.foodres.2010.04.029 [DOI] [Google Scholar]
- 41.Adisakwattana S, Intrawangso J, Hemrid A, Chanathong B, Mäkynen K. Extracts of edible plants inhibit pancreatic lipase, cholesterol esterase and cholesterol micellization, and bind bile acids. Food Technol Biotechnol. 2012;50:11–6. [Google Scholar]
- 42.Narita Y, Inouye K. Inhibitory effects of chlorogenic acids from green coffee beans and cinnamate derivatives on the activity of porcine pancreas α-amylase isozyme I. Food Chem. 2011;127:1532–9. 10.1016/j.foodchem.2011.02.013 [DOI] [Google Scholar]
- 43.Zhang J, Cui JH, Yin T, Sun L, Li G. Activated effect of lignin on α-amylase. Food Chem. 2013;141:2229–37. 10.1016/j.foodchem.2013.05.047 [DOI] [PubMed] [Google Scholar]
- 44.Liu RH. Potential synergy of phytochemicals in cancer prevention: Mechanism of action. In: Norman HA, Go VLW, Butrum RR, editor. International Research Conference on Food, Nutrition, and Cancer, 2004 July 15-16, Washington, DC, USA; 2004. pp. 3479S-85S. [DOI] [PubMed] [Google Scholar]
- 45.Wang H, Du YJ, Song HC. α-Glucosidase and α-amylase inhibitory activities of guava leaves. Food Chem. 2010;123:6–13. 10.1016/j.foodchem.2010.03.088 [DOI] [Google Scholar]
- 46.Quenching of fluorescence. In: Lakowicz JR, editor. Principles of fluorescence spectroscopy. New York, NY, USA.: Kluwer Academic/Plenum Publishers; 1999. pp. 277-330. [Google Scholar]
- 47.van de Weert M, Stella L. Fluorescence quenching and ligand binding: A critical discussion of a popular methodology. J Mol Struct. 2011;998:144–50. 10.1016/j.molstruc.2011.05.023 [DOI] [Google Scholar]
- 48.Gonçalves R, Mateus N, de Freitas V. Study of the interaction of pancreatic lipase with procyanidins by optical and enzymatic methods. J Agric Food Chem. 2010;58:11901–6. 10.1021/jf103026x [DOI] [PubMed] [Google Scholar]
- 49.Skrt M, Benedik E, Podlipnik C, Poklar Ulrih N. Interactions of different polyphenols with bovine serum albumin using fluorescence quenching and molecular docking. Food Chem. 2012;135:2418–24. 10.1016/j.foodchem.2012.06.114 [DOI] [PubMed] [Google Scholar]
- 50.Cai X, Yu J, Xu L, Liu R, Yang J. The mechanism study in the interactions of sorghum procyanidins trimer with porcine pancreatic α-amylase. Food Chem. 2015;174:291–8. 10.1016/j.foodchem.2014.10.131 [DOI] [PubMed] [Google Scholar]
- 51.Kokotos G. Inhibition of digestive lipases by 2-oxo amide triacylglycerol analogues. J Mol Catal, B Enzym. 2003;22:255–69. 10.1016/S1381-1177(03)00041-9 [DOI] [Google Scholar]
- 52.Lüthi-Peng Q, Märki HP, Hadváry P. Identification of the active-site serine in human pancreatic lipase by chemical modification with tetrahydrolipstatin. FEBS Lett. 1992;299:111–5. 10.1016/0014-5793(92)80112-T [DOI] [PubMed] [Google Scholar]
- 53.Guedes IA, de Magalhães CS, Dardenne LE. Receptor–ligand molecular docking. Biophys Rev. 2014;6:75–87. 10.1007/s12551-013-0130-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim JH, Kim HJ, Kim C, Jung H, Kim YO, Ju JY, et al. Development of lipase inhibitors from various derivatives of monascus pigment produced by Monascus fermentation. Food Chem. 2007;101:357–64. 10.1016/j.foodchem.2005.11.055 [DOI] [Google Scholar]
- 55.Böhl M, Czupalla C, Tokalov SV, Hoflack B, Gutzeit HO. Identification of actin as quercetin-binding protein: An approach to identify target molecules for specific ligands. Anal Biochem. 2005;346:295–9. 10.1016/j.ab.2005.08.037 [DOI] [PubMed] [Google Scholar]
- 56.Yang B, Chen F, Hua Y, Huang SS, Lin S, Wen L, et al. Prooxidant activities of quercetin, p-courmaric acid and their derivatives analysed by quantitative structure–activity relationship. Food Chem. 2012;131:508–12. 10.1016/j.foodchem.2011.09.014 [DOI] [Google Scholar]