Summary
Twenty-one patients were treated with ceftolozane-tazobactam for multidrug-resistant Pseudomonas aeruginosa infections, predominantly pneumonia. Thirty- and 90-day mortality rates were 10% and 48%, respectively. Resistance emerged in 3 patients, which was associated with mutations in and/or increased expression of ampC β-lactamase.
Keywords: ceftolozane-tazobactam, MDR Pseudomonas, resistance mechanisms, AmpC beta-lactamase, omega loop
Abstract
Background
Data on the use of ceftolozane-tazobactam and emergence of ceftolozane-tazobactam resistance during multidrug resistant (MDR)-Pseudomonas aeruginosa infections are limited.
Methods
We performed a retrospective study of 21 patients treated with ceftolozane-tazobactam for MDR-P. aeruginosa infections. Whole genome sequencing and quantitative real-time polymerase chain reaction were performed on longitudinal isolates.
Results
Median age was 58 years; 9 patients (43%) were transplant recipients. Median simplified acute physiology score-II (SAPS-II) was 26. Eighteen (86%) patients were treated for respiratory tract infections; others were treated for bloodstream, complicated intraabdominal infections, or complicated urinary tract infections. Ceftolozane-tazobactam was discontinued in 1 patient (rash). Thirty-day all-cause and attributable mortality rates were 10% (2/21) and 5% (1/21), respectively; corresponding 90-day mortality rates were 48% (10/21) and 19% (4/21). The ceftolozane-tazobactam failure rate was 29% (6/21). SAPS-II score was the sole predictor of failure. Ceftolozane-tazobactam resistance emerged in 3 (14%) patients. Resistance was associated with de novo mutations, rather than acquisition of resistant nosocomial isolates. ampC overexpression and mutations were identified as potential resistance determinants.
Conclusions
In this small study, ceftolozane-tazobactam was successful in treating 71% of patients with MDR-P. aeruginosa infections, most of whom had pneumonia. The emergence of ceftolozane-tazobactam resistance in 3 patients is worrisome and may be mediated in part by AmpC-related mechanisms. More research on treatment responses and resistance during various types of MDR-P. aeruginosa infections is needed to define ceftolozane-tazobactam’s place in the armamentarium.
Infections due to multidrug-resistant (MDR)–Pseudomonas aeruginosa are associated with poor outcomes [1–7]. β-lactams are therapeutic mainstays, but development of resistance limits their effectiveness [8, 9]. A signature resistance mechanism in P. aeruginosa is production of AmpC β-lactamase, which hydrolyzes penicillins, monobactams, and oxyimino-cephalosporins (except cefepime) but not carbapenems [10, 11]. Other important β-lactam resistance mechanisms include multidrug efflux pumps and loss of outer membrane porin OprD [12–19]. Acquisition of plasmid-borne extended-spectrum β-lactamases (ESBLs) and carbapenemases is uncommon among P. aeruginosa in the United States [13, 19, 20].
Ceftolozane–tazobactam was recently approved by the US Food and Drug Administration (FDA) for the treatment of complicated intraabdominal and urinary tract infections (cIAIs, cUTIs) [21, 22]. Ceftolozane is an oxyimino-cephalosporin that structurally resembles ceftazidime but has increased activity against P. aeruginosa and decreased susceptibility to AmpC hydrolysis [23, 24]. We previously showed that 92% of 38 meropenem-resistant P. aeruginosa isolates at our center were susceptible to ceftolozane-tazobactam in vitro [25]. Clinical experience with ceftolozane-tazobactam for the treatment of MDR-P. aeruginosa infections is limited. Furthermore, the extent to which ceftolozane-tazobactam resistance may emerge in MDR-P. aeruginosa isolates during treatment is unknown. Our objectives in this study were to describe our experience in treating MDR-P. aeruginosa infections with ceftolozane-tazobactam, assess emergence of resistance, and identify possible resistance mechanisms.
METHODS
Study Design and Definitions
We conducted a retrospective study of patients with MDR-P. aeruginosa infections treated with ceftolozane-tazobactam at the University of Pittsburgh Medical Center from June 2015 to March 2016. MDR was defined by nonsusceptibility to ≥1 agent in ≥3 classes that are typically active against P. aeruginosa [26]. Types of infection were classified according to National Healthcare Safety Network criteria [27]. FDA-approved dosing was defined as compliance with the FDA label dosage for ≥5 days during the first week of therapy, regardless of the site of infection (1.5 g intravenously [IV] every 8 hours with adjustments for renal dysfunction and intermittent hemodialysis [iHD]) [28]. Recent pharmacokinetic (PK) data suggest that 3 g IV every 8 hours may improve target attainment within pulmonary epithelial lining fluid (ELF) [29]. This dosage is being used in a phase 3 trial of ventilator-associated pneumonia (VAP) but it is not currently FDA approved for any indication [30]. PK-derived dosing was defined as compliance with the higher dosing regimen for respiratory tract infections (with renal adjustment) for ≥5 days during the first week of therapy. By these definitions, a patient with a creatinine clearance >50 mL/min treated for pneumonia with a dose of 1.5 g every 8 hours would be labelled as receiving FDA-approved dosing, whereas a patient treated with 3 g every 8 hours would be labelled as receiving PK-derived dosing. Since the phase 3 VAP trial excludes patients receiving any form of renal replacement therapy, we considered dosing for patients who had pneumonia but were on renal replacement therapy as “not defined” [30].
Primary outcome was 30-day all-cause mortality. Secondary outcomes were 90-day all-cause mortality, 30- and 90-day attributable mortality, 90-day clinical failure, recurrent colonization, and emergence of resistance. Mortality was attributed to P. aeruginosa if the patient died with signs and symptoms of infection, microbiologic or histological evidence of an active P. aeruginosa infection, and if other potential causes of death were reasonably excluded. Although attributable mortality is often difficult to ascertain and definitions are controversial, we included it in the outcome analysis in order to compare our data with attributable mortality rates reported in previous studies of pseudomonal infection. Clinical failure was defined as attributable mortality due to P. aeruginosa, persistent signs or symptoms of infection or positive culture despite ≥7 days of ceftolozane-tazobactam, or recurrent P. aeruginosa infection (recurrent signs and symptoms and recurrent culture positivity within 90 days). Combination therapy was defined as receipt of ceftolozane-tazobactam plus ≥1 anti-pseudomonal drug for ≥72 hours. Acute kidney injury (AKI) was defined as ≥1.5-fold increase in serum creatinine from baseline. Ceftolozane-tazobactam resistance was defined as minimum inhibitory concentration (MIC) ≥16 µg/mL (E-test), in accordance with Clinical Laboratory and Standards Institute recommendations [31]. MICs for other agents were determined by MicroScan or disc diffusion.
Whole Genome Sequencing and Analysis
Full details of whole genome sequencing (WGS) and analysis are provided in the Supplementary Methods [32]. Susceptible isolates from 5 patients prior to ceftolozane-tazobactam therapy were selected as ancestral strains for phylogenetic analyses. A P. aeruginosa genome most closely related to the consensus of the isolates (PA_BWHPSA022, as revealed by Mash [33]) was used to determine phylogenetic relationships among isolates. Longitudinal isolates with preexisting (rather than emergent) ceftolozane-tazobactam resistance from a sixth patient were sequenced as controls (this patient is referred to as patient 22). The genome of the founding isolate for each patient was the reference for identifying putative evolved mutations (single nucleotide polymorphisms [SNPs], insertions–deletions [indels], and structural variants) in subsequent isolates by breseq [34]. Raw predicted mutations and filtered lists of mutations are reported in Supplementary Tables 1 and 2. The filtered list, per patient, was curated to highlight genes that were categorically linked to β-lactam resistance in previous studies, including β-lactamases, efflux pumps, porins, and cell wall synthesis machinery [35–37].
Quantitative Reverse Transcription-Polymerase Chain Reaction
DNase-treated RNA was obtained from late-exponential phase cultures in Luria broth at 37°C (RiboPure-Bacteria kit, ThermoFisher Scientific, Waltham, Massachusetts). cDNA was made using qScript cDNAMix (Quanta Biosciences, Gaithersburg, Maryland). Quantitative reverse transcription-polymerase chain reaction (qRT–PCR) of P. aeruginosa genes encoding common β-lactam resistance mechanisms, such as ampC, efflux genes mexB, mexD, mexY, and porin gene oprD, was performed using the Applied Biosystems 7900 system, with established primers (Supplementary Table 2 [38–40]), and the SYBR Green kit (Quanta Biosciences, Maryland). Gene expression was normalized using rspL. Relative expression was calibrated against corresponding baseline ceftolozane-tazobactam susceptible isolates. qRT-PCR for all isolates was performed in at least triplicate on 3 separate days.
Statistical Analyses
Statistical analysis was performed using Stata 13.0 (College Station, Texas) and GraphPad Instat 3 (San Diego, California). Univariate analysis of contingency data was performed by 2-tailed χ2 or 2-tailed Fisher exact tests. P < .05 was considered significant.
RESULTS
Patient Characteristics, Microbiology, and Treatment Regimens
Twenty-one patients were included (Table 1). Nine patients (43%) were transplant recipients (7 lung, 1 lung–kidney, 1 stem cell). Twenty patients (95%) received an anti-pseudomonal antibiotic within 14 days prior to the index culture. Types of infection and susceptibility profiles are shown in Table 1. Eighteen patients (86%) were treated for respiratory tract infections; the remaining 3 patients were treated for bacteremia, cIAI, or cUTI. Initial isolates were MDR but susceptible to ceftolozane- tazobactam (median MIC, 1.5 µg/mL, range, 0.75–4 µg/mL). Fifteen (71%) of the initial isolates were resistant to all anti-pseudomonal β-lactams tested except ceftolozane-tazobactam. Six patients (29%) had coinfections with other pathogens (Table 2).
Table 1.
Demographics, clinical descriptions, and resistance patterns
| Factor | Percent (n) | Age or Score |
|---|---|---|
| Age, median (range) | 58 y (23–91) | |
| Male sex % (n) | 48 (10) | |
| Underlying diseases | ||
| Immunosuppressed | 43 (9) | |
| Organ transplant | 38 (8) | |
| Stem cell transplant | 5 (1) | |
| Ventilator-dependent respiratory failure | 38 (8) | |
| Surgery within 30 days prior to index culture | 33 (7) | |
| Cystic fibrosis | 29 (6)a | |
| Renal failure requiring renal replacement therapy at the time of initiation of ceftolozane-tazobactam | 24 (5) | |
| Cardiovascular disease | 10 (2) | |
| Malignancy | 10 (2) | |
| Chronic obstructive pulmonary disease | 10 (2) | |
| Severity of illness scores | ||
| Simplified acute physiology score-II score, median (range) | 26 (8–49) | |
| Sequential organ failure assessment score, median (range) | 6 (0–17) | |
| Charlson comorbidity index, median (range) | 5 (1–12) | |
| Type of infection | ||
| Respiratory tract | 86 (18) | |
| Pneumoniab | 76 (16) | |
| Purulent tracheobronchitis | 10 (2) | |
| Recurrent bacteremia | 5 (1) | |
| Complicated intraabdominal infection | 5 (1) | |
| Complicated urinary tract infection | 5 (1) | |
| Coinfection with other pathogensc | 29 (6) | |
| Antibiotic resistance | ||
| ≥1 anti-pseudomonal fluoroquinoloned | 95 (20) | |
| Aztreonam | 95 (20) | |
| Cefepime | 90 (19) | |
| ≥1 anti-pseudomonal carbapeneme | 90 (19) | |
| Piperacillin-tazobactam | 81 (17) | |
| Ceftazidime | 76 (16) | |
| ≥1 aminoglycoside | 67 (14) | |
| Colistin | 20 (2/10)f |
aFour of 6 cystic fibrosis patients were lung transplant recipients
bTwo had empyema, which was surgically drained.
cDetails in Table 2.
dCiprofloxacin and/or levofloxacin.
eMeropenem, imipenem, and/or doripenem.
fColistin susceptibility testing is performed upon clinician request (10 isolates).
Table 2.
Clinical Characteristics, Antibiotic Regimens, and Outcome of Patients Treated With Ceftolozane-tazobactam for Multidrug Resistant-Pseudomonas aeruginosa Infections
| Patient | Age in Years (Sex) | Underlying Diseases | Type of Infection | SAPS II, SOFA Scores | Dosing (g every 8 hours) | Duration of Therapy With Ceftolozane- Tazobactam (days) | Anti-Pseudomonal Agents Given With Ceftolozane- Tazobactam for ≥72 Hoursc | Coinfection With Other Pathogens (treatment) | Outcome | Ceftolozane-Tazobactam Minimum Inhibitory Concentration (µg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CrCl (mL/ min) | FDA- Label dose | Actual Dose | FDA or PK-Derived Dosing | Outcome at 30 and 90 Days | Clinical Outcome (success or failure) | Recurrent Infections or Colonization at 90 Days | Initial Isolate | Subsequent Isolate(s) | ||||||||
| 1 | 58 (M) | COPD, VDRF | Pneumonia, empyema | 35, 11 | iHD | Not defined | 0.15 | Not defineda | 29 | Inhaled tobramycin | None | Died within 90 days (attributable) | Failure | Recurrent infections due to resistant isolates | 4 | 32 (14 days after therapy) |
| 2 | 23 (F) | Cystic fibrosis, lung transplant, VDRF | Pneumonia | 23, 6 | >50 | 1.5 | 1.5 | FDA dosing | 14 | Inhaled tobramycin | MRSA pneumonia (linezolid) | Alive | Success | 2 | ||
| 3 | 84 (F) | Dementia | Pneumonia | 25, 7 | >50 | 1.5 | 1.5 | FDA dosing | 17 | Inhaled colistin | MRSA pneumonia (linezolid) | Alive | Success | 4 | ||
| 4 | 70 (M) | Lung transplant | Pneumonia, empyema | 35, 8 | CRRT | Not defined | 1.5 | Not defineda | 14 | Ciprofloxacin, inhaled colistin, inhaled tobramycin | None | Alive | Failure | Recurrent infection | 2 | 2 (6 days after therapy) |
| 5 | 48 (F) | Cystic fibrosis, lung transplant, VDRF | Pneumonia | 19, 3 | >50 | 1.5 | 3 | PK-dosing | 41 | Ciprofloxacin, inhaled colistin | None | Alive | Success | 2 | ||
| 6 | 75 (M) | Dementia, VDRF | Purulent tracheobronchitis | 27, 4 | >50 | 1.5 | 1.5 | FDA dosing | 31 | Inhaled tobramycin | None | Died within 90 days (attributable) | Failure | Recurrent infection | Isolate N/A | Isolate N/A |
| 7 | 58 (F) | Pancreatitis, VDRF | Complicated intraabdominal infection | 26, 8 | 15–29 | 0.75 | 0.75 | FDA dosing | 40 | Inhaled colistin | None | Alive | Success | Colonization with resistant isolates | 4 | 128 (day 8 of therapy); >256, 128, 256 (3, 20, and 41 days after therapy, respectively) |
| 8 | 55 (F) | Cystic fibrosis | Pneumonia | 19, 3 | >50 | 1.5 | 3 | PK-dosing | 42 | Ciprofloxacin, tobramycin | None | Alive | Failure | Recurrent infections due to resistant isolates | 2 | 32 (day 17 of therapy), 64 (19 days after therapy) |
| 9 | 25 (F) | Cystic fibrosis, lung and kidney transplant | Pneumonia | 26, 4 | 30–50 | 0.75 | 1.5 | PK-dosing | 52 | Inhaled ceftazidime, inhaled colistin, ciprofloxacin, meropenem | None | Died within 90 days (not attributable) | Success | 2 | ||
| 10 | 89 (F) | Dementia, VDRF | Pneumonia | 40, 10 | 30–50 | 0.75 | 0.75 | FDA dosing | 14 | Inhaled tobramycin | MRSA pneumonia (vancomycin) | Died within 90 days (attributable) | Failure | Recurrent infection | 2 | 2 (24 days after therapy) |
| 11 | 84 (F) | Mesenteric ischemia | Pneumonia | 43, 15 | 15–29 | 0.375 | 0.375 | FDA dosing | 3 | Inhaled tobramycin | None | Died within 30 days (attributable) | Failure | 1 | ||
| 12 | 91 (F) | Dementia, VDRF | Pneumonia | 42, 8 | 30–50 | 0.75 | 0.75 | FDA dosing | 10 | None | Serratia marcescens pneumonia (did not require additional antibiotics) | Died within 90 days (not attributable) | Success | 1 | ||
| 13 | 59 (F) | Stem cell transplant | Pneumonia | 20, 4 | >50 | 1.5 | 3 | PK-dosing | 13 | Gentamicin | VRE bacteremia (linezolid) | Died within 90 days (not attributable) | Success | 2 | ||
| 14 | 41 (F) | Lung transplant | Pneumonia | 14, 1 | >50 | 1.5 | 3 | PK-dosing | 14 | None | None | Alive | Success | Colonization | 0.5, 1 | 0.5 (20 days after therapy) |
| 15 | 58 (M) | Lung transplant | Pneumonia | 34, 8 | iHD | Not defined | 0.15 | Not defineda | 15 | Inhaled tobramycin | None | Alive | Success | Colonization | 2, 4 | 2, 4 (28 days after therapy) |
| 16 | 58 (M) | Lung transplant | Recurrent bacteremia | 18, 5 | iHD | Not defined | 0.375 | Not definedb | 48 | Ciprofloxacin, inhaled tobramycin | None | Died within 90 days (not attributable) | Success | 1 | ||
| 17 | 23 (M) | Cystic fibrosis | Pneumonia | 8, 0 | >50 | 1.5 | 1.5 | FDA dosing | 10 | Imipenem, inhaled colistin, tobramycin | None | Alive | Success | Isolate N/A | ||
| 18 | 67 (M) | Diabetes mellitus, resection of bladder carcinoma | Complicated urinary tract infection | 14, 4 | >50 | 1.5 | 1.5 | FDA dosing | 10 | None | None | Alive | Success | Colonization | 1 | 0.5, 1 (41 days after therapy) |
| 19 | 39 (M) | Biliary surgery with abscesses, VDRF | Pneumonia | 29, 17 | CRRT | Not defined | 1.5 | Not defineda | 13 | None | VRE bacteremia (daptomycin); Citrobacter fruendii abdominal wound infection (ceftolozane-tazobactam); Candida tropicalis fungemia (caspofungin) | Died within 30 days (not attributable) | Success | 2 | ||
| 20 | 65 (M) | Recent cardiac arrest | Pneumonia | 49, 15 | 30–50 | 0.75 | 0.75 | FDA dosing | 13 | Inhaled tobramycin | None | Died within 90 days (not attributable) | Success | 0.75 | ||
| 21 | 34 (M) | Cystic fibrosis, lung transplant | Purulent tracheobronchitis | 18, 2 | >50 | 1.5 | 1.5 | FDA dosing | 4 | None | None | Alive | Success | NA | ||
Creatinine clearance was calculated using the Cockroft-Gault formula; dosing encompasses the dose used for ≥5 days during the first week of therapy.
Abbreviations: COPD, chronic obstructive pulmonary disease; CrCl, creatinine clearance; CRRT, continuous renal replacement therapy; F, female; FDA, US Food and Drug Administration; iHD, intermittent hemodialysis; M, male; MRSA, methicillin-resistant Staphylococcus aureus; N/A, not available; PK, pharmacokinetic; SAPS-II, simplified acute physiology score; SOFA, sequential organ failure assessment; VDRF, ventilator-dependent respiratory failure; VRE, vancomycin-resistant Enterococcus faecium.
aThese patients were on either intermittent hemodialysis or continuous renal replacement therapy, situations in which there are no PK-derived data for respiratory infections.
bThis patient had primary bacteremia and was receiving intermittent hemodialysis; dosing for this situation is not established.
cNumber of isolates susceptible to the antibiotics used in combination with ceftolozane-tazobactam were as follows: ciprofloxacin (2/5 susceptible), tobramycin (2/2 susceptible), gentamicin (1/1 susceptible), meropenem (1/1 susceptible), imipenem (1/1 resistant), inhaled tobramycin (9/9 susceptible), inhaled colistin (6/6 susceptible), inhaled ceftazidime (1/1 susceptible).
Median duration of therapy was 14 days (range, 3–52 days). Of 18 patients with respiratory tract infections, 5 (28%) received PK-derived dosages and 9 (50%) received FDA-approved dosages. Patients with nonrespiratory tract infections were treated with FDA-approved dosages. Four (22%) of 18 patients with respiratory tract infections were receiving renal replacement therapy, and a patient with primary bacteremia was receiving iHD (Table 2); these are settings in which dosing is not defined. Sixteen (76%) patients received combination anti-pseudomonal therapy for ≥72 hours, including 2, 9, and 5 who received concomitant systemic, inhaled, and both systemic and inhaled agents, respectively (Table 2).
Outcomes
The 30-day mortality rate was 10% (2/21) and the attributable mortality rate was 5% (1/21) (Table 2). Corresponding 90-day rates were 48% (10/21) and 19% (4/21). Attributable 90-day mortality was due to persistent or recurrent MDR-P. aeruginosa pneumonia (patients 1, 6, 10, 11). In patient 11, ceftolozane-tazobactam was discontinued after 3 days due to a rash.
The clinical failure rate of ceftolozane-tazobactam treatment was 29% (6/21). Clinical failures included the 4 patients with attributable deaths at 90 days, and 2 patients with MDR-P. aeruginosa pneumonia who survived to 90 days but developed recurrent pneumonia or suppurative tracheobronchitis (patients 4 and 8). Four patients who were successfully treated for pneumonia (patients 14 and 15), cIAI (patient 7), and cUTI (patient 18) were colonized by MDR-P. aeruginosa within 90 days of the index infection.
Ceftolozane-tazobactam resistance was identified in 3 patients (14%), emerging during recurrent pneumonia that led to death at 90 days (patient 1), airway colonization following intraabdominal infection (patient 7), and suppurative tracheobronchitis following pneumonia (patient 8). Resistance emerged 2 weeks after completion of a 30-day treatment course and on days 8 and 19 of treatment, respectively.
The only variable that was significantly associated with clinical failure was simplified acute physiology score-II (SAPS-II) score (median, 35 for failure and 23 for success; P = .04); there was a trend toward an association between clinical failure and age (median, 72.5 vs 58 years; P = .07). Site of infection, renal failure, combination vs monotherapy, use of inhaled therapy, time to initiation of ceftolozane-tazobactam, presence of coinfections, and FDA-approved or PK-derived dosing were not significantly associated with clinical failure or 90-day mortality. None of these factors were associated with emergence of ceftolozane-tazobactam resistance. Thrombocytopenia occurred in 2 patients (10%) (patients 2 and 10); linezolid, either alone (patient 10) or with concomitant valganciclovir (patient 2), likely contributed to thrombocytopenia. No patient developed AKI attributable to ceftolozane-tazobactam. Ceftolozane-tazobactam was discontinued prematurely in only 1 patient (rash, patient 11).
Molecular Characterization of Longitudinal Isolates
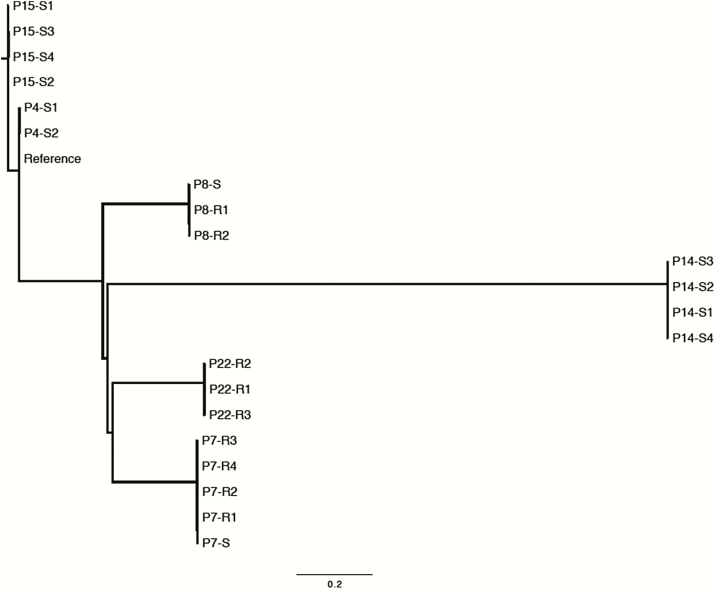
Longitudinal isolates from patients in whom ceftolozane-tazobactam resistance emerged (patients 7 and 8) and did not emerge (patients 4, 14, and 15) underwent WGS (Table 3). Isolates clustered by patient, ruling out a common source of infection or transmission among patients (Figure 1) [41]. Isolates from different patients belonged to distinct multilocus sequence types (Supplementary Table 3). In each patient, infections clearly traced to a single founding strain (Figure 1). The inferred genetic diversity among isolates within patients differed substantially, ranging from 2 to 98 unique SNPs (patients 4 and 8, respectively). More than 100 SNPs were observed in some isolates from patient 7, likely due to homologous recombination of an integrative conjugative element (details below; Supplementary Table 2). Despite the variable genetic diversity, it was possible to identify mutations likely to be associated with gain of ceftolozane-tazobactam resistance that were not found in susceptible isolates.
Table 3.
Ceftolozane-Tazobactam Minimum Inhibitory Concentration and Resistance-Associated Mutations in Longitudinal Isolates
| Patient | Isolate No. | Days from Start of C/Ta | C/T Minimum Inhibitory Concentration (µg/ mL) | Mutation | Annotation | Gene | Description |
|---|---|---|---|---|---|---|---|
| 7 | P7-S | - | 4 | - | - | - | - |
| P7-R1 | 8 | 128 | Δ21 bp | Coding (711–731/1194 nt) | ampC | β-lactamase | |
| P7-R2 | 3 | >256 | Δ21 bp | Coding (711–731/1194 nt) | ampC | β-lactamase | |
| P7-R3 | 20 | 128 | Δ21 bp | Coding (711–731/1194 nt) | ampC | β-lactamase | |
| P7-R4 | 41 | 256 | Δ21 bp | Coding (711–731/1194 nt) | ampC | β-lactamase | |
| Δ57 bp | Coding (693–749/1194 nt) | ampC | β-lactamase | ||||
| 8 | P8-S | - | 0.5 | - | - | - | - |
| P8-R1 | 17 | 32 | G→A | T96I (ACC→ATC) | ampC | β-lactamase | |
| T→C | Intergenic (-43/-106) | ampC ←I → ampR | β-lactamase /HTH-type transcriptional activator AmpR | ||||
| +CATG | Coding (1071/1293 nt) | oprD | Porin D precursor | ||||
| Δ2 bp | Coding (391–392/1293 nt) | oprD | Porin D precursor | ||||
| C→T | G339E (GGG→GAG) | mexB | Multidrug-resistance protein MexB | ||||
| P8-R2 | 61 | 64 | G→A | T96I (ACC→ATC) | ampC | β-lactamase | |
| T→C | Intergenic (-43/-106) | ampC ←I → ampR | Beta-lactamase/HTH-type transcriptional activator AmpR | ||||
| +CATG | Coding (1071/1293 nt) | oprD | Porin D precursor | ||||
| Δ2 bp | Coding (391–392/1293 nt) | oprD | Porin D precursor | ||||
| C→T | G339E (GGG→GAG) | mexB | Multidrug-resistance protein MexB | ||||
| 4 | P4-S1 | - | 2 | Δ1 bp | Coding (1200/1419 nt) | oprD | Porin D precursor |
| Δ2 bp | Coding (470–471/1419 nt) | oprD | Porin D precursor | ||||
| P4-S2 | 6 | 2 | - | - | - | - | |
| 14 | P14-S1 | - | 0.38 | - | - | - | - |
| P14-S2 | 1 | 0.5 | C→T | Q45a (CAG→TAG) | lasR_1 | Transcriptional activator protein LasR | |
| P14-S3 | 34 | 0.38 | - | - | - | - | |
| P14-S4 | 34 | 0.5 | - | - | - | - | |
| 15 | P15-S1 | - | 1.5 | - | - | - | - |
| P15-S2 | 2 | 3 | - | - | - | - | |
| P15-S3 | 43 | 1.5 | - | - | - | - | |
| P15-S4 | 122 | 3 | - | - | - | - | |
| 22 | P22-R1 | NA | 16 | G→T | L279I (CTC→ATC) | ampC | β-lactamase |
| G→A | L819F (CTC→TTC) | acrB | Multidrug efflux pump subunit AcrB | ||||
| P22-R2 | NA | 16 | G→T | L279I (CTC→ATC) | ampC | β-lactamase | |
| G→A | H215Y (CAC→TAC) | ampC | β-lactamase | ||||
| G→A | L819F (CTC→TTC) | acrB | Multidrug efflux pump subunit AcrB | ||||
| C→T | A689T (GCC→ACC) | acrB | Multidrug efflux pump subunit AcrB | ||||
| P22-R3 | NA | 8 | G→T | L279I (CTC→ATC) | ampC | β-lactamase | |
| G→A | H215Y (CAC→TAC) | ampC | β-lactamase |
Isolates from patients 7 and 8 developed resistance to ceftolozane-tazobactam during therapy. Longitudinal isolates from patients 4, 14, and 15 remained ceftolozane-tazobactam susceptible. Isolates from patient 22 were resistant or intermediately susceptible to ceftolozane-tazobactam in the absence of exposure to the drug. Dashes indicate no mutation present.
Abbreviations: C/T, ceftolozane-tazobactam; NA, Not Applicable.
aDays from start of C/T to recovery of isolate.
Figure 1.
Whole genome phylogeny of isolates from patients treated with ceftolozane-tazobactam and isolates with preexisting ceftolozane-tazobactam resistance from a control patient who was not treated with the drug (patient 22 [P22]) [41]. The phylogeny was inferred from all informative single nucleotide polymorphisms in the core genome with FastTree, using the most closely related available reference genome, strain PA_BWHPSA022. All isolates definitively cluster by patient (P). Sensitivity or resistance to ceftolozane-tazobactam is denoted as S or R and timing of isolation is denoted by numbering (eg, for patient 7: P7-S, P7-R1, P7-R2, P7-R3, P7-R4). Distance bar = 0.2 nucleotide differences per phylogenetically informative site.
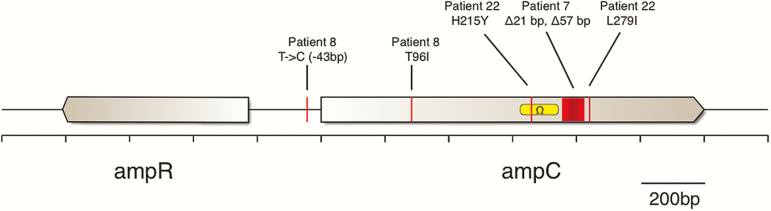
All isolates carried AmpC and OXA-50 β-lactamases but not other β-lactamases, ESBLs, or carbapenemases. However, only ceftolozane-tazobactam–resistant isolates harbored ampC mutations (Table 3 and Figure 2). Resistant isolates from patient 7 had 21- or 57-basepair deletions within ampC (Figure 2). Resistant isolates from patient 8 had point mutations in ampC (resulting in a threonine-to-isoleucine substitution at AmpC amino acid position 96 [T96I]) and the ampR-ampC intergenic region (Figure 2).
Figure 2.
Diagram of mutations occurring in the ampR-ampC genomic region among resistant isolates. Mutation location, type, and proximity to the Ω-loop, known to confer resistance when mutated, is denoted. Orange box: nucleotides deleted in isolates from patient 7. Yellow box: domain encoding the Ω-loop. Abbreviation: bp, basepair.
Resistant isolates from patient 7 also showed evidence of homologous recombination at the site of the integrative conjugative element mentioned above, which introduced >100 SNPs spanning 27 genes (including acrB, encoding a multidrug efflux pump subunit; Supplementary Table 3) [42]. Resistant isolates from patient 8 acquired a small indel mutation in oprD, the porin D precursor [43, 44], and a nonsynonymous mutation G339E in multidrug efflux transporter mexB [45]. Two mutations in oprD were identified in a susceptible isolate (P14-S1) from patient 14. In addition, a mutation introducing a premature stop codon (Q45*) in quorum sensing regulator lasR was found in P14-S2 [46]. None of the isolates that remained susceptible had ampC mutations (Table 3).
Three longitudinal isolates with preexisting ceftolozane-tazobactam resistance, recovered from a patient (patient 22) who was not treated with the drug, were sequenced as controls and compared to the consensus reference isolate (PA_BWHPSA022). ampC (H215Y, L279I) and acrB (A689T, L819F) mutations were identified in each resistant isolate.
Expression of various genes previously implicated in β-lactam resistance was measured by qRT-PCR for isolates from patients 7 and 8. Compared to initial susceptible isolates, ceftolozane-tazobactam–resistant isolates P7-R2 and P8-R2 overexpressed ampC (30-fold and 4.4-fold, respectively; Table 4). In contrast, there was no change in ampC expression by P7-R1 compared to P7-S. Compared to P8-S, isolate P8-R2 also overexpressed efflux pump transporter genes mexY and, to a lesser extent, mexB and mexD (3.2-fold, 1.9-fold, and 1.7-fold, respectively). There were no differences in oprD expression between initial and susceptible isolates from either patient 7 or patient 8.
Table 4.
Expression of β-Lactam Resistance Genes
| Fold Change in Expression Compared to Initial Sensitive Isolatea | ||||
|---|---|---|---|---|
| Protein | Gene Name | Patient 7, MIC (μg/mL) | Patient 8, MIC (μg/mL) | |
| P7-R1, 128 | P7-R2, >256 | P8-R2, 64 | ||
| AmpC β-lactamase | ampC | ↔ 0.8 | ↑ 30 | ↑ 4.4 |
| Multidrug efflux pump | mexB | ↔ 1.2 | ↔ 0.98 | ↑ 1.9 |
| Multidrug efflux pump | mexD | ↔ 1.01 | ↔ 0.90 | ↑ 1.7 |
| Multidrug efflux pump | mexY | ↔ 1.3 | ↔ 1.2 | ↑ 3.2 |
| Porin D | oprD | ↔ 1.1 | ↔ 0.7 | ↔ 0.96 |
Significant difference in expression between sensitive and resistant isolates was defined as >1.5-fold and P < .05 by analysis of variance.
Baseline ampC expression by control strains Pseudomonas aeruginosa PAO1 and ATCC 27853 was at the limit of detection (data not shown), which limited the ability to precisely define the extent to which expression by clinical isolates from patients 7 and 8 (P7-S1, P8-S1) was increased.
Abbreviation: MIC, minimum inhibitory concentration.
aLateral arrow: no change in gene expression; upward arrow: increased gene expression; downward arrow: decreased gene expression.
DISCUSSION
This study reports real-world experience with ceftolozane-tazobactam treatment of MDR-P. aeruginosa infections, which was directed largely against respiratory tract infections rather than FDA-approved indications of cIAI and cUTI. By some measures, treatment appeared to be effective. Thirty-day all-cause mortality, the primary outcome in this study, was only 10%. Moreover, 30- and 90-day attributable mortality rates of 5% and 19%, respectively, were lower than rates of 42%–56% previously reported for MDR-P. aeruginosa infections, including pneumonia in the intensive care unit, hospital-acquired pneumonia, and VAP [47–49]. Ceftolozane-tazobactam was also well tolerated, as premature drug discontinuation was necessary in a single patient. By other measures, however, results were more equivocal. The low 30-day mortality rate was consistent with that predicted by median SAPSII and SOFA (sequential organ failure assessment) scores [50, 51]. Clinical failure of treatment at 90 days, defined as a composite of attributable mortality, or persistent or recurrent infection, was 29%. Most worrisome was our finding that resistance emerged in 3 patients (14%), as quickly as 8 days into treatment. More reports on treatment responses and resistance during various types of MDR-P. aeruginosa infections are needed to put our experience in context and define ceftolozane-tazobactam’s place in the armamentarium.
Several factors may have mitigated ceftolozane-tazobactam responses among our patients. First, our cohort was comprised of patients with a variety of complex medical conditions, including 9 transplant recipients (43%), 8 patients (38%) with ventilator-dependent respiratory failure, 7 patients (33%) who had undergone recent surgery, and 5 patients (24%) who were receiving renal replacement therapy (24%). Second, 16 patients (76%) were treated for pneumonia, a disease characterized by high microbial burdens and unpredictable antibiotic PK at sites of infection. Third, 6 patients (29%) were coinfected with other pathogens, which may have contributed to outcomes. Finally, clinicians often avoided adding antibiotics such as aminoglycosides and colistin, which were also active against infecting isolates but limited by toxicity concerns. Indeed, it is well recognized that antibiotic activity is not the sole determinant of outcomes in patients with MDR-P. aeruginosa infections. The importance of host factors in this study was underscored by the finding that elevated SAPS-II score was the only significant risk factor for clinical failure.
Ceftolozane-tazobactam dosing was not significantly associated with patient outcomes or emergence of resistance, but our study size may have limited the ability to establish relationships. Only 5 (28%) patients with respiratory tract infections received a PK-derived dose, which is double the FDA-approved dose. In healthy volunteers, the FDA-approved dose achieves ELF concentrations that exceed 8 mg/L for >60% of the dosing interval [52]. Recently, however, investigators using Monte Carlo simulations predicted that the probability of ceftolozane-tazobactam pharmacokinetic-pharmacodynamic (PK-PD) target attainment within ELF was 98% with the PK-derived dose compared to 88% with the FDA-approved dose [28]. In a brief report, the higher dosage was effective in treating 3 patients with MDR-P. aeruginosa pneumonia [53]. We currently advocate the PK-derived dosage for treatment of respiratory infections. There are no approved dosing recommendations for patients who receive continuous renal replacement therapy for any type of infection or for patients with pneumonia undergoing iHD.
To our knowledge, this is the first study to use WGS to characterize evolution of antibiotic resistance in longitudinal MDR-P. aeruginosa isolates recovered during the course of infection. Phylogenetic analysis of WGS data demonstrated that ceftolozane-tazobactam resistance evolved independently in index isolates from 2 patients. By detecting mutations and measuring transcription of genes linked to β-lactam resistance, we identified several potential resistance mechanisms. First among these was constitutive overexpression of ampC by resistant isolates (P7-R2, P8-R2) compared to respective index isolates. De-repressed P. aeruginosa mutants account for large proportions of isolates that are broadly resistant to β-lactams [13, 54–56] and archived isolates with preexisting ceftolozane-tazobactam resistance [13]. It is plausible that ampC is induced and/or de-repressed by more efficient binding of the ampR regulatory factor, as has been reported with ampR-ampC intergenic point mutations (as observed in resistant isolates from patient 8) [55, 57].
Other possible ceftolozane-tazobactam resistance determinants included ampC deletions, as seen in patient 7, and amino acid substitutions that impact the β-lactamase Ω-loop (Figure 2). The Ω-loop comprises the substrate binding site and represents a hot spot for mutations that enhance catalytic efficiency and extend the spectrum of β-lactamase activity [15]. Various Ω-loop mutations widen the substrate binding site, thereby facilitating adherence and hydrolysis of β-lactams with bulky side chains such as ceftolozane and other oxyimino-cephalosporins [15, 58]. The H215Y substitution in resistant isolates from control patient 22 fell within the Ω-loop. The T96I substitution in resistant isolates from patient 8 occurred within the H2 helix, which lies close to the serine active site and interacts with the Ω-loop through hydrogen binding [59]. H2 helix mutations can render the active-site serine more pliable, opening the Ω-loop entrance to accommodate larger cephalosporins such as ceftolozane [58]. Longitudinal isolates with retained ceftolozane-tazobactam susceptibility did not carry ampC mutations, whereas ampC mutations were evident in isolates with preexisting ceftolozane-tazobactam resistance. These observations lend credence to the conclusion that such mutations contributed to resistance.
Constitutive overexpression of mexY, mexB, and mexD (which encode cytoplasmic membrane transporters for efflux pumps), presence of a mexB G339E mutation, acquisition of multidrug efflux gene acrB, and acrB mutations were observed in various ceftolozane-tazobactam–resistant isolates. It is unclear if upregulation and/or acquisition of efflux systems may overcome the fact that ceftolozane is a weak efflux substrate [13, 60].
Our study is limited by its single-center, retrospective design and its small sample size. Clinical findings and resistant isolates may not be representative of those from other institutions or countries. Our interpretation of the effectiveness of ceftolozane-tazobactam is limited by the lack of a control group treated with other anti-pseudomonal agents. We also acknowledge that the genetic diversity we described in longitudinal isolates may reflect both bona fide variation in evolutionary dynamics and error in sequencing and bioinformatics analyses.
In conclusion, ceftolozane-tazobactam is an important advance in the treatment of MDR-P. aeruginosa infections, but more clinical data are needed to understand its place in the armamentarium. The emergence of resistance after short courses of therapy in some patients highlights the importance of establishing strict criteria for the drug’s use and the continued need for new antibiotics. Studies are needed to understand the role of combination therapy, define ceftolozane-tazobactam PK during different types of infection, validate dosing regimens derived from PK-PD data, and verify resistance mechanisms.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Financial support. This project was performed by the XDR Pathogen Lab, which is supported by a grant from the University of Pittsburgh Medical Center. V. S. C. is supported by the National Institutes of Health (NIH; AI124302-01 and GM110444). Y. D. is supported by research grants from the NIH (R01AI104895, R21AI123747). R. K. S. is supported by the NIH (K08AI114883). C. J. C. is supported by the US Department of Veterans Affairs (1IO1BX001955) and the NIH (AI111037, AI121555, AI126157) and by an investigator-initiated research award from Merck Sharp & Dohme. M. H. N. is supported by the NIH (AI107290, AI128338).
Potential conflicts of interest. Y. D. has served on advisory boards for Meiji, Achaogen, Allergan, Curetis; has received speaking fees from Merck; and has received research funding from the Medicines Company. C. J. C. has served as a consultant for Merck Sharp & Dohme. The remaining authors: No reported conflicts. The authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Tam VH, Rogers CA, Chang KT, Weston JS, Caeiro JP, Garey KW. Impact of multidrug-resistant Pseudomonas aeruginosa bacteremia on patient outcomes. Antimicrob Agents Chemother 2010; 54:3717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tumbarello M, Repetto E, Trecarichi EM, et al. Multidrug-resistant Pseudomonas aeruginosa bloodstream infections: risk factors and mortality. Epidemiol Infect 2011; 139:1740–9. [DOI] [PubMed] [Google Scholar]
- 3. Zavascki AP, Barth AL, Fernandes JF, Moro AL, Gonçalves AL, Goldani LZ. Reappraisal of Pseudomonas aeruginosa hospital-acquired pneumonia mortality in the era of metallo-beta-lactamase-mediated multidrug resistance: a prospective observational study. Crit Care 2006; 10:R114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hirsch EB, Tam VH. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res 2010; 10:441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. Multidrug-resistant Pseudomonas aeruginosa: risk factors and clinical impact. Antimicrob Agents Chemother 2006; 50:43–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gasink LB, Fishman NO, Weiner MG, Nachamkin I, Bilker WB, Lautenbach E. Fluoroquinolone-resistant Pseudomonas aeruginosa: assessment of risk factors and clinical impact. Am J Med 2006; 119: 526 e19–25. [DOI] [PubMed] [Google Scholar]
- 7. Laupland KB, Parkins MD, Church DL, et al. Population-based epidemiological study of infections caused by carbapenem-resistant Pseudomonas aeruginosa in the Calgary Health Region: importance of metallo-beta-lactamase (MBL)-producing strains. J Infect Dis 2005; 192:1606–12. [DOI] [PubMed] [Google Scholar]
- 8. Carmeli Y, Troillet N, Eliopoulos GM, Samore MH. Emergence of antibiotic-resistant Pseudomonas aeruginosa: comparison of risks associated with different antipseudomonal agents. Antimicrob Agents Chemother 1999; 43:1379–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Quinn JP, Dudek EJ, DiVincenzo CA, Lucks DA, Lerner SA. Emergence of resistance to imipenem during therapy for Pseudomonas aeruginosa infections. J Infect Dis 1986; 154:289–94. [DOI] [PubMed] [Google Scholar]
- 10. Livermore DM, Williams RJ, Lindridge MA, Slack RC, Williams JD. Pseudomonas aeruginosa isolates with modified beta-lactamase inducibility: effects on beta-lactam sensitivity. Lancet 1982; 1:1466–7. [DOI] [PubMed] [Google Scholar]
- 11. Giwercman B, Lambert PA, Rosdahl VT, Shand GH, Høiby N. Rapid emergence of resistance in Pseudomonas aeruginosa in cystic fibrosis patients due to in-vivo selection of stable partially derepressed beta-lactamase producing strains. J Antimicrob Chemother 1990; 26:247–59. [DOI] [PubMed] [Google Scholar]
- 12. Cabot G, Ocampo-Sosa AA, Tubau F, et al. ; Spanish Network for Research in Infectious Diseases Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa isolates from bloodstream infections: prevalence and impact on resistance in a Spanish multicenter study. Antimicrob Agents Chemother 2011; 55:1906–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Castanheira M, Mills JC, Farrell DJ, Jones RN. Mutation-driven β-lactam resistance mechanisms among contemporary ceftazidime-nonsusceptible Pseudomonas aeruginosa isolates from U.S. hospitals. Antimicrob Agents Chemother 2014; 58:6844–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hocquet D, Nordmann P, El Garch F, Cabanne L, Plésiat P. Involvement of the MexXY-OprM efflux system in emergence of cefepime resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2006; 50:1347–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev 2009; 22:161–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jalal S, Wretlind B. Mechanisms of quinolone resistance in clinical strains of Pseudomonas aeruginosa. Microb Drug Resist 1998; 4:257–61. [DOI] [PubMed] [Google Scholar]
- 17. Poole K. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2005; 49:479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents 2015; 45:568–85. [DOI] [PubMed] [Google Scholar]
- 19. Weldhagen GF, Poirel L, Nordmann P. Ambler class A extended-spectrum beta-lactamases in Pseudomonas aeruginosa: novel developments and clinical impact. Antimicrob Agents Chemother 2003; 47:2385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malkocoglu G, Aktas E, Bayraktar B, Otlu B, Bulut ME. VIM-1, VIM-2, and GES-5 carbapenemases among Pseudomonas aeruginosa isolates at a tertiary hospital in Istanbul, Turkey. Microb Drug Resist 2016; 23:1–5. [DOI] [PubMed] [Google Scholar]
- 21. Solomkin J, Hershberger E, Miller B, et al. Ceftolozane/tazobactam plus metronidazole for complicated intra-abdominal infections in an era of multidrug resistance: results from a randomized, double-blind, phase 3 trial (ASPECT-cIAI). Clin Infect Dis 2015; 60:1462–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wagenlehner FM, Umeh O, Darouiche RO. Ceftolozane-tazobactam versus levofloxacin in urinary tract infection—authors’ reply. Lancet 2015; 386:1242. [DOI] [PubMed] [Google Scholar]
- 23. Cluck D, Lewis P, Stayer B, Spivey J, Moorman J. Ceftolozane-tazobactam: a new-generation cephalosporin. Am J Health Syst Pharm 2015; 72:2135–46. [DOI] [PubMed] [Google Scholar]
- 24. van Duin D, Bonomo RA. Ceftazidime/avibactam and ceftolozane/tazobactam: second-generation β-lactam/β-lactamase inhibitor combinations. Clin Infect Dis 2016; 63:234–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Buehrle DJ, Shields RK, Chen L, et al. Evaluation of the in vitro activity of ceftazidime-avibactam and ceftolozane-tazobactam against meropenem-resistant Pseudomonas aeruginosa isolates. Antimicrob Agents Chemother 2016; 60:3227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18:268–81. [DOI] [PubMed] [Google Scholar]
- 27. Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control 2008; 36:309–32. [DOI] [PubMed] [Google Scholar]
- 28. Zerbaxa Package Insert Cubist Pharmaceuticals, Inc; Last revised: 18 May 2015. https://www.merck.com/product/usa/pi_circulars/z/zerbaxa/zerbaxa_pi.pdf. [Google Scholar]
- 29. Xiao AJ, Miller BW, Huntington JA, Nicolau DP. Ceftolozane/tazobactam pharmacokinetic/pharmacodynamic-derived dose justification for phase 3 studies in patients with nosocomial pneumonia. J Clin Pharmacol 2016; 56:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Safety and Efficacy Study of Ceftolozane/Tazobactam to Treat Ventilated Nosocomial Pneumonia (MK-7625A-008) (ASPECT-NP). Available at: https://clinicaltrials.gov/ct2/show/NCT02070757. Accessed July 2016. [Google Scholar]
- 31. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement (M100-S25). CLSI: Wayne, PA, 2015. [Google Scholar]
- 32. Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, Kishony R. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 2015; 10:e0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ondov BD, Treangen TJ, Melsted P, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 2016; 17:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 2014; 1151:165–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cabot G, Bruchmann S, Mulet X, et al. Pseudomonas aeruginosa ceftolozane-tazobactam resistance development requires multiple mutations leading to overexpression and structural modification of AmpC. Antimicrob Agents Chemother 2014; 58:3091–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rahmati S, Yang S, Davidson AL, Zechiedrich EL. Control of the AcrAB multidrug efflux pump by quorum-sensing regulator SdiA. Mol Microbiol 2002; 43:677–85. [DOI] [PubMed] [Google Scholar]
- 37. Jacobs C, Frère JM, Normark S. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible beta-lactam resistance in gram-negative bacteria. Cell 1997; 88:823–32. [DOI] [PubMed] [Google Scholar]
- 38. Oh H, Stenhoff J, Jalal S, Wretlind B. Role of efflux pumps and mutations in genes for topoisomerases II and IV in fluoroquinolone-resistant Pseudomonas aeruginosa strains. Microb Drug Resist 2003; 9:323–8. [DOI] [PubMed] [Google Scholar]
- 39. Juan C, Moyá B, Pérez JL, Oliver A. Stepwise upregulation of the Pseudomonas aeruginosa chromosomal cephalosporinase conferring high-level beta-lactam resistance involves three AmpD homologues. Antimicrob Agents Chemother 2006; 50:1780–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ocampo-Sosa AA, Cabot G, Rodríguez C, et al. ; Spanish Network for Research in Infectious Diseases Alterations of OprD in carbapenem-intermediate and -susceptible strains of Pseudomonas aeruginosa isolated from patients with bacteremia in a Spanish multicenter study. Antimicrob Agents Chemother 2012; 56:1703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 2010; 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vettoretti L, Plésiat P, Muller C, et al. Efflux unbalance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother 2009; 53:1987–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li H, Luo YF, Williams BJ, Blackwell TS, Xie CM. Structure and function of OprD protein in Pseudomonas aeruginosa: from antibiotic resistance to novel therapies. Int J Med Microbiol 2012; 302:63–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pirnay JP, De Vos D, Mossialos D, Vanderkelen A, Cornelis P, Zizi M. Analysis of the Pseudomonas aeruginosa oprD gene from clinical and environmental isolates. Environ Microbiol 2002; 4:872–82. [DOI] [PubMed] [Google Scholar]
- 45. Li XZ, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother 1995; 39:1948–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. D’Argenio DA, Wu M, Hoffman LR, et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol 2007; 64:512–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cao B, Wang H, Sun H, Zhu Y, Chen M. Risk factors and clinical outcomes of nosocomial multi-drug resistant Pseudomonas aeruginosa infections. J Hosp Infect 2004; 57:112–8. [DOI] [PubMed] [Google Scholar]
- 48. Fernández-Barat L, Ferrer M, De Rosa F, et al. Intensive care unit-acquired pneumonia due to Pseudomonas aeruginosa with and without multidrug resistance. J Infect 2017; 74:142–52. [DOI] [PubMed] [Google Scholar]
- 49. Micek ST, Kollef MH, Torres A, et al. Pseudomonas aeruginosa nosocomial pneumonia: impact of pneumonia classification. Infect Control Hosp Epidemiol 2015; 36:1190–7. [DOI] [PubMed] [Google Scholar]
- 50. Le Gall JR, Lemeshow S, Saulnier F. A new simplified acute physiology score (SAPS II) based on a European/North American multicenter study. JAMA 1993; 270:2957–63. [DOI] [PubMed] [Google Scholar]
- 51. Vincent JL, de Mendonça A, Cantraine F, et al. Use of the SOFA score to assess the incidence of organ dysfunction/failure in intensive care units: results of a multicenter, prospective study. Working group on “sepsis-related problems” of the European Society of Intensive Care Medicine. Crit Care Med 1998; 26:1793–800. [DOI] [PubMed] [Google Scholar]
- 52. Chandorkar G, Huntington JA, Gotfried MH, Rodvold KA, Umeh O. Intrapulmonary penetration of ceftolozane/tazobactam and piperacillin/tazobactam in healthy adult subjects. J Antimicrob Chemother 2012; 67:2463–9. [DOI] [PubMed] [Google Scholar]
- 53. Gelfand MS, Cleveland KO. Ceftolozane/tazobactam therapy of respiratory infections due to multidrug-resistant Pseudomonas aeruginosa. Clin Infect Dis 2015; 61:853–5. [DOI] [PubMed] [Google Scholar]
- 54. Berrazeg M, Jeannot K, Ntsogo Enguéné VY, et al. Mutations in β-lactamase AmpC increase resistance of Pseudomonas aeruginosa isolates to antipseudomonal cephalosporins. Antimicrob Agents Chemother 2015; 59:6248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 2009; 22:582–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tam VH, Schilling AN, LaRocco MT, et al. Prevalence of AmpC over-expression in bloodstream isolates of Pseudomonas aeruginosa. Clin Microbiol Infect 2007; 13:413–8. [DOI] [PubMed] [Google Scholar]
- 57. Lodge J, Williams R, Bell A, Chan B, Busby S. Comparison of promoter activities in Escherichia coli and Pseudomonas aeruginosa: use of a new broad-host-range promoter-probe plasmid. FEMS Microbiol Lett 1990; 55(1-2):221–5. [DOI] [PubMed] [Google Scholar]
- 58. Raimondi A, Sisto F, Nikaido H. Mutation in Serratia marcescens AmpC beta-lactamase producing high-level resistance to ceftazidime and cefpirome. Antimicrob Agents Chemother 2001; 45:2331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rodríguez-Martínez JM, Poirel L, Nordmann P. Extended-spectrum cephalosporinases in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2009; 53:1766–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Juan C, Zamorano L, Pérez JL, Ge Y, Oliver A; Spanish Group for the Study of Pseudomonas; Spanish Network for Research in Infectious Diseases Activity of a new antipseudomonal cephalosporin, CXA-101 (FR264205), against carbapenem-resistant and multidrug-resistant Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother 2010; 54:846–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.