Abstract
The Antibacterial Resistance Leadership Group (ARLG) Laboratory Center (LC) leads the evaluation, development, and implementation of laboratory‐based research by providing scientific leadership and supporting standard/specialized laboratory services. The LC has developed a physical biorepository and a virtual biorepository. The physical biorepository contains bacterial isolates from ARLG-funded studies located in a centralized laboratory and they are available to ARLG investigators. The Web-based virtual biorepository strain catalogue includes well-characterized gram-positive and gram-negative bacterial strains published by ARLG investigators. The LC, in collaboration with the ARLG Leadership and Operations Center, developed procedures for review and approval of strain requests, guidance during the selection process, and for shipping strains from the distributing laboratories to the requesting investigators. ARLG strains and scientific and/or technical guidance have been provided to basic research laboratories and diagnostic companies for research and development, facilitating collaboration between diagnostic companies and the ARLG Master Protocol for Evaluating Multiple Infection Diagnostics (MASTERMIND) initiative for evaluation of multiple diagnostic devices from a single patient sampling event. In addition, the LC has completed several laboratory-based studies designed to help evaluate new rapid molecular diagnostics by developing, testing, and applying a MASTERMIND approach using purified bacterial strains. In collaboration with the ARLG’s Statistical and Data Management Center (SDMC), the LC has developed novel analytical strategies that integrate microbiologic and genetic data for improved and accurate identification of antimicrobial resistance. These novel approaches will aid in the design of future ARLG studies and help correlate pathogenic markers with clinical outcomes. The LC’s accomplishments are the result of a successful collaboration with the ARLG’s Leadership and Operations Center, Diagnostics and Devices Committee, and SDMC. This interactive approach has been pivotal for the success of LC projects.
Keywords: antibacterial resistance, diagnostics and devices, biorepository, drug testing, bacterial strains.
The mission of the Antibacterial Resistance Leadership Group (ARLG) is to prioritize, design, and execute clinical research that will reduce the public health threat of antibacterial resistance. The ARLG’s Laboratory Center (LC) has the primary responsibility of developing laboratory‐based research and providing specialized laboratory services and/or technical advice to the ARLG [1]. It comprises scientific leadership and technical components. The LC director and co-director bring extensive experience and scientific leadership in both gram-positive and gram-negative bacterial research in antibiotic resistance and clinical trials. The technical components consist of ARLG investigators and staff with broad expertise in pathogen-specific laboratory approaches, clinical trials, diagnostic platforms, antimicrobial stewardship, and clinical practice that can be tapped on a project-specific basis. Laboratory services include (1) two biorepositories; (2) an early-stage investigator seed grant program, which supports trainee research to generate preliminary data for external grant submissions by facilitating access to laboratory resources; and (3) a molecular and rapid diagnostics component to identify, evaluate, and test novel and rapid molecular diagnostic (RMD) platforms.
STRAIN BIOREPOSITORIES
The ARLG biorepositories were created to aid in the development and evaluation of novel diagnostic tests and laboratory techniques, study mechanisms of resistance, generate preliminary data for study concepts, and support/mentor early-stage investigators pursuing research in the field of antibacterial resistance. While some of the strains are stored in the laboratories of ARLG investigators (ie, the virtual biorepository [VB]), others that are isolated from patients enrolled in ARLG clinical studies are stored in a centralized location (ie, the physical biorepository [PB]).
The PB houses strains that are typically associated with clinical data and are from a variety of ARLG-funded projects. The strains are available to ARLG investigators for subsequent study after approval by the ARLG Executive Committee. Strains in the PB are also considered for inclusion in the VB following subsequent analysis and publication through the ARLG and its investigators.
The VB was created to enrich the laboratory support services with biological resources from existing collections of clinically well-characterized gram-positive and gram-negative bacterial strains published by contributing ARLG investigators. Distinct from typical biobanking approaches, the ARLG sought to make available annotated strains, located at multiple physical locations, via a central virtual catalogue. The LC is the point of contact and has developed procedures for review and approval of requests, guidance during the isolate-selection process, and shipment of isolates from distributing laboratories to requesting investigators. Because of its innovative nature, this approach requires fewer resources than those needed to establish and maintain centralized biorepositories.
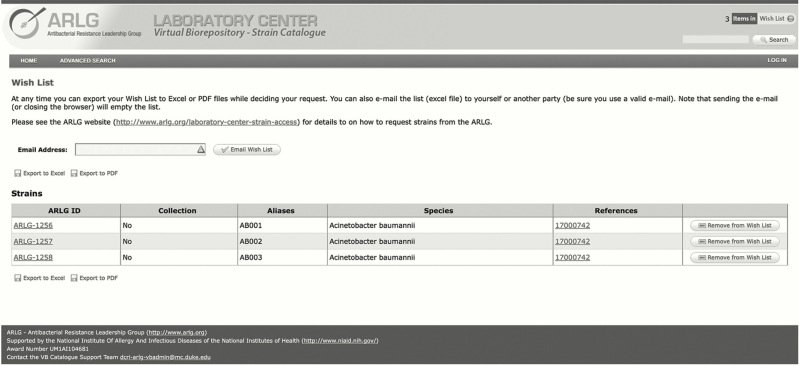
The LC has also launched a novel Web-based VB strain catalogue (https://arlgcatalogue.org) in which strains are searchable by name and aliases, species name, or other characteristics. Depending on either publication or common characteristics, certain strains can be listed as a collection or individual strains can be listed as part of a collection. Strains are labeled with a unique ARLG identifier (ID). Once the ARLG ID is selected in a “Wish List,” the strain-associated data can be exported as a PDF file or Microsoft Excel spreadsheet and emailed through a cart-like function (Figure 1). The VB strain catalogue currently holds genotypic (eg, β-lactamase [bla] genes, genetic background) and phenotypic (eg, minimum inhibitory concentration [MIC]) data for >700 strains from 4 predominant species: Acinetobacter species, Klebsiella pneumoniae, Escherichia coli, and Staphylococcus aureus, including both methicillin-susceptible and methicillin-resistant S. aureus. The majority of strains are multidrug resistant (MDR) (Figure 2). The VB is continually enriched with new data and lists of strains from ARLG-funded and published studies, with an eye on identifying and cataloging strains with significant value to the research community.
Figure 1.
Antibacterial Resistance Leadership Group (ARLG) strain catalogue cart view.
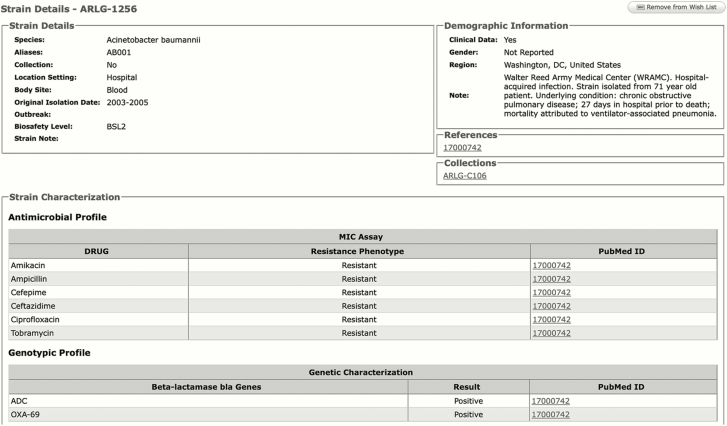
Figure 2.
Antibacterial Resistance Leadership Group (ARLG) catalogue strain detail page. Certain details culled from relevant publications and its supporting data are included on the page.
Strains are available to researchers and diagnostic and/or pharmaceutical companies upon completion and submission of a short questionnaire that captures a rationale for their use (https://duke.qualtrics.com/SE/?SID=SV_56GMAKjMkoo9KT3). The ARLG LC director and co-director, as well as LC team members, provide assistance during the strain identification and request process. In addition, the LC provides a consultative service to aid with project design and strain selection, providing a unique and distinguishing service that separates the ARLG VB from other biorepositories. Importantly, although there are no restrictions on the number of requests submitted by investigators, the number of strains provided is based upon planned use. Strains are provided free of charge, but requestors are responsible for shipping costs. The VB has proven to be a valuable resource for diagnostic companies during the developmental process of platforms by providing scientific and technical guidance in addition to providing access to multiple candidate strains. In addition, the initial contact with diagnostic companies regarding the VB has facilitated their access to the ARLG MASTERMIND (Master Protocol for Evaluating Multiple Infection Diagnostics) initiative [2], the goal of which is to evaluate in parallel multiple diagnostics from single-patient sampling.
PRIMERS
PRIMERS (Platforms for Rapid Identification of Multidrug-Resistant Gram Negative Bacteria and Evaluation of Resistance Studies) is a series of laboratory-based collaborative studies between the ARLG’s LC and Statistical Data Management Center (SDMC) [3] that is focused on developing a MASTERMIND approach to test multiple diagnostic platforms using well-characterized gram-negative pathogens, and determining whether there is a clinical predictive value to genotyping β-lactam resistance determinants. The studies were developed by the two co-directors of the LC (Drs Robert A. Bonomo and Barry N. Kreiswirth) and the director of the SDMC (Dr Scott R. Evans), along with later consultation with the ARLG Diagnostics and Devices Committee [4], with the overall aim of improving empiric therapy and reducing antibiotic usage.
In the initial PRIMERS I study [5], different molecular platforms were compared for their ability to identify the presence of selected β-lactamase bla genes and specific gene mutations among 76 selected MDR strains of E. coli and K. pneumoniae. Using a MASTERMIND approach, the DNA isolated from each strain was evaluated among the following 4 platforms: (1) polymerase chain reaction combined with electrospray ionization mass spectrometry (PCR/ESI-MS); (2) a multiplex diagnostic platform that uses allele-specific fluorescently labeled probes to identify genes, Molecular Beacons (MB); (3) DNA microarrays that detect bla genes (Check-Points); and (4) direct sequencing on an Ion Torrent. The genotyping data were then compared with β-lactam resistance profiles. PRIMERS I evaluated the genotypic-phenotypic comparisons for 14 β-lactam antibiotics, and their performance was estimated with 95% confidence intervals for each platform. The overall performance of each platform was comparable, and genotyping β-lactam resistance correlated with phenotypic results. However, resistance sensitivities did range from >95% for imipenem, ceftazidime, and cefepime to <80% for piperacillin/tazobactam.
As a result of cost, ease of labor, the ability to identify bacterial genus and species, and the ability to expand the number of resistance determinants, the PCR/ESI-MS and MB platforms were selected for additional studies. PRIMERS II was a blinded study using a MASTERMIND approach with 196 susceptible and MDR clinical isolates of E. coli and K. pneumoniae that were genotyped on the two platforms [5]. Evaluating the comparison of phenotypic susceptibility testing to 14 different β-lactam antibiotics vs the detection of 32 genetic targets led to the SDMC’s development of novel interpretative algorithms to estimate susceptibility and resistance sensitivities, referred to as discriminatory summary plots, and to evaluate the susceptibility and resistance predictive values in the context of a known prevalence of resistance.
ARLG support of the PRIMERS studies proved to be a catalyst in addressing sentinel questions regarding the clinical utility and implementation of RMDs in improving empiric therapy. The large strain collections available in the laboratories of Drs Bonomo and Kreiswirth, their access to two different genotyping platforms, and collaboration with the ARLG’s SDMC and Diagnostics and Devices Committee and the ARLG Leadership and Operations Center provided a complementary research team focused on creating a new analytical paradigm by which to evaluate RMDs as a tool in clinical decision making.
PRIMERS III extends this paradigm by comparing the two molecular platforms used in PRIMERS II (PCR/ESI-MS and MB) to determine whether genotyping carbapenem-resistant Acinetobacter species can have a clinical impact on treatment [6]. This study genotyped 200 clinically selected carbapenem-resistant and -susceptible Acinetobacter species isolates for the presence of different OXA carbapenemases (blaOXA-23, -24/40, and -58) as well as blaKPC, -NDM, -VIM, and -IMP. Their resistance and susceptibility predictive values were evaluated. The findings for both platforms indicated that clinicians can be confident >85% of the time when using a carbapenem regimen for a susceptible strain and >95% confident that the molecular detection of an OXA carbapenemase is predictive of carbapenem resistance and that the empiric strategy to change to colistin and tigecycline is clinically warranted.
PRIMERS IV, which addresses MDR Pseudomonas aeruginosa, is being analyzed.
RESEARCH PRIORITIES AND UNMET NEEDS
The PRIMERS studies are a template for the ARLG LC to integrate its scientific leadership and technical components with the SDMC and the ARLG Diagnostics and Devices Committee to address antibacterial resistance from the diagnostic perspective. In each PRIMERS study, evaluation of the predictive value of the rapid molecular typing of various bla genes was compared with the phenotypic determination of the MIC of numerous β-lactam antibiotics. General findings showed that the ability to predict susceptibility and resistance for both the PCR/ESI-MS and MB platforms ranged from approximately 80% to 95%, values that approach but are not equivalent to the Clinical and Laboratory Standards Institute standards used to determine the “appropriate” empiric antibiotic treatment regimen. However, compared with the combination of broad-spectrum antibiotics given as “empiric therapy” without laboratory guidance, the predictive values of both molecular platforms are a significant improvement that could greatly reduce antibiotic use while simultaneously achieving targeted therapy and antimicrobial stewardship.
The findings of the PRIMERS studies could lead to translational studies to implement both the bench and analytic tools that we have developed, or to evaluate whether a targeted molecular approach would improve empiric antibiotic usage in high-risk patient settings, where prophylaxis and treatment against MDR pathogens is standard. For example, perianal swabs from bone marrow and liver transplant unit patients and bronchoalveolar lavages from intubated patients in the intensive care unit are important clinical specimens for which rapid detection of MDR gram-negative pathogens may have significant impact on treatment decisions. In PRIMERS, we have already demonstrated the flexibility of two molecular platforms to target select pathogens and resistance genes and to develop different test panels to evaluate both respiratory and gastrointestinal specimens. In keeping with the focus of the ARLG, future proposed studies will be designed to evaluate how and whether integrating these methods in a clinical microbiology laboratory improves treatment decisions and antibiotic stewardship and to consider their implementation in a clinical trial setting.
A second major direction of the LC will be to build on our already strong working group. PRIMERS took advantage of the multidisciplinary talents within the ARLG and by integrating whole-genome sequencing (WGS), RMD platforms, and gene expression studies using clinically and epidemiologically defined MDR strain collections. We will take advantage of the maturity of ARLG-funded clinical studies CRACKLE (Consortium on Resistance Against Carbapenems in Klebsiella pneumoniae and Other Enterobacteriaceae) [7], CREST (Carbapenem-Resistant Enterobacteriaceae in Solid Organ Transplant Patients) [7], and Blood Culture Identification [4], where isolates have been archived in the ARLG PB and clinical information has been stored at the ARLG Leadership and Operations Center. As an example, WGS of all the carbapenem-resistant Enterobacteriaceae bloodstream isolates from CRACKLE will be analyzed from a phylogenetic perspective and mapped to the rich epidemiological and clinical information to identify putative genetic correlates associated with specific disease presentation, severity, and outcome. In these studies, we will be able to dissect the influences of antibiotic resistance and virulence and identify strains with unique gene expression for future animal and diagnostic studies. Furthermore, the LC will continue to expand the biorepository to also include specimens from patients enrolled in ARLG clinical studies. Enrichment of the biorepositories with biological specimens and microbial isolates will be valuable for designing future efforts.
The LC has also initiated a partnership with BEI Resources (https://www.beiresources.org/) for the development of key antibiotic-resistant strain panels that soon will be made available to researchers. Last, the ARLG is in the process of identifying potential international collaborators with the intent to address the global impact of antimicrobial resistance.
In conclusion, the LC has and will continue to assist in the development of antibacterial-resistance diagnostics, build strategies for antibiotic therapy, and identify host signature markers that could be used as predictors of therapy response and/or disease progression. Laboratory Center highlights and future directions are summarized in Table 1.
Table 1.
Laboratory Center
| Highlights |
| Biorepositories |
| • Established a physical biorepository with isolates from ARLG-funded studies; isolates are associated with clinical data and centralized in a designated laboratory |
| • Established a Web-based virtual biorepository: strain catalogue listing >700 gram-positive and gram-negative bacterial strains published by ARLG investigators |
| • Developed procedures for the review and approval of strain requests |
| Guidance |
| • Provided technical and scientific support to diagnostic companies for the development of diagnostic platforms |
| • Provided >200 virtual biorepository strains to research laboratories and diagnostic companies for basic research and development |
| Collaboration |
| • Established collaboration with BEI Resources for the development of key antibacterial-resistant strain panels |
| • Liaison between diagnostic companies and the ARLG MASTERMIND initiative for in parallel evaluation of multiple diagnostics from single patient sampling |
| Collaboration with ARLG SDMC |
| • Developed a MASTERMIND approach to test multiple diagnostic platforms using well characterized gram-negative pathogens |
| • Established novel analytical strategies integrating microbiologic and genetic data for accurate identification of antimicrobial resistance |
| Future directions |
| • Expand and broaden the number of species and stains with unique genotypic and phenotypic traits |
| • Identify and work with potential international collaborators to understand the global implication of antibacterial resistance |
| • Identify key strains and strain panels to be used as reference standards for research and development and drug testing |
| • Expand the biorepository to include human biological samples to facilitate the development of antibacterial-resistant diagnostics |
| • Apply the LC/SDMC analytical approach to future ARLG
clinical studies to correlate pathogenic markers with disease
severity and clinical outcome |
| • Continue to collaborate with the Diagnostics and Devices Committee on diagnostic studies |
Abbreviations: ARLG, Antibacterial Resistance Leadership Group; LC, Laboratory Center; MASTERMIND, Master Protocol for Evaluating Multiple Infection Diagnostics; SDMC, Statistical and Data Management Center.
Notes
ARLG Laboratory Center. Rebekka Arias, BS; Robert Bonomo, MD; Nancie Deckard, BSN, MS; Carol Hill, PhD; Andrea Hujer, BS; Kristine Hujer, BS; Jacqueline Huvane, PhD; Barry Kreiswirth, PhD; Claudia Manca, PhD; Robin Patel, MD; David Souto, BS; Ephraim Tsalik, MD, MHS, PhD.
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health (NIH).
Financial support. This article was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the NIH (award number UM1AI104681).
Supplement sponsorship. This article appears as part of the supplement “Antibacterial Resistance Leadership Group (ARLG): Productivity and Innovation,” sponsored by the Antibacterial Resistance Leadership Group.
Potential conflicts of interest. R. P. has received grants from BioFire, Check-Points, Curetis, 3M, Merck, Hutchison Biofilm Medical Solutions, Accelerate Diagnostics, Allergan, and The Medicines Company; has served as a consultant to Curetis, Roche, Qvella, and Diaxonhit (monies are paid to Mayo Clinic); holds patent(s) for Bordetella pertussis/parapertussis PCR, device/method for sonication with royalties paid by Samsung to Mayo Clinic, anti-biofilm substance; has served on a data monitoring board for Actelion; and has received travel reimbursement and an editor’s stipend from American Society for Microbiology and IDSA and honoraria from the US Medical Licensing Examination, UpToDate, and the Infectious Diseases Board Review Course. S. R. E. has received grants from NIAID/NIH and Fogarty International Center, and has received personal fees from the American Statistical Association, Society for Clinical Trials, Drug Information Association, US Food and Drug Administration, NIH, City of Hope, Huntington’s Study Group, IMMPACT, PPRECISE, Muscle Study Group, DeGruyter (Statistical Communications in Infectious Diseases), Takeda, Pfizer, Roche, Novartis, Merck, Achaogen, Auspex, Alcon, Chelsea, Mannkind, QRx Pharma, Genentech, Affymax, FzioMed, Amgen, GlaxoSmithKline, Sunovion, Boehringer-Ingelheim, Cubist, AstraZeneca, Teva, Repros, Dexcom, Zeiss, University of Rhode Island, New Jersey Medical School–Rutgers, University of Vermont, Osaka University, and the National Cerebral and Cardiovascular Center of Japan. R. A. B. has received research funding from the NIH and Veterans Affairs Merit Review Board and research grants from Merck, Wockhardt, Allergan, and Roche for preclinical work on β-lactamase inhibitors. All other authors report no potential conflicts. The authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Chambers HF, Bartlett JG, Bonomo RA, et al. Antibacterial resistance leadership group: open for business. Clin Infect Dis 2014; 58:1571–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Patel R, Tsalik EL, Petzold E, et al. Viewpoint: MASTERMIND—bringing microbial diagnostics to the clinic [manuscript published online ahead of print 7 December 2016]. Clin Infect Dis 2016. pii:ciw788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huvane J, Komarow L, Hill C, et al. Fundamentals and catalytic innovation: the Statistical and Data Management Center of the Antibacterial Resistance Leadership Group. Clin Infect Dis 2017; 64(Suppl 1):S18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tsalik EL, Petzold E, Kreiswirth BN, et al. Advancing diagnostics to address antibacterial resistance: the Diagnostics and Devices Committee of the Antibacterial Resistance Leadership Group. Clin Infect Dis 2017; 64(Suppl 1):S41–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Evans SR, Hujer AM, Jiang H, et al. ; Antibacterial Resistance Leadership Group Rapid molecular diagnostics, antibiotic treatment decisions, and developing approaches to inform empiric therapy: PRIMERS I and II. Clin Infect Dis 2016; 62:181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Evans S, Hujer A, Jiang H, et al. Informing antibiotic treatment decisions: evaluating rapid molecular diagnostics (RMDs) to identify susceptibility and resistance to carbapenems against Acinetobacter spp. PRIMERS III [manuscript published online ahead of print 26 October 2016]. J Clin Microbiol 2016. doi:10.1128/JCM.01524-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doi Y, Bonomo RA, Hooper DC, et al. Gram-negative bacterial infections: research priorities, accomplishments, and future directions of the Antibacterial Resistance Leadership Group. Clin Infect Dis 2017; 64(Suppl 1):S24–9. [DOI] [PMC free article] [PubMed] [Google Scholar]