Abstract
Background
Typically, carnivores serve as definitive hosts for Sarcocystis spp. parasites; currently, their role as intermediate hosts is being elucidated. The present study aimed to identify and molecularly characterize Sarcocystis cysts detected in striated muscle of red foxes from different populations in Latvia, Lithuania and Spain.
Methods
Muscle samples from 411 red foxes (Vulpes vulpes) and 269 racoon dogs (Nyctereutes procyonoides) from Latvia, 41 red foxes from Lithuania and 22 red foxes from Spain were examined for the presence of Sarcocystis sarcocysts by light microscopy (LM). Sarcocystis spp. were identified by transmission electron microscopy (TEM) and molecular biology techniques.
Results
Sarcocystis cysts were detected in 11/411 (2.7%) Latvian, 3/41 (7.3%) Lithuanian, and 6/22 (27.3%) Spanish red foxes, however, cysts were not observed in the muscles of racoon dogs. Based on LM, TEM, 18S rDNA, 28S rDNA, ITS1, cox1 and rpoB sequences, Sarcocystis arctica and Sarcocystis lutrae cysts were identified in red fox muscles from Latvia and Lithuania, whereas only S. arctica was detected in Spain. The 18S rDNA, 28S rDNA and ITS1 sequences from the 21 isolates of S. arctica from Latvia, Lithuania and Spain were identical. By contrast, two and four haplotypes were determined based on mtDNA cox1 and apicoplast rpoB sequences, respectively. Polymorphisms were not detected between the two isolates of S. lutrae from Latvia and Lithuania. Based on phylogenetic results, S. arctica and S. lutrae were most closely related to Sarcocystis spp. using predatory mammals as intermediate hosts and to Sarcocystis species with a bird-bird life-cycle.
Conclusions
Based on current knowledge, the red fox and Arctic fox (Vulpes lagopus) could act as intermediate host for the same two Sarcocystis species. Molecular results suggest the existence of two genetic lineages of S. arctica, and such divergence relies on its geographical distribution but not on their intermediate host species.
Keywords: Sarcocystis arctica, Sarcocystis lutrae, Wild carnivores, Haplotypes, Phylogeny, Molecular characterisation
Background
Protozoans of the genus Sarcocystis (Apicomplexa: Sarcocystidae) are worldwide distributed parasites of mammals, birds and reptiles. They are characterized by an obligatory two-host life-cycle, and their transmission is based on prey-predator relationships. Sarcocysts are mainly found in striated muscles of the intermediate host, and sporocysts develop in the small intestine of the definitive host. Some Sarcocystis species are clinically important for humans, domestic and wild animals; pathogenic effects such as abortions, weight loss, encephalitis, and myositis mainly occur in the intermediate hosts [1].
Initially, carnivores were considered the only definitive hosts of Sarcocystis spp.; however, over the last few years, numerous Sarcocystis species employing wild terrestrial carnivores as intermediate hosts have been described, namely S. arctosi from the brown bear (Ursus arctos) [2]; S. ursuri from the black bear (Ursus americanus) [3]; S. kalvikus and S. kitikmeotensis from the wolverine (Gulo gulo) [4]; S. arctica from the Arctic fox (Vulpes lagopus) [5] and S. lutrae from the Eurasian otter (Lutra lutra) [6]. The domestic dog usually acts as a definitive host within the genus Sarcocystis; however, dogs can also serve as intermediate hosts. Infections by S. canis and S. neurona have been reported in a wide variety of mammals, including dogs (reviewed by Dubey et al. [1]). Recently, two more species, S. caninum and S. svanai were described in domestic dogs [7]. The latter species have been genetically characterized by 18S rDNA, ITS1 and rpoB. Sarcocystis species have relatively high host specificity with regards to their intermediate hosts [1]. Besides, reasonably high Sarcocystis infection rates have been reported in some carnivore species [8–11]. Thus, this data confirms that carnivores can also act as intermediate hosts of some Sarcocystis species.
The identification of Sarcocystis species is usually based on the morphology of sarcocysts, DNA sequences analysis and knowledge of parasite life-cycle [12]. Definitive hosts of recently described Sarcocystis species of wild carnivores remain unknown [1]. Comprehensive light microscopy (LM) and transmission electron microscopy (TEM) examination of sarcocysts of the majority Sarcocystis species from carnivores has been performed. Conversely, with the exception of S. neurona, S. arctica and S. lutrae, only limited data on DNA sequences of Sarcocystis species using mammal predators as intermediate hosts is available.
Recently, S. arctica was identified in muscles of red foxes (Vulpes vulpes) collected in the Czech Republic [13]. Here, we provide information on infection rates, morphological and molecular data of Sarcocystis sarcocysts detected in striated muscle of red foxes from different populations in Latvia, Lithuania and Spain.
Methods
Sample collection in Latvia
Between 2013 and 2016, hind leg muscle samples of 411 red foxes and 269 racoon dogs that were legally hunted, road-killed and trapped under state Rabies control and eradication programs were examined. Sampling covered the whole territory of Latvia and was uniformly distributed. Leg muscles were selected, for ease of access and availability of relatively large amounts of material.
Sample collection in Lithuania
In 2015–2016, four muscle types, i.e. diaphragm, heart, hind leg and tongue of 42 red foxes legally hunted in the Central and East Lithuania were examined for Sarcocystis spp. infection under LM.
Sample collection in Spain
Twenty-two red foxes, legally hunted in southwestern Spain in December 2016, were necropsied. Muscle tissue from tongue and forearms were morphologically examined by LM for the presence of sarcocysts.
Light microscopy examination
Sarcocystis prevalence and infection intensity in animals collected from Latvia and Lithuania were evaluated in methylene blue-stained 28 oat-size fragments of muscle (~1 g) under LM using a previously described procedure [14]. In Spain, fresh-squashed muscle samples were examined for the presence of sarcocysts. After squeezing of fresh muscle tissues, sarcocysts were excised with the aid of preparation needles, and then morphologically characterized. The sarcocysts were differentiated according to the size and shape of the cyst, the structure of the cyst wall, and morphometric parameters of the bradyzoites. The isolated cysts were preserved in microcentrifuge tubes containing 96% ethanol and kept frozen at -20 °C for further examination by molecular methods. For molecular analyses, 11 cysts from 11 Latvian red foxes, 4 cysts from 3 Lithuanian red foxes (2 cysts from fox no. 24) and 8 cysts from 3 Spanish red foxes (4 cysts from fox no. 5; 2 cysts from fox no. 7 and 2 cysts from fox no. 11) were isolated.
In addition, sections of fresh muscle tissues from 1 Lithuanian and 22 Spanish foxes were buffered-formalin fixed prior to hematoxylin and eosin (H&E) staining of 5 μm thick and 2.5 cm2 sections.
Electron microscopy examination
Four excised sarcocysts from Spanish foxes, and 1 from a Lithuanian fox were fixed in 2.5% glutaraldehyde and subjected to transmission electron microscopy (TEM) as reported previously [15, 16]. Briefly, samples were post-fixed in 1% buffered osmium tetroxide, and sections were cut on a Leica UC6 ultramicrotome and stained with 4% uranyl acetate and 3% lead citrate.
Molecular analyses
Genomic DNA was extracted from individual sarcocysts using the QIAamp® DNA Micro Kit (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations. Sarcocystis species were characterized at 5 loci, 18S ribosomal DNA (rDNA), 28S rDNA, ITS1 (internal transcribed spacer 1 region), cox1 (mitochondrial gene encoding subunit 1 of cytochrome c oxidase), and rpoB (RNA polymerase B gene of the apicoplast genome). The nearly complete 18S rDNA sequences, partial 28S rDNA sequences, complete ITS1 sequences, partial cox1 sequences and partial rpoB sequences were amplified using primers shown in Table 1.
Table 1.
Primers used for the amplification of five DNA regions
| DNA region | Primer name | Primer sequence (5′-3′) | Reference |
|---|---|---|---|
| 18S rDNA | SarAFa | CTGGTTGATCCTGCCAGTAG | [30] |
| SarBRb | GGCAAATGCTTTCGCAGTAG | [30] | |
| SarCFa | TTTAACTGTCAGAGGTGAAATTCTT | [30] | |
| SarDRb | GCAGGTTCACCTACGGAAA | [30] | |
| 28S rDNA | KL-P1Fa | TACCCGCTGAACTTAAGCAT | [30] |
| KL-P2Rb | TGCTACTACCACCAAGATCTGC | [30] | |
| ITS1 | P-ITSFa | ATTACGTCCCTGCCCTTTGT | [30] |
| P-ITSRb | GCCATTTGCGTTCAGAAATC | [30] | |
| cox1 | SF1a | ATGGCGTACAACAATCATAAAGAA | [31] |
| SR5b | ATATCCATACCTCCATTGCCCAT | [31] | |
| rpoB | RPObFa | TAGTACATTAGAAATCCCTAAAC | [32] |
| RPObRb | TCWGTATAAGGTCCTGTAGTTC | [32] |
aForward primer
bReverse primer
Each PCR reaction mixture contained 12.5 μl DreamTaq PCR Master Mix (2×) (Thermo Fisher Scientific Baltics, Vilnius, Lithuania), 0.05 μg template DNA, 1 μM of each primer, and nuclease-free water to constitute the final 25-μl volume. All amplification reactions were carried out with the same protocol: 5 min at 95 °C, 5 cycles of 45 s at 94 °C, 60 s at 64 °C, 90 s at 72 °C, followed by 30 cycles of 45 s at 94 °C, 60 s at 58 °C, and 70 s at 72 °C and 10 min at 72 °C. The PCR products were visualized using 1.7% agarose gel electrophoresis and purified with exonuclease ExoI and alkaline phosphatase FastAP (Thermo Fisher Scientific). The PCR products were sequenced directly with the 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) using the same forward and reverse primers as for the PCR. The resulting sequences were edited manually when necessary and merged into single sequences representing each of the genetic regions investigated.
To search for highly similar sequences and determining sequence identity values, the newly obtained sequences were compared with those of various Sarcocystis spp. using the Nucleotide BLAST program (megablast option). Multiple alignments obtained using MUSCLE algorithm [17] were loaded into the MEGA7 software [18]. Phylogenetic relationships of the haplotypes were inferred by coalescent simulations using median-joining model implemented in NETWORK 5.0 [19, 20]. The TOPALi v2.5 software [21] was used to select a nucleotide substitution model with the best fit to the aligned sequences dataset and to construct the phylogenetic trees using the Bayesian inference algorithm.
Results
Infection rates and Sarcocystis species identification in red foxes from Latvia, Lithuania and Spain
Sarcocysts were detected in 11/411 Latvian red foxes (2.7%); cysts were not observed in the 294 racoon dogs examined in Latvia. The infection intensity of Sarcocystis spp. sarcocysts in red fox hunted in Latvia varied from 1 to 17 cysts (mean = 5.8, median = 4.0). Examination of diaphragm, heart, leg and tongue muscles of 41 red foxes hunted in Lithuania led to the identification of cysts found in 3 animals (7.3%). Cysts were observed in the hind leg and diaphragm muscle of 1 red fox (isolate NRS2) and only in the diaphragm muscles of two other red foxes (isolates NRS24 and NRS28). The intensity of infection was lower in Lithuanian samples (1 cyst/g in diaphragm muscle and 1–3 cysts/g in leg muscles). Finally, sarcocysts were detected in the forearm muscle tissues of 3 out of 22 red foxes (13.6%) hunted in Spain; however, prevalence of infection increased to 27.3% after examination of H&E-stained slides, with an average of 2.16 cysts/section (range 1–6; median = 1); cysts were not observed in tongue samples. Also, no foci of myositis were observed.
Two morphological types of sarcocysts were observed in fresh muscle samples of red foxes with the aid of LM. Later, by molecular analysis, cysts were identified as S. arctica (Fig. 1a) and S. lutrae (Fig. 1b). Cysts of S. arctica were spindle-shaped, 2565–8414 × 83–204 μm in size, with rounded tip. The cyst wall was 1.6–2.8 μm thick and was characterised by knob-like protrusions on the surface. Septa divided the cyst into compartments, filled with banana-shaped bradyzoites, 6.5–9.3 × 1.6–2.4 (7.2 × 2.0) μm in size (Fig. 1c). Cysts of S. lutrae were spindle-shaped, and measured 1524–3186 × 75–111 μm, with a smooth cyst wall 0.8–2.2 μm thick. Banana-shaped bradyzoites measured 6.4–8.4 × 1.4–2.9 (7.1 × 2.0) μm (Fig. 1d). It should be noted that only one species, either S. arctica or S. lutrae, was detected in each infected animal. Based on the comparison of DNA sequences obtained, 10 foxes from Latvia, 3 from Spain and 2 from Lithuania harboured S. arctica, whereas 1 fox from Latvia and 1 from Lithuania were infected by S. lutrae. Two isolates of S. lutrae were detected in foxes hunted in Central Latvia and Central Lithuania (Fig. 2).
Fig. 1.
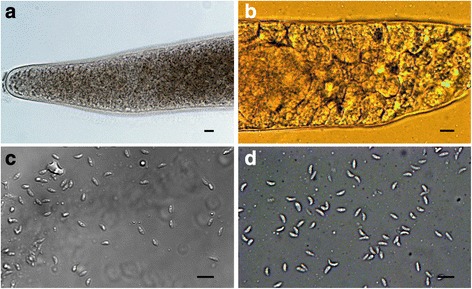
Light microscopy photomicrographs of Sarcocystis arctica (a, c) and S. lutrae (b, d) isolated from fresh muscle samples of the red fox (Vulpes vulpes). a Apical portion of sarcocyst; note knob-like villar protrusions on the cyst wall. b Portion of sarcocyst; note septae and thin, smooth wall with the apparent absence of villar protrusions. c, d Banana-shaped bradyzoites released from a broken sarcocysts. Scale-bars: 10 μm
Fig. 2.
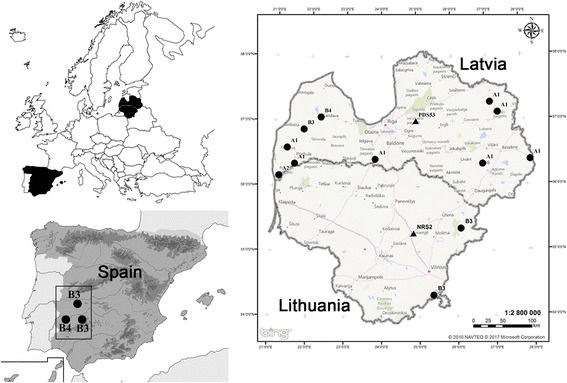
Distribution of S. arctica and S. lutrae in the sampled area and genotypes of S. arctica within cox1 and rpoB. Distribution of S. arctica is represented in black circles and S. lutrae in black triangles
Light and electron microscopy examination
On H&E-stained sections, S. arctica sarcocysts are slender, thin-walled, and present minute and indistinct protrusions (arrowheads) (Fig. 3a); S. lutrae sarcocysts are slender, thin-walled (< 1 μm), and presented well-defined septae (se) grouping bradyzoites (Fig. 3b). By TEM, the S. arctica cyst wall closely resembles type 9c (Fig. 3c); the villar protrusions (vp) were up to 1.3 μm long and 0.5 μm wide; some vp appear anastomosing; the electron-dense layer (edl) had invaginations distributed at irregular distances (arrowheads). The cyst wall (cw) was ~ 1.7–1.8 μm-thick, and the cyst contained up to 6.4 μm-long (n = 5) mature bradyzoites (br). In about 0.7 μm-thick ground substance (gs) very few electron-dense granules were observed. Also, by TEM, S. lutrae cyst wall displayed a simple structure resembling type 1a (Fig. 3d). The parasitophorous vacuolar membrane is about 120 nm-thick and is characterised by a wavy outline, small knob-like blebs with rounded ends (arrows), and the edl also presents invaginations (arrowheads). The cyst wall is up to 0.8 μm in thickness. Ground substance is smooth and projects septae (se) that surround mature up to 6.0 μm-long (n = 3) bradyzoites.
Fig. 3.
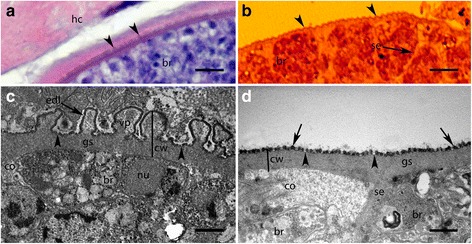
Comparison of light (LM) and transmission electron microscopy (TEM) photomicrographs of Sarcocystis arctica (a, c) and S. lutrae (b, d) from the red fox (Vulpes vulpes). a LM micrograph showing thin-walled cyst, with barely defined villar protrusions (vp, arrowheads); note bradyzoites (br) and muscular host cell (hc). H&E-staining. b LM micrograph showing well-defined thin cyst wall, with minute undulation due to fixation (arrowheads); note bradyzoites (br) and septum (se). H&E-staining. c TEM micrograph showing details of villar protrusions (vp), the total width of cyst wall (cw), ground substance (gs) with few electron-dense granules, and invaginations (arrowheads) of the electron dense layer (edl); note, details of bradyzoites (br), nucleus (nu) and conoid (co). d TEM micrograph is showing details of the simplest structure among cyst walls types within genus Sarcocystis, smooth ground substance (gs), blebs (arrows), invaginations (arrowheads), septum (se) and bradyzoite (br) with defined microtubules in conoid (co). Scale-bars: a, b, 10 μm; c, d, 1 μm
Molecular characterisation of S. arctica
The 18S rDNA (1803 bp long), 28S rDNA (1468 bp), ITS1 (697 bp), cox1 (1053 bp) and rpoB (762 bp) sequences of S. arctica were deposited in GenBank under the accession numbers MF596217–MF596237, MF596240–MF596260, MF596262–MF596282, MF596286–MF596306 and MF596311–MF596331, respectively. Cysts that were isolated from the same animal showed no genetic differences at five loci analysed. The 21 isolates of S. arctica from Latvia, Lithuania and Spain showed 100% sequence identity among each other for 18S rDNA, 28S rDNA and ITS1 sequences. By contrast, 2 distinct haplotypes A and B, differing by 2 SNPs, were determined based on cox1 sequence analysis. In particular, nucleotide A was found in 439 and 924 segregating sites for haplotype A, while nucleotides T and C were identified at the same sites for haplotype B, respectively. Four polymorphic sites were observed within rpoB (Fig. 4), 2 of which were singleton variable sites (137 and 547) and the other 2 were parsimony informative sites (419 and 761). Four different haplotypes (haplotype 1, 2, 3 and 4) of S. arctica were determined based on rpoB sequence analysis. The haplotypes differed from each other by 1–4 substitutions. One mutation was observed between haplotype 1 and haplotype 2, and between haplotype 3 and haplotype 4. Haplotypes 1 and 2 differed from haplotypes 3 and 4 by 3 SNPs on average. Interestingly, based on concatenated cox1 and rpoB sequences, only 4 different genotypes were established (A1, A2, B3 and B4), i. e. isolates having haplotype A within cox1 carried haplotypes 1 or 2 within rpoB, while isolates having haplotype B within cox1 alternatively presented haplotypes 3 and 4 within rpoB (Fig. 4).
Fig. 4.
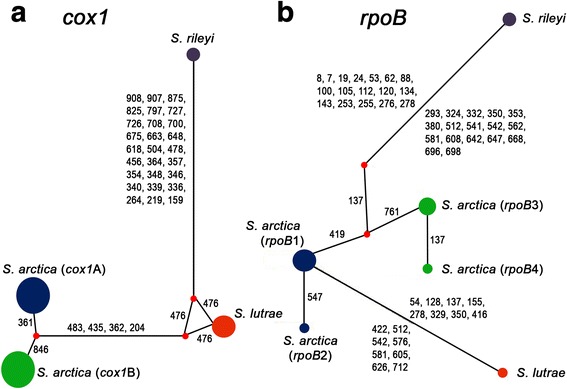
Haplotype networks obtained from cox1 (a) and rpoB (b), respectively. The numbers show the mutation sites splitting haplotypes. Small red dots indicate median vectors which represent hypothesized (often unsampled or extinct ancestral) sequence [20]. Haplotypes of S. arctica are shown in brackets. Blue and green dots indicate different S. arctica haplotypes; same colour groups 1 singleton difference
Four genotypes of S. arctica were identified in Latvia and Lithuania, and their distribution in these 2 neighbouring countries was sporadic, while in Spain only genotypes B3 and B4 were detected (Fig. 2).
Isolates of S. arctica from red foxes in Latvia, Lithuania and Spain were compared with those from the Czech Republic, Norwegian and Alaskan Arctic foxes, and Alaskan wolf (Canis lupus) from the USA at the 18S rDNA, 28S rDNA, ITS1 and cox1 genetic loci. Based on 28S rDNA and ITS1 sequences, S. arctica showed 99.9–100% and 99.2–100% identity, respectively. Two S. arctica haplotypes were identified in cox1, whereas this species had no intraspecific variability at 18S rDNA.
Based on 18S rDNA sequences, S. arctica was identical to S. caninum (GenBank: KM362427) from the domestic dog (Canis familiaris), 99.5% similar to S. svanai (GenBank: KY292487) from the domestic dog, 99.4% similar to S. columbae (GenBank: GU253883) from the wood pigeon (Columba palumbus), and displayed a very high sequence identity to other Sarcocystis spp. using predatory mammals and birds as intermediate hosts. At 28S rDNA, S. arctica was most similar to Sarcocystis sp. ex Accipiter cooperii (GenBank: KY348754), S. lari (GenBank: JQ733509) and S. calchasi (GenBank: FJ232949), infecting birds of the orders Columbiformes and Psittaciformes (approximately 98–99% sequence identity). The newly obtained ITS1 sequences of S. arctica showed 99.5% identity to sequences of S. caninum isolated from muscles of the Rottweiler dog (GenBank: JX993923, JX993924; case C in [7]) and only 88% similarity with S. felis (GenBank: AY190081); even lower rates were observed with sequences from other Sarcocystis spp. employing predatory mammals as intermediate hosts including S. canis, S. kalvikus and S. lutrae. The cox1 sequences of S. arctica showed > 99% similarity to sequences from S. lutrae, S. lari, S. canis and several other Sarcocystis spp. Interestingly, the 462 bp long rpoB sequence of S. caninum (GenBank: KC191641) was identical with haplotype 1 of S. arctica and differed by one SNP (C/T) from other three S. arctica haplotypes determined in this study. Sequences of rpoB of S. arctica also showed identity to S. lari (GenBank: MF596307; 98.2–98.4%), S. campestris (GenBank: GQ851963) from ground squirrels (Sciuridae) (98.1–98.4%), S. lutrae (GenBank: MF596309; 97.8–97.9%), S. svanai from the domestic dog (GenBank: KC191641; 97.6–97.8%) and S. canis (GenBank: KC191642) from the polar bear (Ursus maritimus) (97.4–97.6%).
Molecular characteristics of S. lutrae
Both isolates of S. lutrae from Latvia (PDS53) and Lithuania (NRS2) had identical 18S rDNA (1782 bp) (MF596215, MF596216), 28S rDNA (1492 bp) (MF596238, MF596239), cox1 (1053 bp) (MF596284, MF596285) and rpoB (762 bp) (MF596309, MF596310) sequences. The newly obtained S. lutrae sequences of 18S and 28S rDNA showed 100% identity with those of S. lutrae from the Eurasian otter detected in Norway. Based on cox1, S. lutrae from the red fox had 100% identity with S. lutrae from the Eurasian otter (GenBank: KM657808, KM657809) and the Arctic fox (GenBank: KF601326, KF601327). Unfortunately, only 929 bp of ITS1 of S. lutrae (GenBank: MF596261) from Lithuania were obtained, while ITS1 amplification using the S. lutrae isolate from Latvia was unsuccessful. The ITS1 sequence of S. lutrae from the red fox showed 100% identity with S. lutrae from the European badger (Meles meles) (GenBank: KX431307) from Scotland and had 98.0–100% identity with S. lutrae from the Eurasian otter (GenBank: KM657773–KM657805) studied in Norway.
Based on 18S rDNA, S. lutrae had very high (exceeding 99%) sequence identity with several Sarcocystis species. The 28S rDNA sequences of S. lutrae was 99.4% similar to Sarcocystis sp. ex Accipiter cooperii (GenBank: KY348754), 98.3% similar with S. lari (GenBank: JQ733509), and 98.2% similar with S. turdusi (GenBank: JF975682) from thrushes. Based on ITS1, S. lutrae was characterized by the lowest sequence differences with S. kalvikus (GenBank: GU200661; 3.8–5.2%). Interestingly, at cox1 S. lutrae demonstrated 100% sequence identity with S. lari (GenBank: MF596283) and differed only by one SNP from Sarcocystis sp. ex Accipiter cooperii (GenBank: KY348756) and S. calchasi (GenBank: KU220952). At rpoB, S. lutrae had the highest sequence similarity to S. svanai (GenBank: KC191641; 98.5%), S. campestris (GenBank: GQ851963; 97.7%), S. caninum (GenBank: KC191641; 97.6%), S. arctica (GenBank: MF596311– MF596331; 97.8–97.9%) and S. lari (GenBank: MF596307; 97.5%).
Phylogeny
Phylogenetic results showed that both species identified in the red fox, S. arctica and S. lutrae, were most closely related to Sarcocystis spp. using predatory mammals as intermediate hosts and Sarcocystis species characterized by a bird-bird life-cycle (Fig. 5). Phylogenetic analyses demonstrated that S. arctica could not be discriminated from S. caninum using rpoB and ITS1 sequences. However, based on 28S rDNA and cox1 sequences, S. arctica was grouped with S. lari and S. canis, respectively. It should be noted that 28S rDNA and cox1 sequences of S. caninum are not available on GenBank. The exact phylogenetic position of S. lutrae using 28S rDNA, cox1 and rpoB sequences remains unclear. Based on ITS1, the most variable locus analysed here, S. lutrae was the sister species to S. kalvikus, which both infect mustelids.
Fig. 5.
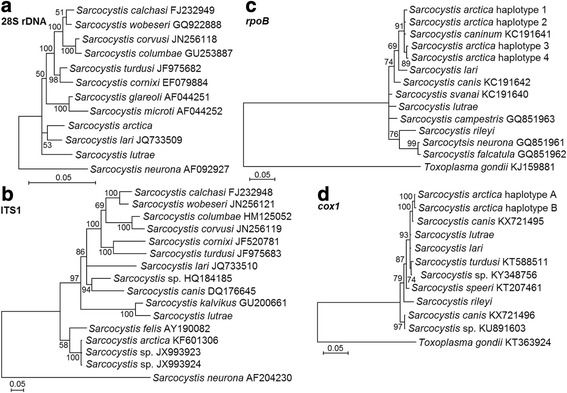
The phylogenetic placement of S. arctica and S. lutrae based on 28S rDNA (a), ITS1 (b), rpoB (c) and cox1 (d) sequences. Sequences for Sarcocystis sp. (GenBank: JX993923, X993924) correspond to S. caninum [7]
Discussion
The racoon dog is naturally distributed in the east of Asia. In the mid-twentieth century, one of the five racoon subspecies, Nyctereutes procyonoides ussuriensis was introduced in eastern Europe and nowadays is successfully expanding to the central part of the continent [22].
In the present study, hind leg muscles of 294 racoon dogs were negative for sarcocysts in Latvia. However, muscular Sarcocystis sp. infection was previously reported in the Japanese racoon dog (N. p. viverrinus). In Japan, sarcocysts were detected in 12 out of 27 animals examined (44.4%) [9]. Prevalence of the Sarcocystis infection varied significantly in different muscles examined in this study and was highest in the tongue (47.6%) and diaphragm (44.4%), the masseter muscles (28.0%), and lowest in the heart (14.8%) and oesophagus (4.0%). Furthermore, the severe meningoencephalitis associated with Sarcocystis sp. asexual development was reported in a free-living, adult racoon dog from Japan [23]. Further studies examining different types of muscle and covering broader geographical regions are needed to verify whether racoon dog populations, located far from their native range, could harbour muscular Sarcocystis infections.
The red fox is one of the main predators of the Canidae in the Northern Hemisphere [24]. In the present study, S. arctica and S. lutrae in red foxes from Latvia, Lithuania and Spain were characterized using LM and TEM, and DNA sequences analysis within five loci. Cysts of Sarcocystis spp. were first found in the muscles of the red fox in Kazakhstan [25]. Briefly, by LM, two morphological types of sarcocyst were distinguished. Cysts (1300–3900 × 130–500 μm in size) having a 2.0–2.8 μm thick and striated cyst wall were named Sarcocystis vulpis. Smaller cysts (25–520 × 40–65 μm in size), with smooth 1.4–2.1 μm thick cyst wall were assigned to Sarcocystis sp. However, in the latest taxonomic review of the genus Sarcocystis, S. vulpis was considered to be species inquirenda [1]. Based on morphological characters, the proposed S. vulpis and Sarcocystis sp. in red foxes from Kazakhstan [25] should correspond to S. arctica and S. lutrae, respectively. In 2009, Sarcocystis sp. cysts were found in one Japanese red fox (Vulpes vulpes japonica) [9]. By LM, cysts were narrow and short (21–710 × 20–47 μm) and by TEM cyst wall was thin (0.4–0.5 μm thick) and had minute undulations. Morphologically, Sarcocystis sp. from the Japanese red fox [9] is similar to S. lutrae. Recently, the Arctic fox and the Alaskan wolf were identified as being an intermediate host of S. arctica [5, 26, 27]. Moreover, based on cox1 comparison it was shown that one Arctic fox from Norway acted as the intermediate host for two Sarcocystis species, S. arctica and S. lutrae [6].
Here, using LM, S. arctica and S. lutrae show thin-walled cysts that can be distinguished within the same host (Fig. 3a, b). Sarcocystis arctica detected in the red fox here presents similar ultrastructure as previously described in the Alaskan wolf and Arctic fox [26, 27] in North America. In addition, S. lutrae sarcocysts from red foxes are morphologically similar to those of other Sarcocystis species detected in carnivores, e.g. S. kalvikus from the wolverine [4], S. arctosi from the brown bear [2], and S. svanai from a domestic dog [7] and the Pampax fox (Lycalopex gymnocercus) from Argentina [11]. Recently, S. lutrae has been detected in the European badger from Scotland [28], but no ultrastructural details were provided. Definitive hosts of S. arctica and S. lutrae remain unknown.
Based on data collected thus far, S. lutrae shows intraspecific diversity only within ITS1, while S. arctica is characterized by some intraspecific variability within 28S rDNA, ITS1, cox1 and rpoB. The clearly distinguished S. arctica haplotypes A and B at cox1 were identified for Norwegian isolates of the Arctic fox, and for Latvian/Lithuanian and Czech isolates from the red fox, whereas Alaskan isolates from two intermediate hosts, the grey wolf and Arctic fox, were only haplotype A, and S. arctica from the Spanish red fox had haplotype B ([5, 13, 26, 27] and present study). At rpoB, two SNPs were determined between haplotype 1 and 3, and only one SNP between haplotype 1 and 2, and between haplotype 3 and 4 (Fig. 4). Primary population genetic data suggest the existence of two genetic lines of S. arctica expanding along the latitudinal cline. Distinguished genetic lines might be associated to different intermediate hosts. However, it is not possible to explain such diversity by historical geographical barriers given the fact that the host Vulpes vulpes is highly vagile and evenly distributed. Thus, the hypothesis that this particular genetic diversity within S. arctica species obeys to a potential adaptive significance [29] cannot be rejected.
Conclusions
In the present study, sarcocysts of S. arctica and S. lutrae infecting red foxes from three different European countries were morphologically and molecularly characterized. Muscle samples of racoon dogs were also investigated, but no sarcocysts were found. LM and TEM provided details to distinguish both Sarcocystis species. Based on nuclear 18S rDNA, 28S rDNA, and ITS1 sequences, the 21 isolates of S. arctica from Latvia, Lithuania and Spain showed a 100% sequence identity among each other. However, 2 and 4 haplotypes were determined within cox1 and rpoB, respectively. Polymorphisms were not detected between the 2 isolates of S. lutrae studied herein. In conclusion, it is being demonstrated that red fox acts as an intermediate host for at least 2 species of Sarcocystis, and it is suggested that the existence of 2 genetic lineages of S. arctica, along with molecular divergence of such species relies on its geographic distribution.
Acknowledgements
The research was supported by the Open Access research infrastructure of the Nature Research Centre under the Lithuanian open access network initiative. Rafael Calero Bernal is funded by the Complutense University of Madrid (Grant CT65/16). Authors of the present work are grateful to hunters who provided samples for study. Also, we are thankful to Maris Nitcis of the Institute of Life Sciences and Technology, Daugavpils University, for help in arranging the map.
Funding
Not applicable.
Availability of data and materials
The sequences generated in the present study were submitted to the GenBank database under the following accession numbers: 18S rDNA sequences (MF596215–MF596237), 28S rDNA sequences (MF596238–MF596260), complete ITS1 sequences (MF596261–MF596282), cox1 sequences (MF596283–MF596306) and rpoB sequences (MF596307–MF596331).
Abbreviations
- br
bradyzoites
- co
conoid
- cox1
cytochrome c oxidase subunit 1 gene
- cw
cyst wall
- edl
electron-dense layer
- gs
ground substance
- H&E
hematoxylin and eosin staining
- hc
muscular host cell
- ITS1
internal transcribed spacer 1
- LM
light microscopy
- mtDNA
mitochondrial deoxyribonucleic acid
- nu
nucleus
- rpoB
RNA polymerase beta-subunit gene
- se
septum
- SNPs
single nucleotide polymorphisms
- TEM
transmission electron microscopy
- vp
villar protrusions
Authors’ contributions
Conceived and designed the laboratory tests: PP, MK, DB and RC. Performed experiments: VK, ER-L, IG, MK and MG. Analysed the data: PP and J-LF. Contributed reagents/materials/analysis tools: IG, PP and MA. Drafted the manuscript: VK, MK, PP, DB, IG, RC and MA. All authors read and approved the final manuscript.
Ethics approval and consent to participate
For this type of formal study consent is not required.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Viktorija Kirillova, Email: viktorija.kirillova@du.lv.
Petras Prakas, Email: prakaspetras@gmail.com.
Rafael Calero-Bernal, Email: rcalerober@gmail.com.
Inese Gavarāne, Email: inese.gavarane@gmail.com.
José Luis Fernández-García, Email: pepelufe@unex.es.
Manuel Martínez-González, Email: manuel.martine.gonzalez@gmail.com.
Eglė Rudaitytė-Lukošienė, Email: egle.rudaityte@gmail.com.
Miguel Ángel Habela Martínez-Estéllez, Email: mahabela@unex.es.
Dalius Butkauskas, Email: dalius@ekoi.lt.
Muza Kirjušina, Email: muza.kirjusina@du.lv.
References
- 1.Dubey JP, Calero-Bernal R, Rosenthal BM, Speer CA, Fayer R. Sarcocystosis of animals and humans. Boca Raton: CRC Press; 2016. [Google Scholar]
- 2.Dubey JP, Rosenthal BM, Sundar N, Velmurugan GV, Beckmen KB. Sarcocystis arctosi sp. nov. (Apicomplexa, Sarcocystidae) from the brown bear (Ursus arctos), and its genetic similarity to schizonts of Sarcocystis canis-like parasite associated with fatal hepatitis in polar bears (Ursus maritimus) Acta Parasitol. 2007;52(4):299–304. doi: 10.2478/s11686-007-0048-6. [DOI] [Google Scholar]
- 3.Dubey JP, Humphreys GJ, Fritz D. A new species of Sarcocystis (Apicomplexa: Sarcocystidae) from the black bear (Ursus americanus) J Parasitol. 2008;94(2):496–499. doi: 10.1645/GE-1406.1. [DOI] [PubMed] [Google Scholar]
- 4.Dubey JP, Reichard MV, Torretti L, Garvon JM, Sundar N, Grigg ME. Two new species of Sarcocystis (Apicomplexa: Sarcocystidae) infecting the wolverine (Gulo gulo) from Nunavut, Canada. J Parasitol. 2010;96(5):972–976. doi: 10.1645/GE-2412.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gjerde B, Schulze J. Muscular sarcocystosis in two arctic foxes (Vulpes lagopus) due to Sarcocystis arctica n. sp.: sarcocyst morphology, molecular characteristics and phylogeny. Parasitol Res. 2014;113(3):811–21. [DOI] [PubMed]
- 6.Gjerde B, Josefsen TD. Molecular characterisation of Sarcocystis lutrae n. sp. and Toxoplasma gondii from the musculature of two Eurasian otters (Lutra lutra) in Norway. Parasitol Res. 2015;114(3):873–86. [DOI] [PubMed]
- 7.Dubey JP, Sykes JE, Shelton GD, Sharp N, Verma SK, Calero-Bernal R, et al. Sarcocystis caninum and Sarcocystis svanai n. spp. (Apicomplexa: Sarcocystidae) associated with severe myositis and hepatitis in the domestic dog (Canis familiaris). J Eukaryot Microbiol. 2015;62(3):307–17. [DOI] [PMC free article] [PubMed]
- 8.Anderson AJ, Greiner EC, Atkinson CT, Roelke ME. Sarcocysts in the Florida bobcat (Felis rufus floridanus) J Wildl Dis. 1992;28(1):16–20. doi: 10.7589/0090-3558-28.1.116. [DOI] [PubMed] [Google Scholar]
- 9.Kubo M, Okano T, Ito K, Tsubota T, Sakai H, Yanai T. Muscular sarcocystosis in wild carnivores in Honshu, Japan. Parasitol Res. 2009;106(1):213–219. doi: 10.1007/s00436-009-1653-0. [DOI] [PubMed] [Google Scholar]
- 10.Verma SK, Calero-Bernal R, Lovallo MJ, Sweeny A, Grigg ME, Dubey JP. Detection of Sarcocystis spp. infection in bobcats (Lynx rufus) Vet Parasitol. 2015;212(3–4):422–426. doi: 10.1016/j.vetpar.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scioscia NP, Olmos L, Gorosábel A, Bernad L, Pedrana J, Hecker YP. Pampas fox (Lycalopex gymnocercus) new intermediate host of Sarcocystis svanai (Apicomplexa: Sarcocystidae) Parasitol Int. 2017;66(3):214–218. doi: 10.1016/j.parint.2017.01.021. [DOI] [PubMed] [Google Scholar]
- 12.Prakas P, Butkauskas D. Protozoan parasites from genus Sarcocystis and their investigations in Lithuania. Ekologija. 2012;58(1):45–58. doi: 10.6001/ekologija.v58i1.2349. [DOI] [Google Scholar]
- 13.Pavlásek I, Máca O. Morphological and molecular identification of Sarcocystis arctica sarcocysts in three red foxes (Vulpes vulpes) from the Czech Republic. Parasitol Int. 2017;66(5):603–605. doi: 10.1016/j.parint.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Kutkienė L, Sruoga A, Butkauskas D. Sarcocystis sp. from white-fronted goose (Anser albifrons): cyst morphology and life cycle studies. Parasitol Res. 2006;99(5):562–565. doi: 10.1007/s00436-006-0196-x. [DOI] [PubMed] [Google Scholar]
- 15.Prakas P, Butkauskas D, Rudaitytė E, Kutkienė L, Sruoga A, Pūraitė I. Morphological and molecular characterization of Sarcocystis taeniata and Sarcocystis pilosa n. Sp. from the sika deer (Cervus nippon) in Lithuania. Parasitol Res. 2016;115(8):3021–3032. doi: 10.1007/s00436-016-5057-7. [DOI] [PubMed] [Google Scholar]
- 16.Trupkiewicz JG, Calero-Bernal R, Verma SK, Mowery J, Davison S, Habecker P, et al. Acute, fatal Sarcocystis calchasi-associated hepatitis in roller pigeons (Columba livia f. Dom.) at Philadelphia Zoo. Vet Parasitol. 2016;216:52–8. [DOI] [PubMed]
- 17.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phylogenetic Network Software. http://www.fluxus-engineering.com/sharenet.htm. Accessed 4 Sept 2017.
- 20.Bandelt H-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 21.Milne I, Wright F, Rowe G, Marshall DF, Husmeier D, McGuire G. TOPALi: Software for automatic identification of recombinant sequences within DNA multiple alignments. Bioinformatics. 2004;20(11):1806–1807. doi: 10.1093/bioinformatics/bth155. [DOI] [PubMed] [Google Scholar]
- 22.Kauhala K, Kowalczyk R. Invasion of the raccoon dog Nyctereutes procyonoides in Europe: history of colonization features behind its success, and threats to native fauna. Curr Zool. 2011;57(5):584–598. doi: 10.1093/czoolo/57.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubo M, Kawachi T, Murakami M, Kubo M, Tokuhiro S, Agatsuma T, et al. Meningoencephalitis associated with Sarcocystis spp. in a free-living Japanese raccoon dog (Nyctereutes procyonoides viverrinus) J Comp Pathol. 2010;143(2–3):185–189. doi: 10.1016/j.jcpa.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 24.Ginsberg JR, Macdonald DW. Foxes, wolves, jackals, and dogs. An action plan for the conservation of canids. Broadview: Kelvyn Press, Inc.; 1990. [Google Scholar]
- 25.Pak SM, Sklyarova ON, Dymkova ND. Sarcocysts (Sporozoa, Apicomplexa) of some wild mammals. Izvestiya Akademii Nauk Kazakhskoi SSR Seriya Biologicheskaya. 1991;5:35–40. [Google Scholar]
- 26.Calero-Bernal R, Cerqueira-Cézar CK, Verma SK, Mowery J, Carmena D, Beckmen K, et al. Sarcocystis arctica (Apicomplexa: Sarcocystidae): ultrastructural description and its new host record, the Alaskan wolf (Canis lupus) Parasitol Res. 2016;115(7):2893–2897. doi: 10.1007/s00436-016-5067-5. [DOI] [PubMed] [Google Scholar]
- 27.Cerqueira-Cézar CK, Thompson PC, Verma SK, Mowery J, Calero-Bernal R, Antunes Murata FH, et al. Morphological and molecular characterization of Sarcocystis arctica-like sarcocysts from the Arctic fox (Vulpes lagopus) from Alaska, USA. Parasitol Res. 2017;116(7):1871–1878. doi: 10.1007/s00436-017-5462-6. [DOI] [PubMed] [Google Scholar]
- 28.Lepore T, Bartley PM, Chianini F, Macrae AI, Innes EA, Katzer F. Molecular detection of Sarcocystis lutrae in the European badger (Meles meles) in Scotland. Parasitology. 2017;144(11):1426–1432. doi: 10.1017/S0031182017000762. [DOI] [PubMed] [Google Scholar]
- 29.Avise JC. Phylogenetic units and currencies above and below the species level. In: Purvis A, Brooks T, Gittleman J, editors. Phylogeny and conservation. Cambridge: Cambridge University Press; 2005. pp. 76–100. [Google Scholar]
- 30.Kutkienė L, Prakas P, Sruoga A, Butkauskas D. The mallard duck (Anas platyrhynchos) as intermediate host for Sarcocystis wobeseri sp. nov. from the barnacle goose (Branta leucopsis) Parasitol Res. 2010;107(4):879–888. doi: 10.1007/s00436-010-1945-4. [DOI] [PubMed] [Google Scholar]
- 31.Gjerde B. Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int J Parasitol. 2013;43(7):579–591. doi: 10.1016/j.ijpara.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Wendte JM, Miller MA, Nandra AK, Peat SM, Crosbie PR, Conrad PA, et al. Limited genetic diversity among Sarcocystis neurona strains infecting southern sea otters precludes distinction between marine and terrestrial isolates. Vet Parasitol. 2010;169(1–2):37–44. doi: 10.1016/j.vetpar.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences generated in the present study were submitted to the GenBank database under the following accession numbers: 18S rDNA sequences (MF596215–MF596237), 28S rDNA sequences (MF596238–MF596260), complete ITS1 sequences (MF596261–MF596282), cox1 sequences (MF596283–MF596306) and rpoB sequences (MF596307–MF596331).


