Abstract
Defining the microbial etiology of culture-negative prosthetic joint infection (PJI) can be challenging. Metagenomic shotgun sequencing is a new tool to identify organisms undetected by conventional methods. We present a case where metagenomics was used to identify Mycoplasma salivarium as a novel PJI pathogen in a patient with hypogammaglobulinemia.
Keywords: prosthetic joint infection, Mycoplasma, metagenomics.
CASE REPORT
A 52-year-old man was referred to our Department of Orthopedic Surgery and Orthopedic Infectious Disease Focus Group for evaluation of a chronic right knee prosthetic joint infection (PJI). His history was significant for follicular B-cell lymphoma diagnosed 15 years prior, in remission after treatment with chemotherapy including rituximab; seronegative inflammatory arthritis treated with leflunomide, hydroxychloroquine, and prednisone 5 mg daily; and common variable immune deficiency (CVID) with hypogammaglobulinemia treated with intravenous immune globulin. He had undergone a right total hip arthroplasty, left total hip arthroplasty, and right total knee arthroplasty (TKA) at an outside institution sequentially 3 years prior due to the inflammatory arthritis of the respective joints. Six months after the TKA he developed pain, swelling, and eventually drainage from multiple sinus tracts around the knee. Cultures of synovial fluid aspirates were negative from other institutions. Over the ensuing 2 years he was prescribed multiple oral antibiotic courses without significant improvement, after which he was referred to us for further evaluation and treatment.
At the time of referral, the patient reported ongoing knee pain and sinus drainage, but no fevers or chills. Physical examination revealed a small effusion and multiple draining sinuses over the anterior and medial portion of the tibia. Laboratory evaluation was significant for erythrocyte sedimentation rate (ESR) of 53 mm/hour (normal range, 0–22 mm/hour), C-reactive protein (CRP) of 71.6 mg/L (normal, <8.0 mg/L), hemoglobin 11.3 g/dL, total immunoglobulin G 646 mg/dL (normal range, 767–1590 mg/dL), immunoglobulin A <1 mg/dL, and immunoglobulin M <5 mg/dL. Synovial fluid analysis revealed 28756 cells/mm3 with 93% neutrophils; aerobic and anaerobic bacterial cultures were negative (see Supplementary Materials for full details). Radiographs showed a radiolucent line at the bone- cement interface of the tibial component, with reactive sclerotic changes, suggestive of septic loosening.
His inflammatory arthritis treatment was discontinued and he was kept off any additional antibiotics in anticipation of surgery. Five months later, he underwent resection of his TKA as part of a planned 2-stage exchange arthroplasty. Purulent material and necrotic tissue were observed during surgery. Pathology revealed acute and chronic inflammation. Aerobic and anaerobic cultures of the implant sonicate fluid and 5 periprosthetic tissue specimens were negative. Fungal and mycobacterial cultures were also negative. An antibiotic-loaded nonarticulating spacer containing 3 g of vancomycin, 6.6 g of gentamicin, and 150 g of amphotericin B in one 40-g batch of cement was placed. The following day, a pedicled right medial gastrocnemius flap and skin graft were placed to repair the anterior knee defect resulting from sinus tract excision. The patient completed a 6-week course of empiric intravenous vancomycin and cefepime.
After completion of antibiotics, his ESR and CRP remained elevated, at 40 mm/hour and 93.5 mg/L, respectively. A knee aspiration was therefore performed, revealing 2288 cells/mm3 synovial fluid with 80% neutrophils. A Mycoplasma hominis polymerase chain reaction (PCR) performed on the synovial fluid was negative, as were fungal and aerobic and anaerobic bacterial cultures. A computed tomographic scan revealed a mild left lower lobe lung infiltrate and mild right axillary lymphadenopathy. Recurrent follicular lymphoma or a secondary effect from the patient’s CVID was considered as possible causes, but further studies were not conducted as treatment was considered contraindicated in the setting of active infection. ESR and CRP remained elevated (44 mm/hour and 51.3 mg/L, respectively) 5 months later. He then underwent right knee reimplantation. Three tissue specimens were submitted for aerobic and anaerobic bacterial, fungal, and mycobacterial culture and were negative. No acute inflammation was noted histopathologically.
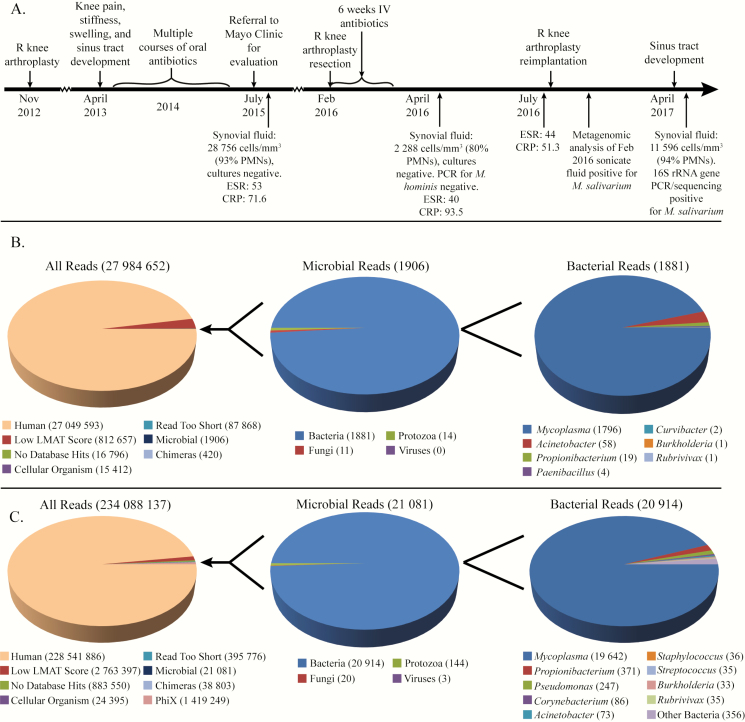
After reimplantation, the sonicate fluid from his original arthroplasty extraction, which had been kept at −80°C, was analyzed using metagenomics. The MolYsis Basic kit was used for microbial enrichment prior to DNA extraction and sequencing on an Illumina HiSeq 2500 instrument [1]. This came back positive for Mycoplasma salivarium, with 1796 reads being from this organism (0.0064% of all sequenced reads, 94.2% of microbial reads; see Figure 1B) as identified by the Livermore Metagenomic Analysis Toolkit (LMAT), a k-mer based taxonomic classifier (see Supplementary Materials for full details of analysis) [2]. The remaining 5.8% of microbial reads were from organisms commonly observed as contaminants with this method. A second stored aliquot of sonicate fluid underwent DNA extraction without microbial enrichment and was also strongly positive for M. salivarium, with 19642 reads being from this organism (0.0084% of all reads, 93.2% of microbial reads; see Figure 1C). When the reads from the initial and repeat sequencing sets were aligned to the M. salivarium ATCC 23064 reference genome, they covered 32.5% and 84.2% of the genome, respectively. Sonicate fluid, as well as purified DNA, was also subjected to partial 16S ribosomal RNA (rRNA) gene amplification and sequencing with previously-discribed primers [3]. The resultant sequences were identified as being from M. salivarium with the strongest match showing 99.8% identity to M. salivarium strain PG20. Formalin-fixed, paraffin-embedded tissue specimens were retrieved from the explantation and implantation surgeries and tested using 16S rRNA gene PCR with previously-discribed primers (3), but no PCR product was produced. Attempts to culture the stored sonicate fluid using A8 agar plates and SP4 broth to selectively enrich for Mycoplasma species were unsuccessful.
Figure 1.
Timeline of events and taxonomic assignment of reads by the LMAT. A, Timeline of clinical course with relevant laboratory findings listed. Taxonomic classification of reads by LMAT [2] from sonicate fluid with (B) and without (C) microbial DNA enrichment using the MolYsis Basic kit prior to DNA extraction. “Reads too short” indicates reads with <30 k-mers (k = 20). Abbreviations: CRP, C-reactive protein (mg/L); ESR, erythrocyte sedimentation rate (mm/hour); IV, intravenous; LMAT, Livermore Metagenomics Analysis Toolkit; PCR, polymerase chain reaction; PMNs, polymorphonuclear leukocytes; R, right.
The patient returned for evaluation, 2 months after reimplantation, and reported that the knee was doing well; there no were signs of infection. However, ESR and CRP remained elevated at 35 mm/hour and 84.4 mg/L, respectively. Nine months after reimplantation, the patient reported spontaneous development of a sinus tract at the border of his gastrocnemius flap. Synovial fluid was aspirated and 16S rRNA gene amplification and sequencing performed using previously described primers (3) was positive for M. salivarium. The patient is being treated with doxycycline.
DISCUSSION
Prosthetic joint infections are devastating complications of joint replacement surgery that affect approximately 2% of arthroplasties and often require repeat surgeries and prolonged intravenous antibiotic treatment. These infections can be caused by a wide range of pathogens, including gram-negative and gram-positive aerobic bacteria, anaerobic bacteria, and fungi, and can also be polymicrobial. Identification of the pathogens by culture can be challenging for multiple reasons, including antecedent antibiotic administration, fastidious nature of the organisms, or growth in a biofilm state, all of which likely contribute to 15%–20% of PJIs being classified as culture negative. This has significant implications for treatment as such cases require broad-spectrum antibiotics to cover the wide range of potential pathogens.
Mycoplasmas are the smallest known bacteria and are notable for a lack of cell wall, rendering cell wall–targeting antibiotics ineffective. Mycoplasma species are among the many organisms capable of causing PJIs, with M. hominis and Mycoplasma pneumoniae previously identified as PJI pathogens [4, 5]. This is the first case we are aware of in which M. salivarium has been identified as a PJI pathogen. Mycoplasma salivarium is a common commensal found in the saliva of healthy individuals. Case reports have reported it as a cause of various infections, including septic arthritis, empyema, and abscesses [6–8]. Most, but not all, of these individuals were immunocompromised (eg, hypogammaglobulinemia, receiving chemotherapy for leukemia or lymphoma). Hypogammaglobulinemia in particular has been associated with septic arthritis caused by Mycoplasma species, including M. salivarium [6, 9]. Because of this association, the patient’s synovial fluid had been tested for M. hominis.
The patient received empiric treatment with vancomycin and cefepime as intravenous antibiotics, which are not active against Mycoplasma species due to their lack of a cell wall. However, gentamicin, which was incorporated into the antibiotic spacer, may have activity against M. salivarium. Without isolation of the organism, we cannot know for sure whether it was susceptible to gentamicin. However, no genes conferring resistance to gentamicin were detected when the metagenomic sequences were searched against 2 antibiotic resistance gene databases (Resfams and the Comprehensive Antibiotic Resistance Database). Analysis for resistance mutations revealed an A2058T mutation of the 23S rRNA gene (Escherichia coli numbering) that has been associated with moderate levels of macrolide resistance in M. pneumoniae [10], suggesting that macrolide therapy should be avoided.
Metagenomic shotgun sequencing involves sequencing all of the DNA (or RNA in some instances) present in a sample and identifying which organisms the nucleic acid came from to determine which potential pathogens are present. Microbial enrichment methods (MolYsis Basic) were initially used to overcome the often overwhelming amount of human DNA in the sample, but due to concern that this method would lyse Mycoplasma species, which lack a cell wall, unenriched processing was pursued, which also showed a strong signal (Figure 1C). Other case reports have demonstrated its value in clinical scenarios of difficult-to-diagnose infections; however, incorporation into routine clinical diagnostics faces many hurdles, including optimization of methods, cost, and time needed to carry out these methods.
This case demonstrates a new PJI pathogen that is otherwise difficult to detect using conventional methods. Mycoplasma salivarium should be considered in the differential diagnosis in patients with culture-negative PJI, particularly those with hypogammaglobulinemia. We also highlight another example of the potential for metagenomic shotgun sequencing to detect unexpected or difficult-to-detect pathogens as well as guide therapy based on gene content analysis.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Financial support. This work was supported by the National Institutes of Health (award number R01AR056647).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Thoendel M, Jeraldo PR, Greenwood-Quaintance KE, et al. Comparison of microbial DNA enrichment tools for metagenomic whole genome sequencing. J Microbiol Methods 2016; 127:141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ames SK, Hysom DA, Gardner SN, Lloyd GS, Gokhale MB, Allen JE. Scalable metagenomic taxonomy classification using a reference genome database. Bioinformatics 2013; 29:2253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gomez E, Cazanave C, Cunningham SA, et al. Prosthetic joint infection diagnosis using broad-range PCR of biofilms dislodged from knee and hip arthroplasty surfaces using sonication. J Clin Microbiol 2012; 50:3501–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Han Z, Burnham CA, Clohisy J, Babcock H. Mycoplasma pneumoniae periprosthetic joint infection identified by 16S ribosomal RNA gene amplification and sequencing: a case report. J Bone Joint Surg Am 2011; 93:e103. [DOI] [PubMed] [Google Scholar]
- 5. Sneller M, Wellborne F, Barile MF, Plotz P. Prosthetic joint infection with Mycoplasma hominis. J Infect Dis 1986; 153:174–5. [DOI] [PubMed] [Google Scholar]
- 6. So AK, Furr PM, Taylor-Robinson D, Webster AD. Arthritis caused by Mycoplasma salivarium in hypogammaglobulinaemia. Br Med J 1983; 286: 762–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ørsted I, Gertsen JB, Schønheyder HC, Jensen JS, Nielsen H. Mycoplasma salivarium isolated from brain abscesses. Clin Microbiol Infect 2011; 17:1047–9. [DOI] [PubMed] [Google Scholar]
- 8. Baracaldo R, Foltzer M, Patel R, Bourbeau P. Empyema caused by Mycoplasma salivarium. J Clin Microbiol 2012; 50:1805–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Steuer A, Franz A, Furr PM, Taylor-Robinson D, Webster AD, Hughes GR. Common variable immunodeficiency presenting as a Mycoplasma hominis septic arthritis. J Infect 1996; 33:235–7. [DOI] [PubMed] [Google Scholar]
- 10. Pereyre S, Goret J, Bébéar C. Mycoplasma pneumoniae: current knowledge on macrolide resistance and treatment. Front Microbiol 2016; 7:974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.