Abstract
Objectives
Recent commercialization of methods for in situ hybridization using Z-pair probe/branched DNA amplification has led to increasing adoption of this technology for interrogating RNA expression in formalin-fixed, paraffin-embedded (FFPE) tissues. Current practice for FFPE block storage is to maintain them at room temperature, often for many years.
Methods
To examine the effects of block storage time on FFPE tissues using a number of RNA in situ probes with the Advanced Cellular Diagnostic’s RNAscope assay.
Results
We report marked reductions in signals after 5 years and significant reductions often after 1 year. Furthermore, storing unstained slides cut from recent cases (<1 year old) at –20°C can preserve hybridization signals significantly better than storing the blocks at room temperature and cutting the slides fresh when needed.
Conclusions
We submit that the standard practice of storing FFPE tissue blocks at room temperature should be reevaluated to better preserve RNA for in situ hybridization.
Keywords: RNA in situ hybridization, Branched DNA amplification, Formalin-fixed, paraffin-embedded tissues
Molecular analyses of RNA expression using formalin-fixed, paraffin-embedded (FFPE) tissue have become widely used both for research and clinical purposes. In situ detection of RNA provides single-cell-level localization of gene expression in tissues that often results in important insights into normal biological function or disease processes, as well as enhanced diagnostic certainty for pathologists in clinical tissue samples. Furthermore, recent advancements have resulted in markedly increased sensitivity of RNA detection on tissues or whole cells, at times at the single molecule level.1-3 Recently, commercially offered reagents and probes have become available to interrogate expression of many types of individual RNA species in situ in FFPE tissues.3,4 Both RNAscope from Advanced Cellular Diagnostics (ACD, Newark, CA) and ViewRNA from Affymetrix (Santa Clara, CA) are commercial kits and probes for RNA in situ hybridization (RISH) assays that rely on dual binding of two separate probes present in close proximity (“Z-pair” probes) to produce a template for signal amplification using a branched DNA process. The requirement for two separate probes to hybridize simultaneously, and the elimination of any repetitive sequence elements, results in a marked improvement in specificity, while the branched DNA hybridization process allows a simultaneous enhancement in sensitivity.1,3 In addition, the reactions can be visualized either as chromogenic signals, suitable for standard light microscopes, or as fluorescent signals suitable for fluorescence microscopy. Furthermore, both of these commercially available technologies are offered for use either by a manual approach or with automated immunohistochemistry (IHC) stainers5,6 that are commonly used in Clinical Laboratories Improvement Act (CLIA)–certified pathology laboratories. Hybridization signals can be quantified by either manual “eyeball” methods or by relatively unbiased and objective means based on image analysis,7,8 both of which can be useful for making relative expression level comparisons.
Given the specificity of nucleic acid hybridization for virtually any RNA species, these technologies are greatly augmenting prior capabilities for the detection of gene expression in normal and diseased tissues. Some examples of the usefulness of RISH include the detection and localization in tissues or cells of the following: viral RNA such as human papillomavirus RNA in diagnostic tissue biopsy specimens of the cervix and head and neck region6,9; noncoding RNAs (eg, lncRNAs10,11 and vlinc RNAs12); messenger RNA (mRNA) splice variants useful for predicting response to specific therapies13,14; cytokine expression (eg, interleukin 6) since cytokines are often rapidly secreted, which may preclude effective tissue localization by antibodies15; subcellular localization and trafficking of mRNA or other RNAs16; and validation of expression array or RNAseq data in situ in tissues to determine which cells express specific RNA species of interest.17
In addition to localization and quantification, RNA expression levels in situ may be used as prognostic and/or predictive biomarkers in cancer research studies.7,18 The study of prognostic biomarkers generally requires the use of older tissue blocks to study patient samples that have long-term follow-up data. Furthermore, many studies rely on using archival tissue blocks of various ages, even without a specific focus on evaluating prognosis. The use of archival tissue blocks raises the question of RNA stability, which, if affected adversely in relation to block age, could result in biased results and lead to inappropriate conclusions about RNA expression in clinical and research FFPE specimens. Indeed, many studies of solution-based assays have shown the adverse effects of tissue block age on RNA quality and levels. Examples include reverse transcription polymerase chain reaction (RT-PCR), capillary electrophoresis (eg, providing RNA integrity numbers [RIN]), microarrays, and, more recently, branched DNA-based applications as well as RNAseq19-25 on RNA extracted from FFPE tissues. Yet, only a single study to date has specifically addressed the question of the effects of tissue block age on RISH using Z-pair/branched DNA methods.7
Bordeaux et al7 evaluated RISH using RNAscope technology for two different mRNA probes (ERS1 and UBC) and reported little effect of block age on signal levels if specimens were obtained between 1994 and 2003, but those that were obtained from 1975 to 1993 showed diminished signals. However, in that study, all of the tissue blocks used were at least 5 years old or older (up to 7-8 years depending on when hybridizations were done),7 such that there is no systematic information in the literature examining the effects of tissue block age on tissues from 0 to 7 years old using Z-pair/branched DNA in situ hybridization technology. In the present study, we examined the effects of tissue block age on FFPE tissues from radical prostatectomy specimens from patients with prostate cancer using a number of RNA in situ probes, using both standard tissue sections and tissue microarrays (TMAs). We then examined quantitatively the effects of block age in a controlled TMA experiment consisting of tissues from prostate cancer patient-derived xenografts (PDXs) handled under identical conditions. Finally, we carried out an additional prospective quantitative TMA-based experiment to test the effects of storing unstained FFPE slides under various conditions to determine whether loss of in situ hybridization signals could be prevented using a simple strategy of cold slide storage.
Materials and Methods
Prostatectomy Specimens and TMA Construction
This study was approved by our institutional internal review board. Radical retropubic prostatectomy specimens (RRPs) were handled in the Johns Hopkins Hospital CLIA-certified and College of American Pathologists–certified pathology laboratory using standardized techniques. To construct “new case” TMAs, RRPs were injected with 10% neutral buffered formalin and microwaved as previously described26,27 or were sectioned fresh and pinned and floated in neutral buffered formalin for up to 4 hours, prior to additional sectioning and automated tissue processing, followed by processing into FFPE blocks. For more than 20 years, our CLIA-certified laboratory has been using commercially acquired 10% neutral buffered formalin (current material is from Cardinal Health, Dublin, OH), which preserves pH between 6.9 and 7.1. Using these specimens, all of which were collected within 10 months of specimen submission, a number of “new case” TMA blocks were produced. Gleason scores ranged from 6 to 9, and pathologic stages ranged from organ confined (T2N0) to those with pelvic lymph node positivity (T3bN1). The tumor with the highest grade was used for tissue sampling for TMA construction. Patient ages ranged from 50 to 70 years. Carcinoma tissue and matched normal-appearing prostatic tissues were punched multiple times (between three and five random punches within regions of interest) from each case. Control FFPE tissues from various other normal tissues (n = 30 TMA cores) were also placed throughout the TMA. For “older case” TMAs, a similar method was employed for RRP specimen handling with a range of tissue block ages (between 3 and 13 years relative to the “new case” TMAs). Pathologic slides from RRP specimens or from prostate cancer xenografts were used as templates for constructing high-density TMAs (cores, 0.6 mm in diameter) using a manual tissue puncher/arrayer designed by Beecher Instruments (Estigen, Tartu, Estonia). For standard tissue sections (n = 25 patients), a representative tissue block containing the highest grade tumor was used for sectioning. Cases were taken at random that were from specimens with different tissue block ages as indicated in the Results.
Xenograft Tissue Fixation and Processing
Prostate cancer LuCaP PDXs (LCap 23.1, 35, 70, and 96)28-31 were fixed in 10% neutral buffered formalin (phosphate buffered; Fisher, Pittsburgh, PA) for 16 to 24 hours prior to routine processing into FFPE blocks. All FFPE blocks were stored at room temperature for time points indicated in the Results section.
In Situ Hybridization for RNA on FFPE and Frozen Sections
For all experiments except those on TMAs, RISH was performed using the RNAscope 2.0 FFPE Brown Reagent Kit (ACD) as previously described with exceptions indicated below.13-15,32 Briefly, FFPE tissue slides were baked at 60°C for 1 hour followed by deparaffinization in 100% xylene twice for 5 minutes each and two changes of 100% alcohol. The slides were treated with endogenous peroxidase-blocking pretreatment reagent and then incubated for 15 minutes in a boiling 1× Pretreat 2 reagent (ACD) and then treated with protease digestion buffer (III, Cat. 322337) for 30 minutes at 40°C. The slides were incubated with a RNAscope target probes as follows: PPIB (or peptidyl prolyl isomerase B, also known as cyclophilin B, probe region 139-989, NCBI reference sequence accession NM_000942.4), TP63 (Homo sapiens tumor protein p63, probe region 209-1174, NCBI reference sequence accession NM_001114982.1), ERG (Homo sapiens V-ets erythroblastosis virus E26 oncogene homolog [avian], probe region 2933-3913, NCBI reference sequence accession NM_001136154), PTEN (Homo sapiens phosphatase and tensin homolog, target region 758-3263, NCBI reference sequence accession NM_000314.4), PCA3 (Homo sapiens prostate cancer antigen 3 [nonprotein coding], noncoding RNA, probe region 1683-2796, NCBI reference sequence accession NR_015342.1), MYC (Homo sapiens v-myc myelocytomatosis viral oncogene homolog [avian] (MYC), probe region 536-1995, NCBI reference sequence accession NM_002467.4), AR exon 1 (Homo sapiens androgen receptor, exon 1, cat. 401211), and 5.8S rRNA (Homo sapiens 5.8s ribosomal RNA, probe region 2-59, NCBI reference sequence accession J01866.1) for 2 hours at 40°C, followed by signal amplification. 3,3′-Diaminobenzidine (DAB) was used for colorimetric detection for 10 minutes at room temperature. For RNAscope using frozen tissues, tissue sections were fixed in prechilled 10% neutral buffered formalin for 15 minutes at 4°C. Slides were dehydrated in 50%, 70%, and 100% alcohol for 5 minutes each. The slides were treated with endogenous peroxidase-blocking pretreatment reagent and then incubated for 5 minutes in a boiling water bath and then treated with protease digestion buffer for 10 minutes at room temperature. The slides were incubated with RNAscope target probes for 2 hours at 40°C, followed by signal amplification. DAB was used for colorimetric detection for 10 minutes at room temperature. For experiments using TMAs shown in Figures 5 and 6, RNAscope 2.5 kit was used according to the manufacturer’s recommendations, except that protease III (ACD, cat. 322337) was used and amplification steps 5 and 6 were extended to 1 hour (from 30 minutes) and 30 minutes (from 15 minutes), respectively.
Cell Culture and Preparation of FFPE Cell Lines
LNCaP and VCaP prostate cancer cells were obtained from the American Type Culture Collection (Manassas, VA). PC-3 and DU-145 prostate cancer cells were obtained from the National Cancer Institute (Frederick, MD). All cell line identities were verified using short tandem repeat profiling of nine genomic loci with the Powerplex 1.2 system (Promega, Madison, WI). Cells were maintained at 37°C and 5% CO2, and they supplemented with RPMI 1640 or Dulbecco’s modified Eagle’s medium with 10% serum (Corning, Fisher Scientific, Hampton, NH). For FFPE cell line blocks, cells were harvested, fixed in neutral buffered formalin, and processed into FFPE blocks as previously described.33
RNA Interference Knockdown (MYC) and Transfections
Cells were transfected using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA). Pools containing four small interfering RNA (siRNAs) against MYC (L-003282; Dharmacon, Lafayette, CO) were transfected at a final concentration of 50 nmol/L. As a control in each transfection experiment, cells were transfected with siCONTROL Non-Targeting siRNA pool 1 (D-001810; Dharmacon). A molecular clone of human PTEN driven by the cytomegalovirus promoter (PTEN NM_000314 Human cDNA Clone; Origene, Rockville, MD) was transfected in the PTEN null PC-3 cells using Oligofectamine according to the manufacturer’s directions (Invitrogen) as described.33 Transfected cells were harvested after 48 hours and prepared for cell blocks as described above.
Semiquantitative Scoring of Standard Slides
Standard whole slides (eg, not TMA slides) for the FFPE tissue block age study were assessed using a combined score in which an average visually determined intensity (scored on a 0-4 scale) was multiplied by extent of staining (percentage of tissue stained) across the slide for benign (TP63 or PTEN) or carcinoma (MYC, PCA3), such that scores ranged from 0 to 400.
Image Analysis of TMAs
TMA slides were scanned using an Aperio ScanScope CS slide scanner (Leica Biosystems) using the ×20 objective (0.49 m per pixel resolution). The resulting whole slide image was segmented into individual core images using the TMALab module in eSlideManager (Leica Biosystems, Wetzlar, Germany). The TMA core images were imported into the TMAJ image manager (http://tmaj.pathology.jhmi.edu),34-36 and histologic diagnoses were rendered. For the TMAs, regions of interest were created using the lasso mask and annotations tool in TMAJ/FrIDA (an ImageJ plugin for TMAJ)37 to avoid regions of necrosis and poor-quality staining. In TMAJ/FrIDA, hue, saturation, and value (HSV) color space segmentation was used to define the “brown” (DAB; ie, positive IHC/RISH) and “blue” (hematoxylin) masks. Area percent of staining was defined as the total “brown” area in the region of interest divided by the total tissue area in the region of interest as determined by the software. For the RRP TMAs, identical thresholds for identifying DAB were employed for all slides in a given experiment. For the PDX TMAs, HSV color space segmentation was used to optimally define the DAB for each slide.
Data Analysis
Standard slide and TMA data were analyzed by generating descriptive tables and performing statistical testing as indicted for specific comparisons in the Results section using STATA 13 for Microsoft Windows (StataCorp LP, College Station, TX). For standard slides using manual scoring, we used analysis of variance (ANOVA) to compare the scoring results by the categorical variables of tissue block age groups. For experiments comparing “old case” and “new case” TMAs using image analysis, we also used ANOVA as well as the Student t test for pairwise comparisons, after natural log transformations of area scores. For PDX image analysis, data quartiles of staining for each probe set were tabulated and compared using the extended Mantel-Haenszel (Cochran-Mantel-Haenszel) stratified test of association.
Results
Specificity of RISH Using Commercial ACD Probes and Staining Kits
We previously evaluated the specificity of ACD-based RISH staining using FFPE blocks of prostate tissues with a number of separate probe sets, including those targeting IL6,15,38TP6332 (referred to as p63), AR exon 1, and AR splice variant V7.13,14 In the present study, we further examined TP63, ERG, PCA3, PTEN, and MYC. Expression of TP63 was localized nearly exclusively to the basal compartment of the prostate (as is known for p63 protein39,40) Image 1 HSV, with typical prostatic adenocarcinomas being almost completely negative for hybridization signals (not shown). Furthermore, we employed a probe set against ERG and demonstrated its specificity by showing that strong signals were present in approximately 50% of cases tested, and signal was detected only in prostatic tumor cells (as expected)41-43 (Supplementary Figure 1A; all supplemental materials can be found at American Journal of Clinical Pathology online). These positively staining cases correlated with positive staining for ERG protein by IHC (not shown), which itself is well known to correlate with ETS rearrangements such as those resulting in the TMPRSS2-ERG fusion gene product.41-43 In prostate cancer cell lines prepared as FFPE blocks, ERG was positive in VCaP cells and negative in DU-145 cells as expected44,45 (Supplementary Figures 1B and 1C). Similarly, PCA3 signals were also found to be specific as signals were present nearly exclusively in tumor cells and high-grade prostatic intraepithelial neoplasia (PIN) lesions, with very low or no signals found in normal-appearing epithelium46,47Image 2. PTEN specificity was demonstrated by lack of signals in PC-3 prostate cancer cells, which contain a homozygous PTEN deletion,48 compared with strong signals in PC-3 cells transfected with a human PTEN complementary DNA–containing expression vector (Supplementary Figure 2). In prostate tissue, PTEN mRNA was present in virtually all cells in benign prostatic tissues, as expected Image 3. The specificity of MYC was demonstrated by using FFPE mouse tissues that overexpress human MYC in the prostate,49 which showed robust signals, compared with wild-type mice that were negative for human MYC (Supplementary Figures 3A and 3B), and by siRNA knockdown of MYC in human PC-3 cells in FFPE blocks (Supplementary Figures 3C and 3D). MYC mRNA, which is known to be overexpressed in at least 80% of human prostate cancer tissues,36 compared with matched benign samples, was overexpressed in most prostate carcinomas (I. Kulac and A. M. De Marzo, unpublished data). Thus, between prior work and the current study, we demonstrated high specificity of in situ signals for RNA for eight different probe sets using ACD RNAScope technology, and we conclude that it is generally a robust and reliable method for localizing RNA in FFPE tissues.
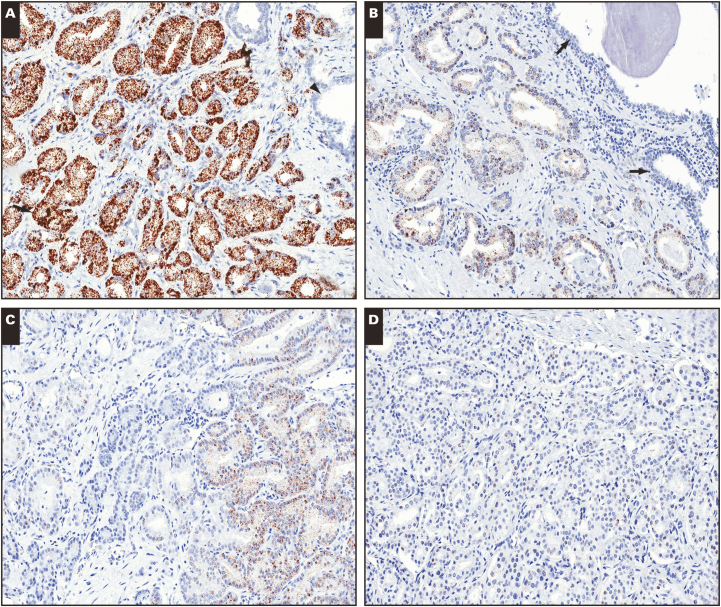
Image 1.
RNA in situ hybridization for TP63 in tissues from formalin-fixed, paraffin-embedded (FFPE) blocks stored for different lengths of time. A, FFPE block from less than 1 year of age showing strong signals for TP63 probe set in prostatic basal cells (arrows). B, Another FFPE block from less than 1e year of age showing similar strong basal cell signals. Note absence of luminal cell or stromal cell signals. C, FFPE block that is 5 years of age showing markedly reduced signals. D, FFPE block that is 8 years old also showing greatly diminished signals. (×100)
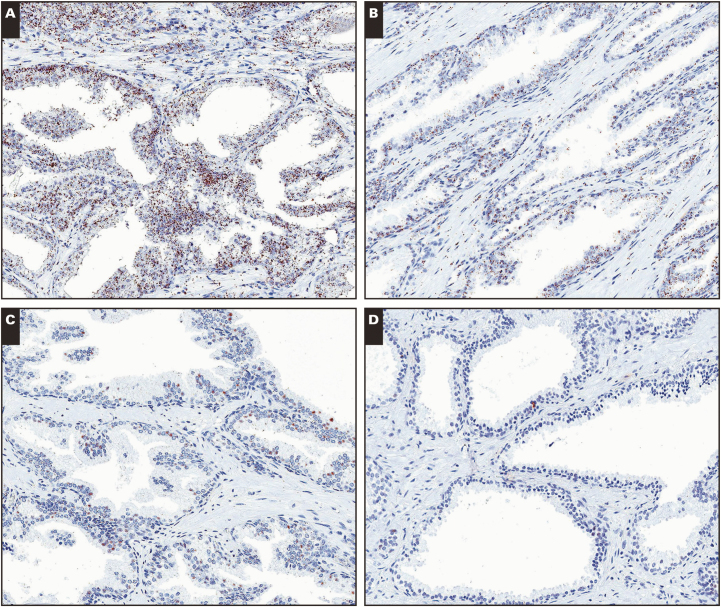
Image 2.
RNA in situ hybridization for PCA3 on tissues from formalin-fixed, paraffin-embedded (FFPE) blocks stored for different lengths of time. A, FFPE block from less than 1 year of age showing strong signals for PCA3 probe set specifically in tumor cells (arrow) but not in normal cells (arrowhead) or stroma. B-D, Markedly reduced signals (arrows) are seen in FFPE blocks containing carcinoma from 7 years (B), 9 years (C), and 11 years (D) of age. (×100)
Image 3.
RNA in situ hybridization for PTEN in tissues from formalin-fixed, paraffin-embedded (FFPE) blocks stored for different lengths of time. A, FFPE block from less than 1 year of age showing strong signals for PTEN probe set in all cells. B-D, Markedly reduced signals are seen in FFPE blocks from 6 years (B), 7 years (C), and 10 years (D) of age. (×100)
Reduced RISH Signal Levels Are Associated With Tissue Block Age
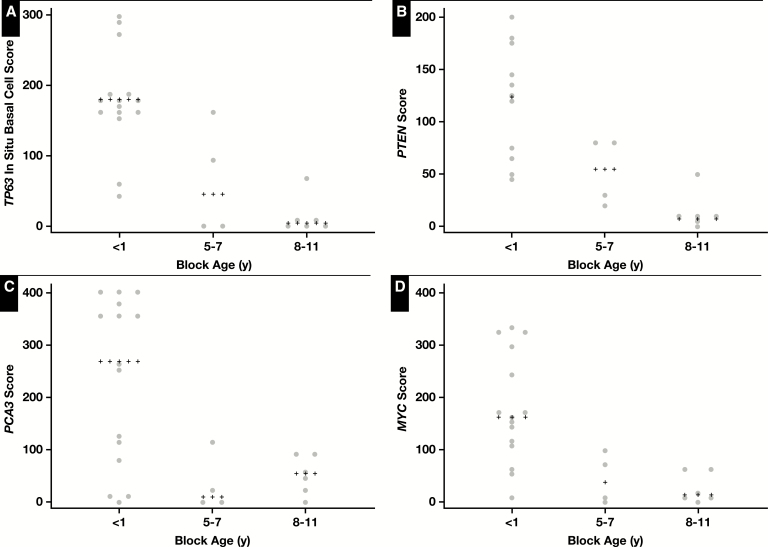
In the studies mentioned above using slides from prostatectomies, we noticed substantial variability in RISH signals from case to case. This led us to consider whether tissue block age could be associated with some of the staining heterogeneity. Therefore, we undertook a study to perform RISH on 25 standard FFPE tissue blocks from prostatectomy specimens with different block ages selected at random. We used tissues that were handled in the same clinical laboratory setting with highly similar protocols over time (including fixation and processing), including those that were less than 1 year old (0 years) and those that were from 5 to 11 years old. We chose four probe sets and performed manual scoring. Probe sets were chosen such that two of the RNAs were expected to be present in all normal-appearing prostatic tissues (TP63 and PTEN), and two were expected to be overexpressed in most of the prostatic adenocarcinoma lesions (MYC and PCA3). Using a combined score that takes into account the overall intensity and extent of staining across the slide for benign tissue (TP63 or PTEN) or carcinoma (MYC, PCA3), we found lower signals using all four probe sets in standard slides from tissue blocks that were 5 years or older compared with those that were less than 1 year old at the time of hybridization. Images 1 to 3 show representative results of in situ hybridization for TP63, PTEN, and PCA3 from standard slides of different block ages showing a marked decrease in signals in the older blocks. Figure 1 shows the manual scoring data of these results, revealing lower signals that were statistically significant for each probe set when stratifying cases into three groups (<1 year, 5-7 years, and 8-11 years) (ANOVA; P = .0001 for TP63, P = .0005 for PTEN, P = .0018 for MYC, and P = .0064 for PCA3).
Figure 1.
Semiquantitative scoring of RNA in situ hybridization signals for TP63 (A), PTEN (B), PCA3 (C), and MYC (D) in relation to formalin-fixed, paraffin-embedded (FFPE) block age. Scores are shown for each probe set where each data point represents the score for a single FFPE slide. Median values are indicated by plus signs.
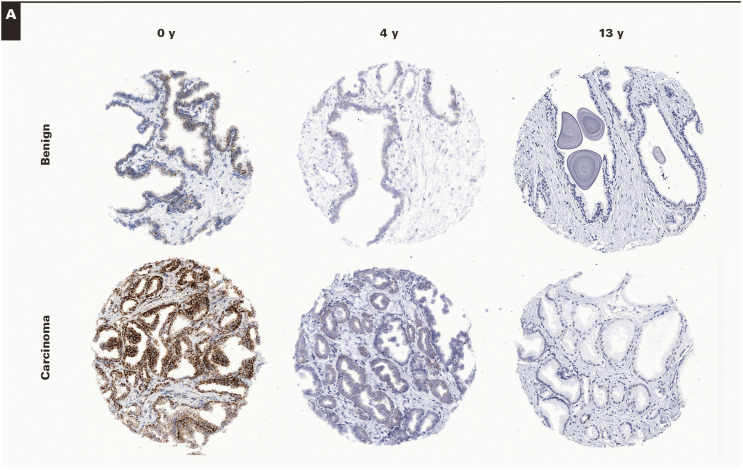
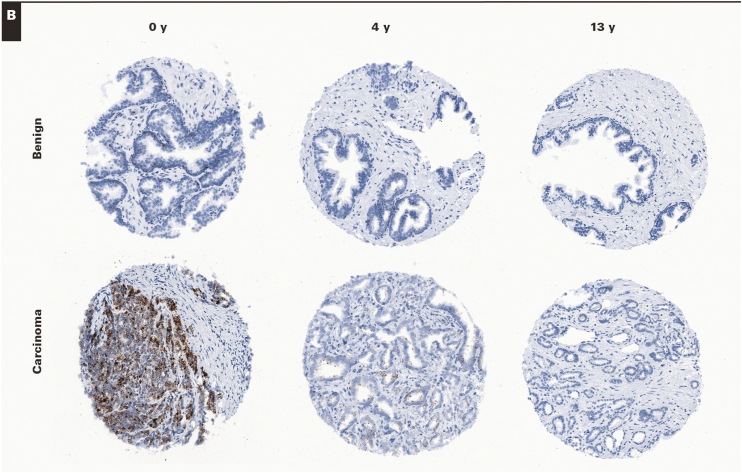
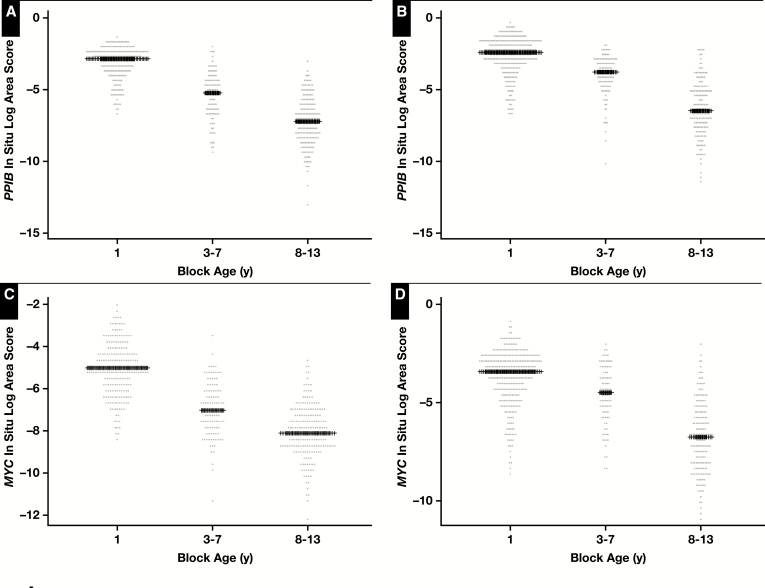
To confirm and extend these findings we generated TMAs from RRP specimens that were considered “new case” TMA (eg, tissue blocks <6 months old) and performed RISH on these as well as TMAs taken from similarly treated specimens in which the tissue block ages varied from 3 years to 13 years. For this, we used the same MYC probe set as above as well as a probe set for PPIB, which is generally used as a “housekeeping” gene probe set recommended by the manufacturer. Each of the TMA slides were digitally scanned, and the composite TMA spot images were imported into TMAJ. TMA spots were assigned a diagnosis (benign or carcinoma), and regions of interest were circled for image analysis. TMA images were viewed by a pathologist (J.A.B.), who was blinded to the TMA design in terms of block age. Supplementary Table 1 shows the distribution of cases and TMA spots by tissue block age and by diagnosis (benign or carcinoma). Image analysis was performed using FrIDA; a given threshold for brown staining was set, and this threshold was used for all subsequent TMA spots in a given slide and for each of the slides stained with each probe set. Image 4 shows representative images of hybridizations with MYC and PPIB probe sets for both benign and carcinoma TMA spots stratified by tissue block age. This process allowed for an unbiased estimate of overall brown staining on each TMA slide. Figure 2 shows the result of the quantitative image analysis. For both probe sets, there was a clear and highly statistically significant decrease in signals in relation to tissue block age for both tissue types analyzed (Figure 2) (eg, for PPIB, the median area score [median of the area fraction of the region of interested stained] was 0.063 for the 0-year time point, 0.01 for the 3- to 7-year time period, and 0.001 for the 8- to 13-year time period). These results are consistent with the findings from the standard tissue sections and show that computerized image analysis of TMAs is a useful method for quantification. Using these same TMAs, although we did not formally quantify the results, we also examined probe sets for PTEN and TP63, and similar overall results with visually observable marked decreases with tissue block age were found (see Supplementary Figure 4 for examples).
Image 4.
RNA in situ hybridization on RPR tissue microarrays (TMAs) in relation to formalin-fixed, paraffin-embedded (FFPE) block age. Probe sets for PPIB (A). (×100) Probe sets for MYC (B) were hybridized to FFPE TMAs in which tissue block ages varied from 0 to 13 years. Representative spots for both benign and carcinoma are shown from TMA spots containing samples from block ages as indicated. Note the decrease in signals in relation to tissue block age. Also, note that, as expected, MYC levels are much lower in benign samples than in carcinoma and that MYC signals in benign epithelium are present mostly in basal cells. (×100)
Figure 2.
Quantitative scoring of signals for PPIB and MYC in relation to tissue microarray (TMA) block age. Natural log-transformed area fraction scores are shown for each probe set where each data point represents the log-transformed score for a single TMA spot for a given diagnosis type (benign in A and C, carcinoma in B and D). Median values are indicated by plus signs. P < .0001 for differences across block age groups for each probe set for each tissue type (analysis of variance). P < .0001 for pairwise comparisons between each block age group (eg, <1 year vs 3-7 years, <1 year vs 8-13 years, 3-7 years vs 8-13 years) for each probe set for each diagnostic category.
TMAs for Block Age-Related RISH Signal Reduction Study
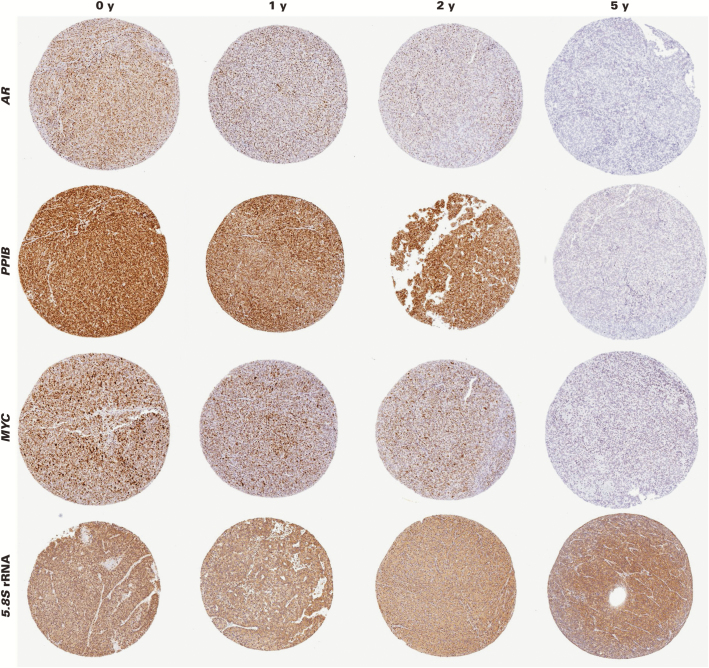
We further examined the effects of tissue block age on RISH using RNAscope by generating a set of four novel TMAs using the well-characterized human prostate cancer LuCaP PDXs (LuCaP 23.1, 35, 70, and 96). Each of these PDXs (eg, LUCaP 23.1) has been serially propagated in immune-compromised mice, and tumor tissue has been harvested and processed under the same conditions. FFPE tissue blocks were stored at room temperature for differing lengths of time. For each of the four TMAs, FFPE tissue blocks of each LuCaP PDX line were included from four time points as follows: (1) less than 1 year old (0 years), (2) approximately 1 year old (1 year), (3) 2 years old, and (4) 5 years old. For each time point, FFPE blocks from three different tumors of each PDX line were used in a given TMA. For each TMA, we performed RISH for four probe sets (n = 16 total TMA slides), including MYC, PPIB, AR, and 5.8S rRNA. We chose these probe sets because MYC is expressed in most prostate cancers; PPIB is an established positive control gene4; AR is well known to be expressed in most prostate cancers, including these PDXs50; and 5.8S rRNA is highly abundant, transcribed by RNA polymerase I, and may behave differently in terms of stability than a “typical” RNA polymerase II transcribed mRNA gene. The specificity of the in situ hybridization using RNAscope for AR exon 1 was established using a series of positive and negative control cell lines.13,14 The specificity of 5.8S rRNA was established herein by its ubiquitous expression across many tissues examined and the expected pattern of staining within cells, including nucleolar accumulation and abundant cytoplasmic dot-like foci consistent with the pattern expected of ribosomes Image 5.
Image 5.
RNA in situ hybridization on prostate cancer xenografts in relation to formalin-fixed, paraffin-embedded (FFPE) block age. Probe sets for AR, PPIB, MYC, and 5.8S ribosomal RNA were hybridized to FFPE slides from the LUCaP 35 xenograft stored in FFPE blocks at room temperature for the indicated lengths of time. Representative tissue microarray spots are shown. Note markedly reduced signals by 5 years of age. (×50)
As indicated above for the RRP TMAs, each TMA slide was digitally scanned, the composite TMA spot images were imported into TMAJ, and image analysis was performed such that a given threshold for brown staining was set, and this threshold was used for all subsequent TMA spots for that particular probe set. Areas of necrosis or obvious poor staining quality/artifacts were excluded. This process allowed for an unbiased estimate of overall brown signal levels on each TMA slide. TMA images were viewed by a pathologist (J.A.B.), who was blinded to the TMA design in terms of block age. For quantification, we used the total area of brown staining divided by the total tissue area (eg, the area fraction of staining) as determined by the software.
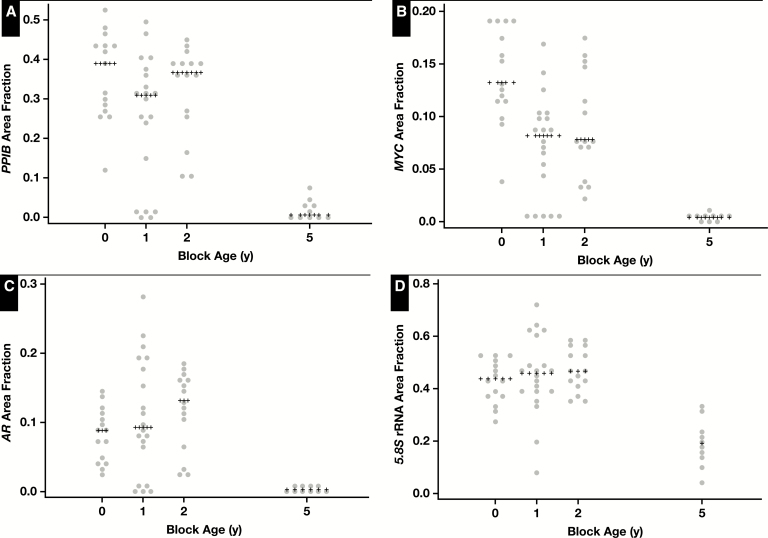
By visual inspection, it was clear that there was generally a marked reduction in RISH signal going from blocks that were less than 1 year old to blocks that were 5 years old, with the least reduction in the 5.8S rRNA probe set. Image 5 shows representative images for RISH signals for all four probe sets from the LuCaP 35 PDXs at the indicated time points. Figure 3 shows the results of the image analysis for each set of in situ probes across block age for the LuCaP 23.1 PDX tissues, and Supplementary Figure 5 shows the results for the other three xenografts. When expression of a given gene was examined across the different PDXs, there were differences in the apparent level of signals depending on which PDX was examined. Therefore, we used the quartiles of the area fraction of staining from each TMA for each probe set to combine data across the four TMAs for a given probe set for analysis. Table 1 shows these results along with results of statistical analysis showing a statistically significant decrease in signal with increasing block age for each of the probe sets.
Figure 3.
Quantitative scoring of signals for PPIB, MYC, AR, and 5.8S ribosomal RNA in relation to formalin-fixed, paraffin-embedded block age for the LUCaP 23.1 xenograft. Scores are shown for each probe set where each data point represents the score for a single tissue microarray spot. Median values are indicated by plus signs.
Table 1.
Relation Between In Situ Hybridization Signals and Block Age in Xenograft Tissues by Image Analysisa
| Characteristic | Block Age, y | |||
|---|---|---|---|---|
| 0 | 1 | 2 | 5 | |
| PPIB | ||||
| Quartile | ||||
| 1 (lowest) | 2 (3.4) | 4 (7.6) | 12 (22.6) | 39 (65.0) |
| 2 | 12 (20.3) | 12 (22.6) | 22 (41.5) | 10 (16.7) |
| 3 | 18 (30.5) | 19 (35.9) | 14 (26.4) | 6 (10.0) |
| 4 (highest) | 27 (45.8) | 18 (34.0) | 5 (9.4) | 5 (8.3) |
| P < .0001, χ2 | ||||
| MYC | ||||
| Quartile | ||||
| 1 (lowest) | 7 (11.7) | 5 (9.4) | 9 (17.0) | 36 (60.0) |
| 2 | 11 (18.3) | 14 (26.4) | 24 (45.3) | 7 (11.7) |
| 3 | 17 (28.3) | 19 (35.9) | 9 (17.0) | 12 (20.0) |
| 4 (highest) | 25 (41.7) | 15 (28.3) | 11 (21.0) | 5 (8.3) |
| P < .0001, χ2 | ||||
| AR | ||||
| Quartile | ||||
| 1 (lowest) | 3 (5.1) | 7 (13.2) | 6 (11.3) | 41 (69.5) |
| 2 | 11 (18.6) | 14 (26.4) | 25 (47.2) | 6 (10.2) |
| 3 | 20 (33.9) | 18 (34.0) | 10 (18.9) | 8 (13.6) |
| 4 (highest) | 25 (42.3) | 14 (26.4) | 12 (22.6) | 4 (6.8) |
| P < .0001, χ2 | ||||
| 5.8S | ||||
| Quartile | ||||
| 1 (lowest) | 9 (15.3) | 5 (9.4) | 19 (35.9) | 23 (39.7) |
| 2 | 14 (23.7) | 16 (30.2) | 17 (32.1) | 9 (15.5) |
| 3 | 17 (28.8) | 20 (37.7) | 11 (20.8) | 8 (13.8) |
| 4 (highest) | 19 (32.2) | 12 (22.6) | 6 (11.3) | 18 (31.0) |
| P < .0292, χ2 | ||||
aValues are presented as number (column percentages) in each quartile across all three patient-derived xenographs examined.
Frozen Tissues Preserve RISH Signals
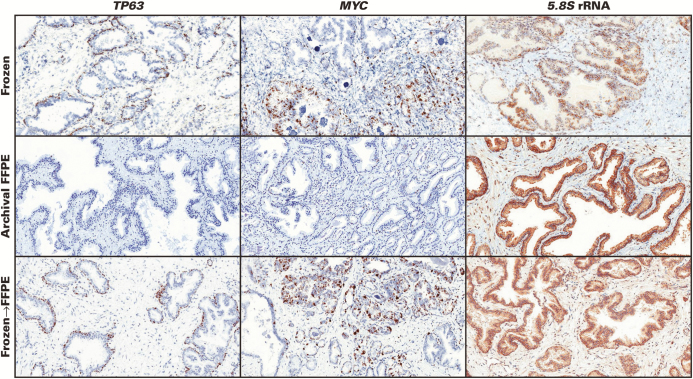
We next questioned whether the reduction of RISH signals with block age is an inherent property of tissue aging or whether such degradation in signal could be prevented. For this, we performed RISH for the same four probe sets using tissues from RRP specimens that were 10 years old but were originally snap frozen at the time of prostatectomy (n = 3 cases each with four probe sets, including PPIB, TP63, PTEN, and 5.8S rRNA) and compared these with FFPE blocks taken from neighboring areas of the same 10-year-old RRP cases but that were stored at room temperature as per standard protocol. Compared with the 10-year-old FFPE tissues that showed very weak signals for PPIB, TP63, and PTEN, the frozen sections from the snap-frozen samples contained much more robust signals, comparable to recent case FFPE samples (eg, <1 year old) Image 6. The signals for 5.8S rRNA were generally preserved, even in the 10-year-old FFPE blocks. To determine the effect of tissue processing, the 10-year-old frozen tissue blocks used for RISH were fixed in formalin, processed into FFPE, and then subjected to RISH for the same four probe sets. The signals were quite similar in these cases to their frozen-section counterparts (Image 6), indicating that the FFPE process itself does not cause reductions in RNAscope hybridization signals. Supplementary Table 2 shows the scores of the manual assessments results from these 36 slides.
Image 6.
Fresh-frozen tissues preserve RNAscope signals and formalin-fixed, paraffin-embedded (FFPE) processing itself does not reduce signals. Top row, prostate tissues that were snap frozen and stored at –80°C for 10 years. Center row, tissues from the same cases as top panel but that were stored as FFPE blocks for 10 years. Bottom row, tissues from the top panel were fixed in formalin and paraffin embedded and then subject to RNA in situ hybridization. Top left, frozen section of benign region for TP63 probe. Center left, same case as in top row but stored in FFPE block for 10 years showing completely lost signals. Bottom left, frozen tissue from top row was subjected to formalin fixation and paraffin embedding and shows retained signals, similar to the frozen section. Center column shows a case with both carcinoma and benign glands. Most carcinoma glands are positive for the MYC probe. Right column shows a case with benign glands that show strong signals in all three conditions, indicating very little loss of signal for the 5.8S ribosomal RNA probe in FFPE block stored for 10 years. (×100)
Storing Unstained FFPE Slides at –20°C Preserves RISH Signals
Freezing FFPE unstained slides at –20°C has been shown to preserve protein staining in a manner comparable to leaving tissue in FFPE blocks at room temperature, which is considered the standard best practice for protein preservation (eg, compared with leaving unstained slides at room temperature).51,52 Furthermore, storage of tissue blocks at 4°C helps to preserve RNA when solution-based analyses are used.20 Thus, we sought to determine whether cold storage (maintaining unstained slides at –20°C) for prolonged periods of time could preserve RISH signals. For this, we used unstained sections from three different tissue blocks that were stored at –20°C shortly after processing of tissues into FFPE (within 2 months) and compared these with unstained sections that were cut from the same FFPE blocks 5 years later after the blocks had been stored at room temperature. Supplementary Figure 6 shows a clear preservation of RISH signals over the 5 years, with much stronger signals for the two probe sets tested (MYC and PTEN) using unstained sections stored unstained at –20°C compared with unstained sections that were cut from the blocks stored for 5 years at room temperature.
To more formally test whether storage of slides at –20°C preserves signals compared with storing them at room temperature, we performed a prospective study in which we generated an additional “new case” TMA using tissues from new/recent cases (≤3 months old). For this TMA, we sectioned deep into the block and numbered each of the TMA slides. Then, for a group of four adjacent slides, we stored them under different conditions according to the following sequence: 1 = room temperature, 2 = room temperature with desiccant (manufacturer’s recommendation), 3 = –20°C, and 4 = –20°C with desiccant. We then performed RISH for each of the probe sets at different time points (1 month, 6 months, 1 year, and 2 years). At each time point, the staining with a given probe set (eg, PPIB) was performed under identical conditions in a single batch that included the adjacent slides stored under the four different conditions. For each time point, we digitally scanned the TMA slides, segmented the core images, imported the data and images into TMAJ/FrIDA, and performed image analysis in a blinded fashion with regard to storage conditions. For the image analysis, we quantified total brown area of staining using the same thresholds for the determination of brown staining for all TMA spots for all slides in this study. This unbiased assessment showed very little difference at 1 month, but differences became apparent at 6 months Image 7 and Supplementary Figure 7) and were highly significantly different for three of the four probe sets at 1 year and 2 years after block generation (Supplementary Figure 7 and Supplementary Table 3). These results clearly show that –20°C storage is superior to storage of slides at room temperature and that desiccant does not appear to add value after the 1-month time period. Also, the positive effects of using desiccant appear to be limited to early time points and to room temperature storage, with little effect at –20°C. Interestingly, as in the other tested scenarios, the 5.8S rRNA signals were much less affected by time of storage and temperature, suggesting that this transcript appears to be more robust in terms of degradation over time compared with the three mRNAs that were studied herein.
Image 7.

Representative images from prospective slide storage tissue microarray (TMA) study. Slides stored at room temperature for the indicated times are shown on the left, and slides stored at –20°C for indicated times are shown on the right. TMA spots are from adjacent slides, so histology is highly similar. Note very little difference at 1 month but a marked increase in signals in TMA spots stored at –20°C for 2 years. (×200)
Discussion
Localization of RNA expression in situ can provide a number of important insights into basic biology, disease mechanisms, and pathology diagnostics. Improvements in genomics-based approaches such as RNAseq are producing rapid growth in the number of RNA targets that investigators are seeking to localize in situ, especially in archival tissues that are formalin fixed and paraffin embedded. Recent enhancements in technology for in situ hybridization of RNA using commercially available reagents, such as those using “Z-pair” probes and branched DNA amplification, are rapidly being adapted to address this need. For example, as listed on the manufacturer’s websites to date (as of April 2017), since 2011 there have been 702 publications using RNAscope. Many of these studies have been conducted with archival FFPE tissues, either those that are from clinical surgical pathology specimens or research tissue blocks. Since it is known that RNA quality decreases with tissue block age in blocks stored at room temperature when using solution-based assays, it is important to systematically study the effects of tissue block age on this Z-pair/branched DNA in situ hybridization technology. In the present study, we show that the signal intensity and extent of RISH (using ACD’s RNAscope) correlated inversely with FFPE tissue block age, such that there were highly significant and marked reductions in hybridization signals after 5 years, and the reduction in signal often began to occur by 1 year of age. This finding was consistent across tissue blocks stored at room temperature (the current gold standard of storage) from two different institutions, using different tissue fixation and processing protocols. Furthermore, we found that storage of unstained slides at –20°C can preserve the signals, providing a relatively simple solution. Current studies are under way to determine whether storage of the FFPE blocks at –20°C provides a similar level of protection. While not formally included in this study, prior work in our laboratory using ViewRNA from Affymetrix (between 5 and 6 years ago) indicated that there were also tissue block age effects for a number of probe sets using RRP FFPE tissue blocks (eg, probe sets for MYC, PTEN, PCA3, and ERG; Q. Zheng and A. M. De Marzo, unpublished data). This is not surprising given that the underlying technology of using Z-pair probes and branched DNA amplifications is similar in both commercial applications. However, additional studies will be required to determine fully the extent of the effects of FFPE block age on Affymetrix ViewRNA.
It is well documented that storing slides at room temperature is detrimental for antibody immunoreactivity for a number of antigens53-56 compared with leaving the tissue in the FFPE blocks and storing them at room temperature. To preserve staining in such unstained slides, DiVito et al56 have advocated coating slides with paraffin to simulate leaving slides in the protective confines of the paraffin block, and the best results were obtained if these slides were prevented from undergoing further oxidation by storing them under nitrogen vapor. Others have also advocated coating slides with paraffin.57 However, we have found some difficulties with paraffin removal prior to staining from the paraffin-dipped slides, and it has been reported that this step does not necessarily help preserve epitopes, but it can result in a slight decrease in signals.58 Furthermore, freezing FFPE unstained slides at –20°C has been shown to preserve protein immunoreactivity similar to tissues kept in FFPE blocks at room temperature.51,52 Thus, in our study, we did not attempt to preserve RISH signals using paraffin slide dipping but rather opted for cold storage at –20°C.
While most agree that leaving tissue in FFPE blocks is superior to cutting the blocks and subsequently storing the cut sections at room temperature,59 a number of studies have reported decreased immune staining of at least some IHC-based markers with FFPE tissue block age.60,61 Thus, while we did not examine protein staining by IHC in this study, future studies should address whether storing unstained slides or FFPE blocks as in our study (at –20°C) can also preserve protein immunogenicity. Our preliminary data so far suggest that storing FFPE blocks at –20°C is also helpful for preserving RNAscope in situ signals (Q. Zheng and A. M. De Marzo, unpublished data). While the precise chemical mechanisms of RNA alterations occurring after prolonged FFPE storage at room temperature are still under investigation and may involve oxidative damage, hydrolysis, or both, it is clear that whatever the nature of the chemical processes resulting in the decreased RNA signals using Z-pair/branched DNA technology in situ, cold storage can prevent it. It will also be of interest to determine whether cold storage of FFPE slides or blocks also results in improved measures when examining solution-based RNA quality such as that of RT-PCR, capillary electrophoresis (eg, RIN number), and new methods being developed (eg, paraffin-embedded RNA metric62).
We acknowledge that our study has a number of limitations. For example, given the cost and time involved, we could only evaluate a limited number of probe sets, and thus it is not clear precisely how these findings will relate to other RNA targets in vivo. For example, mRNA stability is related to a number of factors, including the sequence, portion of the RNA examined (eg, 5′ or 3′ regions), and level and extent of protein binding; thus, the detected signals for different gene products may also be differentially affected by storage conditions in FFPE blocks as well. Nevertheless, we did find marked reductions in signals across a number of different mRNA targets and across tissues fixed and processed at two separate institutions using different protocols, which suggests that our findings are likely to be reasonably generalizable. Interestingly, while there was some degradation in signals, the probe targeting the 5.8S rRNA showed much less of a reduction in signal with storage time. This raises the question regarding whether some RNAs, particularly those tightly bound to a number of proteins, such as ribosomal RNAs, are better preserved in tissues over time. In fact, it has been shown that ribosomal RNAs are more resistant to degradation occurring over time in postmortem samples.63 This also raises the issue of using RIN numbers, which largely reflect rRNA levels, as the main gold standard when assessing RNA quality as a whole, which has indeed recently been challenged.62,64 Additional studies will be needed to determine whether general classes of RNA have more stability in FFPE tissues blocks with storage time.
In summary, our findings indicate that many studies using RISH may be significantly affected by FFPE tissue block age and that if this feature is not carefully considered in study designs, it could introduce bias into many results. For example, for RNAs with relatively low expression, older FFPE blocks may appear to be negative (eg, false negative) when in fact signals would be present in newer blocks (eg, true positive). Likewise, for genes with high expression, the expression would appear to be much lower than what was detected when older FFPE blocks are employed. This issue is especially a potential critical pitfall for studies examining the prognostic significance of in situ–based RNA expression that intrinsically use older tissue blocks or those studies in which tissue block age was not considered and tissues from different ages were employed without attention to this detail. Our prospective study clearly showed that a relatively simple solution of storage of unstained cut slides in the cold (–20°C) preserves RISH signals in unstained FFPE slides, and this is superior to leaving the tissues in FFPE blocks stored at room temperature. Since most of the millions of archival FFPE blocks in thousands of pathology laboratories and research laboratories throughout the world are stored at room temperature (including human and animal tissues), our findings challenge the conventional wisdom. If these results are replicated in other studies using different tissue types and probe sets, it may be appropriate to reconsider the longstanding tradition of storing archival tissue blocks at room temperature. A simple alternative (cold storage of unstained slides) may help to better preserve invaluable RNA-based information currently locked in massive FFPE archives.
Supplementary Material
Acknowledgments:
We thank Kristen Lecksell and Karen Fox-Talbot for help with slide scanning and the members of the Oncology Tissue Services Core for efforts related to histology.
Footnotes
This work was supported by the US Department of Defense Prostate Cancer Research Program (W81XWH-14-2-0182 and W81XWH-14-2-0183), Prostate Cancer Biorepository Network, and National Cancer Institute/National Institutes of Health (Prostate SPORE P50CA58236, 5P30CA006973).
References
- 1. Player AN, Shen LP, Kenny D et al. Single-copy gene detection using branched DNA (bDNA) in situ hybridization. J Histochem Cytochem. 2001;49:603-612. [DOI] [PubMed] [Google Scholar]
- 2. Itzkovitz S, van Oudenaarden A. Validating transcripts with probes and imaging technology. Nat Methods. 2011;8(suppl):S12-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kwon S. Single-molecule fluorescence in situ hybridization: quantitative imaging of single RNA molecules. BMB Rep. 2013;46:65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang F, Flanagan J, Su N et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anderson CM, Zhang B, Miller M et al. Fully automated RNAscope in situ hybridization assays for formalin-fixed paraffin-embedded cells and tissues. J Cell Biochem. 2016;117:2201-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kerr DA, Arora KS, Mahadevan KK et al. Performance of a branch chain RNA in situ hybridization assay for the detection of high-risk human papillomavirus in head and neck squamous cell carcinoma. Am J Surg Pathol. 2015;39:1643-1652. [DOI] [PubMed] [Google Scholar]
- 7. Bordeaux JM, Cheng H, Welsh AW et al. Quantitative in situ measurement of estrogen receptor mRNA predicts response to tamoxifen. PLoS One. 2012;7:e36559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Palmer MV, Thacker TC, Waters WR. Analysis of cytokine gene expression using a novel chromogenic in-situ hybridization method in pulmonary granulomas of cattle infected experimentally by aerosolized Mycobacterium bovis. J Comp Pathol. 2015;153:150-159. [DOI] [PubMed] [Google Scholar]
- 9. Bishop JA, Ma XJ, Wang H et al. Detection of transcriptionally active high-risk HPV in patients with head and neck squamous cell carcinoma as visualized by a novel E6/E7 mRNA in situ hybridization method. Am J Surg Pathol. 2012;36:1874-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Prensner JR, Iyer MK, Sahu A et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Castellanos-Rubio A, Fernandez-Jimenez N, Kratchmarov R et al. A long noncoding RNA associated with susceptibility to celiac disease. Science. 2016;352:91-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. St Laurent G, Vyatkin Y, Antonets D et al. ; FANTOM Consortium Functional annotation of the vlinc class of non-coding RNAs using systems biology approach. Nucleic Acids Res. 2016;44:3233-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Antonarakis ES, Lu C, Wang H et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guedes LB, Morais CL, Almutairi F et al. Analytic validation of RNA in situ hybridization (RISH) for AR and AR-V7 expression in human prostate cancer. Clin Cancer Res. 2016;22:4651-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu SH, Zheng Q, Esopi D et al. A paracrine role for IL6 in prostate cancer patients: lack of production by primary or metastatic tumor cells. Cancer Immunol Res. 2015;3:1175-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gervasi NM, Scott SS, Aschrafi A et al. The local expression and trafficking of tyrosine hydroxylase mRNA in the axons of sympathetic neurons. RNA. 2016;22:883-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Treutlein B, Brownfield DG, Wu AR et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vassilakopoulou M, Togun T, Dafni U et al. In situ quantitative measurement of HER2mRNA predicts benefit from trastuzumab-containing chemotherapy in a cohort of metastatic breast cancer patients. PLoS One. 2014;9:e99131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cronin M, Pho M, Dutta D et al. Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am J Pathol. 2004;164:35-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. von Ahlfen S, Missel A, Bendrat K et al. Determinants of RNA quality from FFPE samples. PLoS One. 2007;2:e1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dunn TA, Fedor H, Isaacs WB et al. Genome-wide expression analysis of recently processed formalin-fixed paraffin embedded human prostate tissues. Prostate. 2009;69:214-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Müller BM, Kronenwett R, Hennig G et al. Quantitative determination of estrogen receptor, progesterone receptor, and HER2 mRNA in formalin-fixed paraffin-embedded tissue—a new option for predictive biomarker assessment in breast cancer. Diagn Mol Pathol. 2011;20:1-10. [DOI] [PubMed] [Google Scholar]
- 23. Hedegaard J, Thorsen K, Lund MK et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One. 2014;9:e98187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zheng Q, Peskoe SB, Ribas J et al. Investigation of miR-21, miR-141, and miR-221 expression levels in prostate adenocarcinoma for associated risk of recurrence after radical prostatectomy. Prostate. 2014;74:1655-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luder Ripoli F, Mohr A, Conradine Hammer S et al. A comparison of fresh frozen vs. formalin-fixed, paraffin-embedded specimens of canine mammary tumors via branched-DNA assay. Int J Mol Sci. 2016;17:E724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zha S, Gage WR, Sauvageot J et al. Cyclooxygenase-2 is up-regulated in proliferative inflammatory atrophy of the prostate, but not in prostate carcinoma. Cancer Res. 2001;61:8617-8623. [PubMed] [Google Scholar]
- 27. Ruijter ET, Miller GJ, Aalders TW et al. Rapid microwave-stimulated fixation of entire prostatectomy specimens. Biomed-II MPC study group. J Pathol. 1997;183:369-375. [DOI] [PubMed] [Google Scholar]
- 28. Ellis WJ, Vessella RL, Buhler KR et al. Characterization of a novel androgen-sensitive, prostate-specific antigen-producing prostatic carcinoma xenograft: LuCaP 23. Clin Cancer Res. 1996;2:1039-1048. [PubMed] [Google Scholar]
- 29. Corey E, Quinn JE, Buhler KR et al. LuCaP 35: a new model of prostate cancer progression to androgen independence. Prostate. 2003;55:239-246. [DOI] [PubMed] [Google Scholar]
- 30. Linja MJ, Porkka KP, Kang Z et al. Expression of androgen receptor coregulators in prostate cancer. Clin Cancer Res. 2004;10:1032-1040. [DOI] [PubMed] [Google Scholar]
- 31. Sun X, Frierson HF, Chen C et al. Frequent somatic mutations of the transcription factor ATBF1 in human prostate cancer. Nat Genet. 2005;37:407-412. [DOI] [PubMed] [Google Scholar]
- 32. Tan HL, Haffner MC, Esopi DM et al. Prostate adenocarcinomas aberrantly expressing p63 are molecularly distinct from usual-type prostatic adenocarcinomas. Mod Pathol. 2015;28:446-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lotan TL, Gurel B, Sutcliffe S et al. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res. 2011;17:6563-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Faith DA, Isaacs WB, Morgan JD et al. Trefoil factor 3 overexpression in prostatic carcinoma: prognostic importance using tissue microarrays. Prostate. 2004;61:215-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fedor HL, De Marzo AM. Practical methods for tissue microarray construction. Methods Mol Med. 2005;103:89-101. [DOI] [PubMed] [Google Scholar]
- 36. Gurel B, Iwata T, Koh CM et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21:1156-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cornish TC, Morgan J, Gurel B et al. FrIDA: an open source framework for image dataset analysis. Arch Pathol Lab Med. 2008;132:856. [Google Scholar]
- 38. Sfanos KS, Canene-Adams K, Hempel H et al. Bacterial prostatitis enhances 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP)–induced cancer at multiple sites. Cancer Prev Res (Phila). 2015;8:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Signoretti S, Waltregny D, Dilks J et al. P63 is a prostate basal cell marker and is required for prostate development. Am J Pathol. 2000;157:1769-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parsons JK, Gage WR, Nelson WG et al. P63 protein expression is rare in prostate adenocarcinoma: implications for cancer diagnosis and carcinogenesis. Urology. 2001;58:619-624. [DOI] [PubMed] [Google Scholar]
- 41. Furusato B, Gao CL, Ravindranath L et al. Mapping of TMPRSS2-ERG fusions in the context of multi-focal prostate cancer. Mod Pathol. 2008;21:67-75. [DOI] [PubMed] [Google Scholar]
- 42. Park K, Tomlins SA, Mudaliar KM et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Furusato B, Tan SH, Young D et al. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis. 2010;13:228-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tomlins SA, Rhodes DR, Perner S et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644-648. [DOI] [PubMed] [Google Scholar]
- 45. Mertz KD, Setlur SR, Dhanasekaran SM et al. Molecular characterization of TMPRSS2-ERG gene fusion in the NCI-H660 prostate cancer cell line: a new perspective for an old model. Neoplasia. 2007;9:200-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Popa I, Fradet Y, Beaudry G et al. Identification of PCA3 (DD3) in prostatic carcinoma by in situ hybridization. Mod Pathol. 2007;20:1121-1127. [DOI] [PubMed] [Google Scholar]
- 47. Warrick JI, Tomlins SA, Carskadon SL et al. Evaluation of tissue PCA3 expression in prostate cancer by RNA in situ hybridization—a correlative study with urine PCA3 and TMPRSS2-ERG. Mod Pathol. 2014;27:609-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vlietstra RJ, van Alewijk DC, Hermans KG et al. Frequent inactivation of PTEN in prostate cancer cell lines and xenografts. Cancer Res. 1998;58:2720-2723. [PubMed] [Google Scholar]
- 49. Hubbard GK, Mutton LN, Khalili M et al. Combined MYC activation and pten loss are sufficient to create genomic instability and lethal metastatic prostate cancer. Cancer Res. 2016;76:283-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Corey E, Vessella RL. Xenograft models of human prostate cancer. In: Chung LW, Isaacs WB, Simons JW, eds. Prostate Cancer: Biology, Genetics and the New Therapeutics. 2nd ed. Totowa, NJ: Springer; 2007:3-31. [Google Scholar]
- 51. Wester K, Wahlund E, Sundström C et al. Paraffin section storage and immunohistochemistry: effects of time, temperature, fixation, and retrieval protocol with emphasis on p53 protein and MIB1 antigen. Appl Immunohistochem Mol Morphol. 2000;8:61-70. [PubMed] [Google Scholar]
- 52. Cornish TC, De Marzo AM. Tissue microarrays in cancer research. In: Yegnasubramanian S, Isaacs WB, eds. Modern Molecular Biology: Approaches for Unbiased Discovery in Cancer Research. New York, NY: Springer; 2010:157-184. [Google Scholar]
- 53. Jacobs TW, Prioleau JE, Stillman IE et al. Loss of tumor marker-immunostaining intensity on stored paraffin slides of breast cancer. J Natl Cancer Inst. 1996;88:1054-1059. [DOI] [PubMed] [Google Scholar]
- 54. Mirlacher M, Kasper M, Storz M et al. Influence of slide aging on results of translational research studies using immunohistochemistry. Mod Pathol. 2004;17:1414-1420. [DOI] [PubMed] [Google Scholar]
- 55. Fergenbaum JH, Garcia-Closas M, Hewitt SM et al. Loss of antigenicity in stored sections of breast cancer tissue microarrays. Cancer Epidemiol Biomarkers Prev. 2004;13:667-672. [PubMed] [Google Scholar]
- 56. DiVito KA, Charette LA, Rimm DL et al. Long-term preservation of antigenicity on tissue microarrays. Lab Invest. 2004;84:1071-1078. [DOI] [PubMed] [Google Scholar]
- 57. Gelb AB, Freeman VA, Astrow SH. Evaluation of methods for preserving PTEN antigenicity in stored paraffin sections. Appl Immunohistochem Mol Morphol. 2011;19:569-573. [DOI] [PubMed] [Google Scholar]
- 58. Karlsson C, Karlsson MG. Effects of long-term storage on the detection of proteins, DNA, and mRNA in tissue microarray slides. J Histochem Cytochem. 2011;59:1113-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Manne U, Myers RB, Srivastava S et al. Re: loss of tumor marker-immunostaining intensity on stored paraffin slides of breast cancer. J Natl Cancer Inst. 1997;89:585-586. [DOI] [PubMed] [Google Scholar]
- 60. Vis AN, Kranse R, Nigg AL et al. Quantitative analysis of the decay of immunoreactivity in stored prostate needle biopsy sections. Am J Clin Pathol. 2000;113:369-373. [DOI] [PubMed] [Google Scholar]
- 61. Toubaji A, Sutcliffe S, Chaux A et al. Immunohistochemical expression of minichromosome maintenance complex protein 2 predicts biochemical recurrence in prostate cancer: a tissue microarray and digital imaging analysis-based study of 428 cases. Hum Pathol. 2012;43:1852-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chung JY, Cho H, Hewitt SM. The paraffin-embedded RNA metric (PERM) for RNA isolated from formalin-fixed, paraffin-embedded tissue. Biotechniques. 2016;60:239-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sidova M, Tomankova S, Abaffy P et al. Effects of post-mortem and physical degradation on RNA integrity and quality. Biomol Detect Quantif. 2015;5:3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Björkman J, Švec D, Lott E et al. Differential amplicons (δamp)—a new molecular method to assess RNA integrity. Biomol Detect Quantif. 2016;6:4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.