Abstract
In the human killer cell immunoglobulin-like receptors, KIR2DL2, and KIR2DL3, a triad of amino acids in the D1 domain interact to stabilize protein structure. Substitution of any one of these residues caused significant loss of cell surface expression. Although KIR2DS4 and KIR2DS5, two homologous receptors, differ for this triad, flow cytometry analysis of NK and T cell lines transfected with stimulatory KIR genes KIR2DS4 (allele *001) and KIR2DS5 (allele *002) demonstrated cell surface expression. For KIR2DS5, restoration of the triad sequence increased surface expression. Further studies of the receptor encoded by KIR2DS5*002 showed both mature and immature protein isoforms upon gel electrophoresis coupled with surface biotinylation or deglycosylation. In contrast, the KIR2DS5*001 allelic product was not expressed on the cell surface of either NK or T cells and exhibited only a single immature isoform upon gel electrophoresis. Site-directed mutagenesis demonstrated that absence of the KIR2DS5*001-encoded protein at the cell surface was imparted primarily by two amino acid polymorphisms in the D2 domain. Analysis using molecular dynamics simulations suggested that the substitution of a proline for a serine at residue 111 or the substitution of a serine for a phenylalanine at residue 164 caused destabilization of the domain structure and intracellular retention. A third polymorphism at residue 174 impacted the level of KIR2DS5 surface expression. This is the first description at a stimulatory KIR locus of the impact of specific amino acid variations on receptor maturation and the level of surface expression.
Keywords: Natural killer cells, Cell surface molecules, Human
Introduction
Bridging the innate and adaptive immune systems, NK cells express a number of cell surface receptors which either inhibit or stimulate their cytotoxicity (Lanier 2005). NK cells are prevented from killing healthy cells by a subset of killer cell immunoglobulin-like receptors (KIR) that recognize specific class I HLA ligands. For example, inhibitory receptors KIR2DL2 and KIR2DL3 recognize a subset of HLA-C molecules and inhibit killing of cells expressing those ligands (Colonna et al. 1993). Other receptors in the KIR family presumably stimulate killing when their ligands are present. Some of these stimulatory KIR may recognize HLA-peptide complexes with low affinity, although much is to be learned about their specificity (Katz et al. 2001, 2004; Saulquin et al. 2003; Stewart et al. 2005; Winter et al. 1998). The balance of inhibitory and activating signals received by the NK cell via KIR and its other receptors determines the fate of nearby cellular targets.
The 15 highly homologous receptors in the KIR family are type I transmembrane glycoproteins with either two (2D) or three (3D) extracellular immunoglobulin-like domains (D0, D1, D2; Boyington et al. 2000; Fan et al. 2001; Lanier 2005; Saulquin et al. 2003). Inhibitory or stimulatory properties are controlled by the presence of ITIM in their intracellular segments (inhibitory KIR possessing long (L) cytoplasmic tails) or the association with an adaptor protein (e.g., DAP12) carrying ITAM (stimulatory KIR with short (S) cytoplasmic tails).
From two (KIR2DS3) to over 30 (KIR3DL3) allelic products are encoded by each KIR locus (Robinson et al. 2005). Available KIR-HLA crystal structures (Boyington et al. 2000; Fan et al. 2001) do not readily suggest the functional importance of allelic diversity in ligand recognition but allelic variation in KIR3DL1 has been shown to modulate the inhibitory response of NK cells to various HLA ligands (Yawata et al. 2006). It is also clear that the level of cell surface expression of the receptors is altered by some alleles. Several loci (e.g., KIR2DL4, KIR2DS4) include alleles that encode alternatively spliced or truncated receptors that may be secreted rather than cell-membrane-bound (Goodridge et al. 2007; Maxwell et al. 2002). Other alleles (e.g., KIR2DL2*004, KIR3DL1*004) encode misfolded receptors which remain within the cell (Pando et al. 2003; VandenBussche et al. 2006). Still other alleles control high (e.g. 3DL1*001, 2DL5B*003) vs. low to absent (e.g. 3DL1*005; 2DL5B*002) cell surface expression (Vilches et al. 2000a; Yawata et al. 2006).
In addition to allelic diversity, the KIR gene complex on chromosome 19 includes variable numbers of the 15 inhibitory and stimulatory genes (Parham 2005a). All haplotypes appear to carry several inhibitory genes, including ubiquitous framework loci, KIR3DL3, KIR3DL2, and KIR2DL4, but the number of stimulatory genes can vary from one to several. Furthermore, in haplotypes designated as group A, alleles encoding a secreted product of the sole stimulatory gene, KIR2DS4, define haplotypes encoding no membrane-bound short-cytoplasmic-tailed KIR (Hsu et al. 2002; Middleton et al. 2007a). These differences can impact the activity and specificity of NK cells and provide one mechanism for differences in human immune responsiveness (Khakoo and Carrington 2006; Parham 2005b).
Previously, we reported that a D1 domain polymorphism encoded by KIR2DL2*004 results in the receptor product being detectable intracellularly but not on the cell surface (VandenBussche et al. 2006). An arginine/threonine interchange at residue 41 was shown to impact surface expression. R41 in the expressed receptor interacts with residues D47 and Y77 to maintain beta strand contacts critical to structural integrity of the receptor. This interaction was critical to both KIR2DL2 and its allelic partner, KIR2DL3. Site-directed mutation of D47 to L47 to eliminate the interaction with R41 significantly reduced surface expression of the KIR2DL3*001-encoded protein. D47 is conserved among all D1–D2 (type 1) KIR receptors except for receptors encoded by KIR2DS4 and KIR2DS5. All KIR2DS4 alleles encode a receptor with N47 in place of D47; all KIR2DS5 alleles encode H47 at this position. Other key residues, R41 and Y77, are retained in both stimulatory receptors. We therefore hypothesized that the polymorphism at residue 47 may disrupt the surface expression of the two stimulatory receptors in the same fashion as the receptor encoded by KIR2DL2*004. We found that the level of KIR2DS5 surface expression was impacted by residue 47 but that variation at several residues in the D2 domain of KIR2DS5 ultimately had a more profound effect.
Materials and methods
DNA constructs
These studies have been reviewed and approved by the Georgetown University Institutional Review Board. All expression vectors were created from pEF-DEST51 (Invitrogen, Carlsbad, CA) by Gateway Technology (Invitrogen) using the pENTR/D-TOPO (Invitrogen) entry vector. KIR2DS1*002 and KIR2DS4*001 cDNA were amplified from a normal human peripheral blood cDNA pool obtained from Biochain Institute, Inc. (Hayward, CA). KIR2DS5*001 cDNA was obtained from Origene Technologies, Inc. (Rockville, MD). To create C-terminally V5-tagged KIR2DS4 and KIR2DS5 constructs, the appropriate cDNA was amplified with a glycine codon replacing the stop codon using the following primers: 2DS4-ATG-F (5′-CACCATGTCGCTCATGGTCATCAT-3′), 2DS5-ATG-F (5′-CACCATGTTGCTCATGGTCAT CAG-3′) with reverse primer 2DS4/5-R (5′-TCCTG CGTATGACACCTCCT-3′). N-terminally HA-tagged KIR2DS4 and KIR2DS5 constructs were created by site-directed mutagenesis which inserted nucleotides encoding YPYDVPDYA between the regions encoding for the signal peptide and the D1 domain. Wild type KIR2DS5*002 and mutants of KIR2DS4 and KIR2DS5 were created by site-directed mutagenesis using QuikChange II (Stratagene, La Jolla, CA). All DNA constructs were prepared using the HiSpeed Plasmid Maxi Kit (QIAGEN, Valencia, CA).
Cell lines, culture, and transfection
The NKL cell line was a gift of Dr. Francisco Borrego (NIAID, Rockville, MD). The Jurkat T cell clone E6-1 was obtained from the American Type Culture Collection (Manassas, VA). All cells were cultured in RPMI 1640 supplemented with 2 mM L-glutamine, 10 mM HEPES, 1 mM sodium pyruvate, and 10% FBS. Cultures of NKL cells were supplemented with rIL-2 (30 U/mL). NKL cells (10×106) were transiently transfected in triplicate with 10 μg of the appropriate constructs. pmaxGFP (2 μg; Amaxa Inc., Gaithersburg, MD) was used as a transfection efficiency control. Transfection was performed with a Nucleofector II instrument (Amaxa Inc.) with Solution V and programs O-017 (NKL) and S-016 (Jurkat).
Immunoprecipitation and Western blotting
Approximately 18 h after transfection with constructs encoding C-terminally V5-tagged KIR, cells were washed in PBS. If indicated, surface proteins were biotinylated using NHS-LC-Biotin (Pierce Biotechnology, Inc., Rockford, IL) according to the manufacturer’s protocol. Cells were lysed in 0.5% NP-40 diluted in PBS containing protease inhibitors (Protease Inhibitor Set III; EMD Biosciences, San Diego, CA). Proteins were immunoprecipitated with 2 μg V5-specific Ab (Invitrogen) bound to either Protein G Plus-Agarose (EMD Biosciences) or GammaBind Plus Sepharose (GE Healthcare Amersham Biosciences, Piscataway, NJ).
To deglycosylate KIR, 10× PNGase F denaturation buffer was added to washed cells and the cells lysed. The lysates were digested with either PGNase F (New England Biolabs, Ipswich, MA) or endoglycosidase H (Roche Diagnostics Corporation, Roche Applied Science, Indianapolis, IN) according to manufacturer’s instructions. Beta actin was detected with a mAb (Sigma-Aldrich, St. Louis, MO) and served as a loading control.
All protein samples were reduced and denatured in 2× Laemmli buffer (Sigma-Aldrich, St. Louis, MO) and electrophoresed on 4–15% polyacrylamide Tris–HCl Ready gels (Bio-Rad, Hercules, CA). Separated proteins were blotted to Hybond-ECL nitrocellulose membrane (Amersham Biosciences) or iBlot gel transfer nitrocellulose (Invitrogen) and the membrane was blocked for an hour using 5% (w/v) nonfat dry milk (SACO Foods, Inc. Middleton, WI). For detection of proteins with a V5 tag including deglycosylated proteins, a 1/5,000 dilution of a V5-specific Ab (Invitrogen) was used with a 1/15,000 dilution of secondary Ab directed against the mouse H and L chains (Jackson Immunoresearch, West Grove, PA). The blot containing deglycosylated proteins was stripped using Restore Western blot buffer (Pierce, Rockford, IL) and restained with an Ab to beta actin (clone AC-15; Sigma-Aldrich) to control for the amount of sample loaded on the gel. For the detection of immunoprecipitated protein with a V5 tag, a 1/5,000 dilution of a V5-specific Ab (Invitrogen) was used with a 1/20,000 dilution of secondary Ab directed against the mouse kappa L chain (Serotec, Oxford, UK). For detection of biotinylated proteins, a 1/1,000 dilution of streptavidin-HRP (Amersham Biosciences) was used. Protein bands were detected using ECL chemiluminescent detection (Amersham Biosciences). NKL cells transfected with an empty vector served as a negative control.
Flow cytometry
Approximately 18 h post-transfection, purified mouse monoclonal HA-specific Ab (Sigma-Aldrich, St. Louis, MO) with PE-conjugated anti-mouse IgG (Beckman Coulter, Fullerton, CA) was used for extracellular staining at 4°C. KIR2DS4 was also detected with a PE-conjugated CD158i-specific mAb: clone FES172 (Beckman Coulter, Marseille Cedex 9, France; Moretta et al. 2001) or clone 5F2 (Santa Cruz Biotechnology, Inc. Santa Cruz, CA; Katz et al. 2001). Following extracellular staining, cells were fixed and permeabilized with Cytofix/ Cytoperm (BD Pharmingen, Franklin Lakes, NJ) in preparation for intracellular staining. After blocking with 5% (w/v) nonfat dry milk, cells were stained with FITC-conjugated V5-specific Ab (Invitrogen) at room temperature. Image-iT FX (Invitrogen) was used to improve the signal-to-noise ratio. Stained cells were analyzed on a Becton Dickinson FACSort (San Jose, CA) with FCS Express 2 software (De Novo Software, Thornhill, Canada). MFI of PE was measured after gating on those cells positive for V5 staining. Statistical analysis for differences in expression as measured by the comparison of the ratio of extracellular MFI to intracellular MFI of the entire population was performed using Student’s unpaired t-tests. Results were normalized to a positive control, either KIR2DS1 or the wild type KIR2DS5 encoded by allele *002. Negative controls were empty vector, vector carrying a KIR insert encoding a V5, but not an HA, tag, and transfectants expressing V5-tagged KIR2DS1 (as a control for the CD158i mAb). Each flow cytometry assay was performed in triplicate. The results described derive from two or more transfections performed on different days; representative data are shown in the figures.
Molecular modeling
As the crystal structure is not available for KIR2DS4 and KIR2DS5, a homology model was constructed using the program Modeller (Fiser et al. 2000). The input alignment for the Modeller was obtained with ClustalW (Thompson et al. 1994) based on the sequence of the KIR2DS2 (PDB: 1M4K; Saulquin et al. 2003). The model was refined further by brief molecular dynamics (MD) simulations and energy minimization for 10,000 cycles with a consistent valence force field using the Discover module of Insight II (Accelrys Inc., San Diego, CA). We again compared the homology models with the KIR2DS2 structure to ensure that the residue side chains were in the proper orientations after refinement. The quality of the refined model was checked with Procheck (Laskowski et al. 1993).
In order to see the consequences of allelic variation, various rotomers of the substituted amino acids were evaluated and optimized. MD simulations (3 ns) were performed for the molecules encoded by the two KIR2DS5 alleles with a distant-dependent dielectric constant using the Sander module of the Amber 8.0 simulation package (University of California, San Francisco, CA) with the PARM98 force-field parameter. Simulations were performed with 0.001-ps time steps, and the temperature set at 300°K. The Shake algorithm (Hanson et al. 2003) was employed to keep all bonds involving hydrogen atoms rigid. Weak coupling temperature and pressure coupling algorithms (Berendsen et al. 1984) were used to maintain constant temperature and pressure, respectively. Electrostatic interactions were calculated with the Ewald particle mesh method (Darden et al. 1993) with a dielectric constant at 1Rij and a non-bonded cut-off of 12 Å for the real part of electrostatic interactions and for van der Waals interactions. Simulated structures were energy-minimized using the consistent valence force field with the default partial atomic charges available in Discover version 3.0 of Insight II. The dielectric constant was set at ε=4 to account for the dielectric shielding found in proteins. Each minimization was carried out in two steps, first using steepest descent minimization for 200 cycles and then using conjugate gradient minimization until the average gradient fell below 0.01 kcal/mol. Minimization was performed for 10,000 iterations. Structural analyses were done using Insight II (Accelrys Inc, San Diego, CA) and Sybyl 8.0 (Tripos, Inc., St. Louis, MO) molecular modeling programs.
Phylogenic analysis
An unrooted phylogenetic tree was constructed using the neighbor-joining method with PAUP version 4 (Sinauer Associates, Sunderland, MA) based on the amino acid sequence alignments of KIR2DS5 allelic products.
Results
The level of KIR2DS5 cell surface expression is impacted by residue 47
In order to determine whether the replacement of D47 with N47 or H47 disrupted KIR2DS4 or KIR2DS5 surface expression through receptor misfolding, molecular modeling was used to analyze the possible effects of this polymorphism on the D1 domains. In the KIR2DL2/2DL3 D1 domain, D47 interacts with R41 and Y77 since mutation of any one of these three residues disrupts the KIR structure and significantly decreases receptor surface expression (VandenBussche et al. 2006). To examine the D1 domain of the two stimulatory receptors, models were based on the KIR2DS2 crystal structure because of its relatively high resolution compared to other available KIR structures (Saulquin et al. 2003). The receptors modeled based on KIR2DS2 are encoded by KIR2DS4*001 (N47) and KIR2DS5*002 (H47), alleles chosen because they are frequent in individuals carrying these loci (Hou et al. 2008; Middleton et al. 2007a, b; Yawata et al. 2006). KIR2DS4*001 also encodes the only transmembrane protein specified by that locus; the remainder of the allelic products are thought to be secreted (Maxwell et al. 2002). When the KIR2DS4 and KIR2DS5 structures were modeled based on the KIR2DS2 crystal structure, the hydrogen bond patterns were found to be similar at the R41 site in that R41 appears to form a H-bond with N47 in KIR2DS4 (or H47 in KIR2DS5) and Y77 (data not shown).
To test KIR surface expression, receptors were expressed in NKL (NK cell lineage) and Jurkat (T cell lineage), two cell lines that lack surface expression of all KIR. Although Jurkat cells do not express the adaptor DAP12, previous studies have shown surface expression of transfected stimulatory KIR in its absence (e.g., Campbell et al. 1998). Thus, Jurkat provided an alternative cell type to test stimulatory KIR surface expression since the mechanism of expression appears to differ from NKL. KIR2DS1 (encoded by frequently observed allele KIR2DS1*002, Hou et al. 2008) was expressed as a control since it encodes a well-characterized DAP12-associated stimulatory KIR molecule with high amino acid sequence similarity to KIR2DS4 and KIR2DS5. By flow cytometry, mAbs specific for KIR2DS4 stained NKL transfectants expressing KIR2DS4, but not KIR2DS1 (Fig. 1a); detection of the C-terminal V5 tags measured total KIR protein expression. Because an Ab specific for KIR2DS5 was not available, expression was measured with KIR carrying an N-terminal HA tag and a C-terminal V5 tag. The negative control was a vector encoding KIR2DS1 without an HA tag. KIR2DS5 (*002)-expressing NK cells were surface-stained with HA-specific Ab as were the HA-tagged KIR2DS1 expressing cells (Fig. 1b). Jurkat cells also showed expression of both KIR2DS4 (data not shown) and KIR2DS5 (described below). These data suggest that both KIR2DS4 and KIR2DS5 (*002) are expressed at the cell surface.
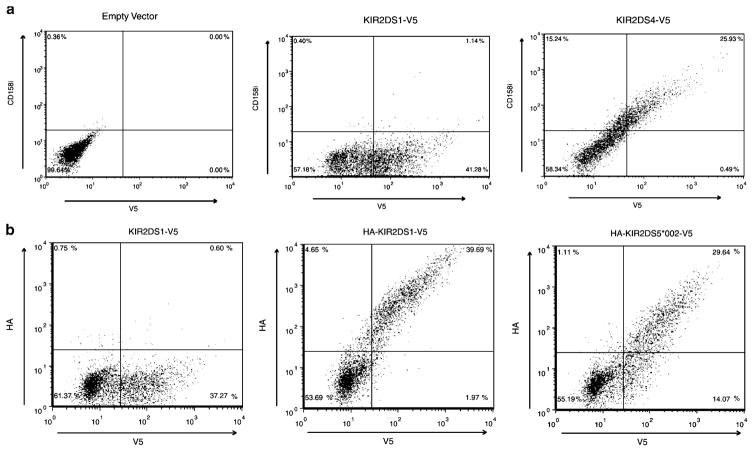
Fig. 1.
KIR2DS4 and KIR2DS5 are expressed on the surface of NKL cells. Representative flow cytometric analysis of NKL transfectants expressing C-terminally V5-tagged KIR constructs. Total KIR expression was measured with an FITC-conjugated antibody to the V5 tag after the externally stained cells were fixed and permeabilized (X axis). a Representative scatter plots of vector without a KIR insert (left), KIR2DS1 (center) and KIR2DS4 (right) surface expression showing that cells expressing KIR2DS4, but not KIR2DS1, were stained by a PE-conjugated KIR2DS4 (CD158i)-specific mAb, clone FES172 (Y axis). Approximately 26% of KIR2DS4-transfected cells were found in the upper right quadrant of the plot on the right compared to approximately 1% in the center plot. b Representative scatter plots of negative control (left), KIR2DS1 (middle) and KIR2DS5 (*002; right) surface expression. PE positive extracellular expression was detected with an antibody to the HA-tagged KIR (Y axis). Approximately 30% (KIR2DS5 (*002) and 40% (KIR2DS1) of the V5 positive cells exhibited HA surface staining. The negative control expressed a V5-tagged KIR2DS1 molecule without an HA tag
In order to further evaluate our hypothesis that an alteration at residue 47 may impact receptor expression on the cell surface, site-directed mutagenesis was used to alter the N47 found in KIR2DS4 and the H47 found in KIR2DS5 to D47 observed in the expressed KIR2DL2/ 2DL3. A further mutant introducing L47 was also created because this mutation of KIR2DL2/2DL3 also reduced surface expression (VandenBussche et al. 2006). Alteration of residue 47 in KIR2DS4 had no significant impact on the level of surface expression as analyzed by flow cytometry (data not shown). In contrast, using an HA-specific Ab, the D47 mutation significantly increased KIR2DS5 surface expression in both NKL (Fig. 2) and Jurkat (data not shown) while substitution of L47 further reduced KIR expression in NKL. This suggests that the histidine at residue 47 in KIR2DS5, while not abrogating expression, does reduce surface expression compared to the aspartic acid found in other D1 domain KIRs.
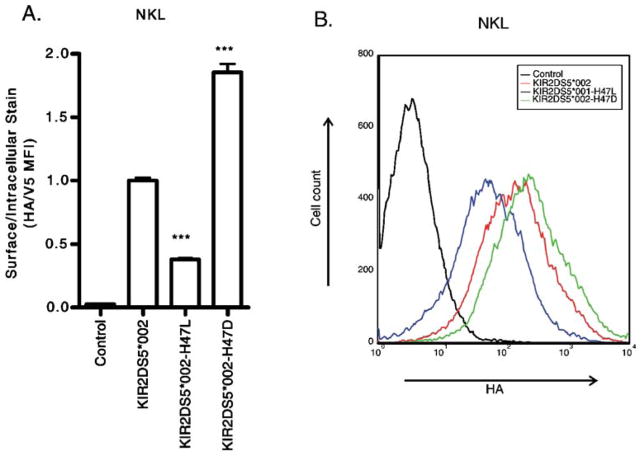
Fig. 2.
Alteration of H47 in KIR2DS5 affects surface expression. A representative flow cytometric analysis of NKL cells transfected with constructs encoding N-terminally HA-tagged, C-terminally V5-tagged KIR. The negative control was a cell transfected with a KIR construct without an HA tag. a In NKL, replacement of H47 in KIR2DS5 (*002) with L47 reduced surface expression while alteration to D47 significantly increased surface expression of KIR2DS5. b The same data shown as a histogram of the HA staining of the transfectants after gating on the V5-positive populations. By Student’s t-test compared to wild type for a representative experiment performed in triplicate: ***p-value<0.001
KIR2DS5 allelic products vary in surface expression
When the amino sequences of KIR2DS5 allelic products are compared to one another, KIR2DS5 (*001) is the most divergent sequence at this locus (Robinson et al. 2005). In order to evaluate the impact of this allelic diversity, NKL and Jurkat cells were transfected with an expression vector encoding KIR2DS5 (*001) for comparison with the surface-expressed KIR2DS5 (*002). Cells expressing the KIR2DS5*001 construct were stained intracellularly for V5 yet demonstrated no detectable HA surface staining on either cell line by flow cytometry (Fig. 3a). Extracellular expression was comparable to a negative control cell expressing a KIR2DS gene without an HA tag. This suggested that the KIR2DS5*001 encoded protein was synthesized but not localized to the cell surface.
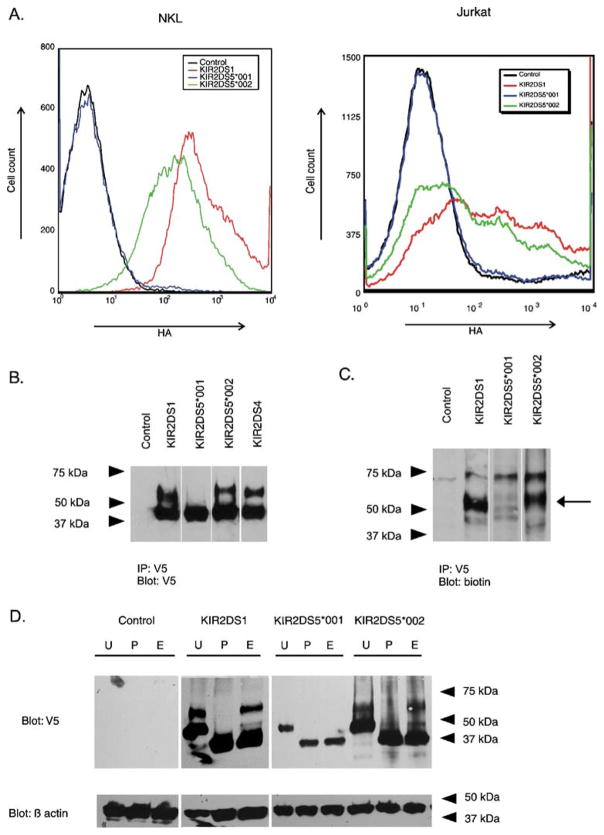
Fig. 3.
Flow cytometry and gel electrophoresis of stimulatory KIR show differences in cell surface expression and in the presence of receptor isoforms. a Representative flow cytometric analysis of transient transfectants expressing N-terminally HA-tagged, C-terminally V5-tagged KIR constructs. The tags facilitated the measurement of surface expression (HA) and total KIR protein (V5). The histogram shows the fluorescence intensity (HA) for receptors encoded by KIR2DS5*001 and KIR2DS5*002 in NKL (left) and in Jurkat (right) compared to a similarly tagged KIR2DS1 positive control after gating on the V5-positive populations. The negative control expressed a KIR2DS molecule without an HA tag. b Isolation of C-terminally V5-tagged KIR by immunoprecipitation from transfected NKL cells was followed by gel electrophoresis and Western blotting with a V5-specific Ab. Cells transfected with empty vector served as a negative control. Two isoforms were detected for KIR2DS1, KIR2DS4, and KIR2DS5 (*002). A single isoform with mobility similar to the lower molecular weight band exhibited by the other transfectants was detected for cells transfected with KIR2DS5*001. c Transfected cells were lysed after surface biotinylation. Proteins were isolated by immunoprecipitation and characterized by gel electrophoresis and Western blotting. A probe specific for biotinylated protein detected the higher molecular weight isoforms of KIR2DS1 and KIR2DS5 (*002) as indicated by the arrow. d NKL cells were transfected with KIR constructs carrying a V5-tag and the lysates were mock digested (U) or digested with PNGase F (P) or endoglycosidase H (E). Lysates were analyzed by Western blotting using the V5-specific Ab. Control indicates NKL cells transfected with an empty vector. Beta actin served as a loading control. PNGase F digested both isoforms of KIR2DS5 (*002) and KIR2DS1 to a core protein of one molecular mass. The lower molecular mass forms, but not the higher molecular mass forms, of KIR2DS5 (*002) and KIR2DS1 were susceptible to endoglycosidase H digestion. In contrast, the single isoform observed for KIR2DS5 (*001) was susceptible to both treatments
Biotinylation and glycosylation patterns suggest the cellular location of KIR2DS5
Previously we found that the amount of surface-expressed KIR2DL2/2DL3 could be correlated with the proportion of receptor that was terminally glycosylated (VandenBussche et al. 2006). To examine if a similar phenomenon was present for KIR2DS5, NKL cells transiently expressing KIR2DS5 (encoded by alleles *001 or *002) or positive control KIR2DS1with C-terminal V5 tags were surface biotinylated and the cell lysates were immunoprecipitated with a V5-specific Ab. Protein electrophoresis followed by Western blotting with the V5-specific Ab demonstrated two specific bands for positive control KIR2DS1 at ~65 kDa and ~45 kDa (Fig. 3b). Probing for biotin detected the higher molecular mass form of KIR2DS1, suggesting that only this isoform is present at the cell surface (Fig. 3c). The receptor encoded by KIR2DS5*002 showed a banding pattern similar to KIR2DS1 with ~67 kDa and ~47 kDa isoforms; Fig. 3c shows that the higher molecular weight band was biotinylated for KIR2DS5 (*002). The same analysis of the receptor encoded by KIR2DS5*001 only detected the lower band at ~47 kDa when blotted with V5-specific Ab. Probing for biotin did not detect this band indicating that the ~47 kDa isoform was not significantly present on the cell surface.
To demonstrate that the specific bands found for the stimulatory KIR are differentially glycosylated isotypes of the same core protein, C-terminally V5-tagged KIR2DS5 or KIR2DS1 were expressed in NKL cells. Cell lysates were mock digested or digested with either PNGase F or endoglycosidase H (Fig. 3d). PNGase F cleaves N-glycans entirely from asparagine residues, leaving only the core protein. Endoglycosidase H only cleaves N-glycans that have not been fully glycosylated in the Golgi apparatus. Therefore, mature receptor proteins that have undergone post-translational modification in the Golgi apparatus are resistant to endoglycosidase H digestion while immature proteins that have not reached the Golgi remain susceptible to the same digestion; both forms of the receptor are susceptible to digestion by PNGase F. Cellular lysates were digested with PNGase F or endoglycosidase H. Both the higher and lower molecular mass isoforms of positive control KIR2DS1 could be digested with PNGase F to a core protein of ~39 kDa (Fig. 3d). However, only the lower molecular mass isoform was susceptible to endoglycosidase H digestion, suggesting that this was an immature form of KIR2DS1 found in the endoplasmic reticulum. The higher molecular mass isoform was resistant to digestion with endoglycosidase H and likely represents the mature form of KIR2DS1 expressed on the cellular surface. Both isoforms of KIR2DS5 (*002) and the lower molecular weight isoform of KIR2DS5 (*001) were susceptible to PNGase. Only the lower molecular weight bands were susceptible to endoglycosidase H digestion. This suggests, like KIR2DS1, that the higher molecular weight isoform encoded by KIR2DS5*002 likely represents the mature form of this receptor while the lower band represents the immature, intracellular form. KIR2DS5*001 only encodes an immature form likely found in the endoplasmic reticulum because it is fully susceptible to endoglycosidase H digestion. Detection of only an immature isoform of KIR2DS5 (*001) is consistent with the lack of KIR2DS5 (*001) surface expression, since the immature receptor is not expressed on the surface. The molecular weight of all three core proteins following PNGase F digestion appeared to be the same based on gel mobility, consistent with molecular weights predicted by their amino acid sequence. This suggests that KIR2DS5 (*002) might be more heavily glycosylated compared to KIR2DS1 since its mature isoform migrated more slowly during gel electrophoresis.
Several polymorphic amino acids contribute to loss of expression of the KIR2DS5*001 receptor
The polypeptide encoded by KIR2DS5*001 differs from that encoded by KIR2DS5*002 by four amino acids: one in the signal peptide (L/S (*001/*002) at residue -20) and three in the D2 extracellular domain (P/S at residue 111, S/F at residue 164, and A/T at residue 174; Table 1). To evaluate which of the four amino acids might contribute to expression differences, 14 mutant constructs altering from one to three amino acids which differ between the two allelic products were created and expressed in NKL cells. Western blots of whole lysates probed with V5-specific Ab were obtained from transfectants expressing wild type alleles of KIR2DS5 (*001 or *002) or each of the 14 mutants (Fig. 4a). Two isoforms were detected for KIR2DS5 (*002) and positive control KIR2DS1 as expected; a single band of the lower molecular weight isoform was detected for KIR2DS5 (*001). Although the bands were faint for two of the mutants, the upper molecular weight isoform was observed in the three mutants which retained both S111 and F164 of the expressed KIR2DS5*002 allele (i.e., singly substituted mutants 5 and 8 and the double mutant 11, Table 1). All of the other mutants showed only the lower isoform. These data suggest that residues S111 and F164 in the D2 domain of KIR2DS5 together are critical for terminal glycosylation and surface expression. In the KIR2DS5*002-encoded protein, alteration of one or both of these residues resulted in only an immature isoform, likely retained within the cell.
Table 1.
Differences in full length amino acid sequences encoded by KIR2DS5*001, KIR2DS5*002, and 14 mutant constructs
| Codon | −20 | 111 | 164 | 174 |
|---|---|---|---|---|
| KIR2DS5*001 (not expressed on surface) | L | P | S | A |
| KIR2DS5*002 (expressed on surface) | S | S | F | T |
| Mutant 1 | S | – | – | – |
| Mutant 2 | – | S | – | – |
| Mutant 3 | – | – | F | – |
| Mutant 4 | – | – | – | T |
| Mutant 5 | – | S | F | T |
| Mutant 6 | S | – | F | T |
| Mutant 7 | S | S | – | T |
| Mutant 8 | S | S | F | – |
| Mutant 9 | – | S | – | T |
| Mutant 10 | S | – | F | – |
| Mutant 11 | – | S | F | – |
| Mutant 12 | S | – | – | T |
| Mutant 13 | S | S | – | – |
| Mutant 14 | – | – | F | T |
Dash indicates sequence is identical to the sequence of KIR2DS5*001.
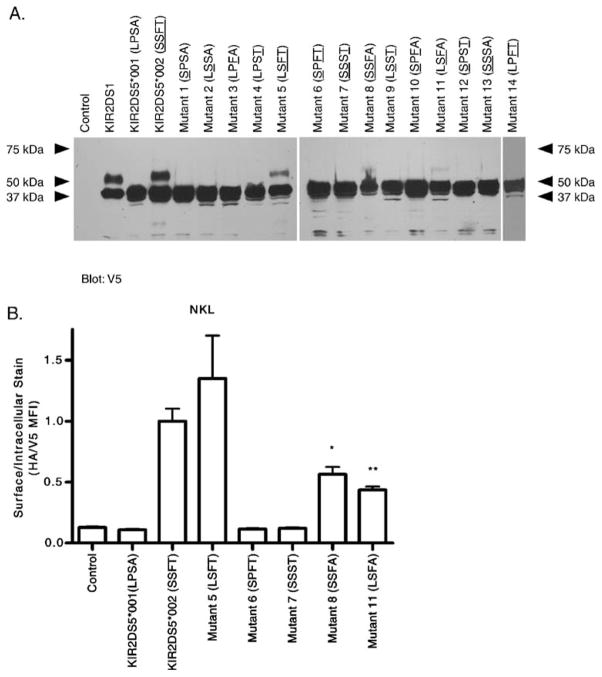
Fig. 4.
Mutagenesis of four codons that differ between two KIR2DS5 alleles identifies residues altering cell surface expression. NKL cells were transfected with constructs encoding V5-tagged nonexpressed KIR2DS5 (*001 with L-20, P111, S164, A174), expressed KIR2DS5 (*002 with S-20, S111, F164, T174) and mutants of KIR2DS5 altering codons shown in Table 1. The mutants are labeled with the four polymorphic amino acids present; residues found in the expressed KIR2DS5 (*002) are underlined. a Lysates from each transfectant were electrophoresed under denaturing conditions and proteins analyzed by Western blotting using a V5-specific Ab. Two isoforms were exhibited by KIR2DS5 (*002) and positive control KIR2DS1; only the lower molecular weight isoform was present in the lane containing lysate from KIR2DS5 (*001). Mutants 5, 8, and 11 which carry S111 and F164 of KIR2DS5 (*002) exhibited two isoforms with molecular weights identical to KIR2DS5 (*002) although the upper molecular weight bands were faint in mutants 8 and 11. The remaining mutants, like KIR2DS5 (*001), only exhibited the lower molecular weight isoform. Cells transfected with empty vector served as a negative control. b Representative flow cytometric analysis of transfectants expressing a subset of the mutant constructs. Mutants with both S111 and F164 (mutants 5, 8, 11) were expressed on the cell surface as is KIR2DS5 (*002). The levels of expression of KIR2DS5 (*002) and mutant 5 which shares T174 were similar while the surface expression of mutants 8 and 11 with A174 were lower than KIR2DS5 (*002). The levels of mutants 8 and 11 were not significantly different from one another. Mutants 6 and 7, lacking S111 or F164, were not expressed. A vector carrying a KIR insert without the external HA tag served as a negative control. By Student’s t-test compared to wild type KIR2DS5 (*002) for a representative experiment performed in triplicate: *, 0.05–0.01; **, p-value 0.01–0.001
To further confirm surface expression, flow cytometry was performed with NKL transfectants expressing KIR2DS5*001, KIR2DS5*002, or each of a subset of KIR2DS5 mutants (single substitutions of KIR2DS5*002 and the double mutant altering both residues −20 and 174 to that found in the protein encoded by KIR2DS5*001; Fig. 4b). Expressed KIR were N-terminally HA-tagged and C-terminally V5-tagged. The results parallel those observed in the Western blot in that only mutants which retain both S111 and F164 (mutants 5, 8, 11) were detected at the cell surface. In addition to S111 and F164 being critical to surface expression, residue 174 also appeared to play some role in surface expression because mutants 8 and 11 encoding both S111 and F164 but carrying the *001 variation at residue 174 (A174) were approximately half of KIR2DS5 (*002) expression levels. Restoration of T174 in a mutant which also carries S111 and F164 with the KIR2DS5*001-encoded leader sequence returned expression to the KIR2DS5 (*002) level. This parallels the intensity of the upper band observed in the Western blot in Fig. 4a. Thus, T174 impacts the level on the surface but does not abrogate surface expression while the amino acid variation in the signal peptide did not appear to impact expression level. Taken together, these data suggest that S111 and F164 together are critical for surface expression; alteration of one or both residues to that found in the KIR2DS5*001 allelic product eliminated surface expression.
Molecular modeling predicts the impact of polymorphism
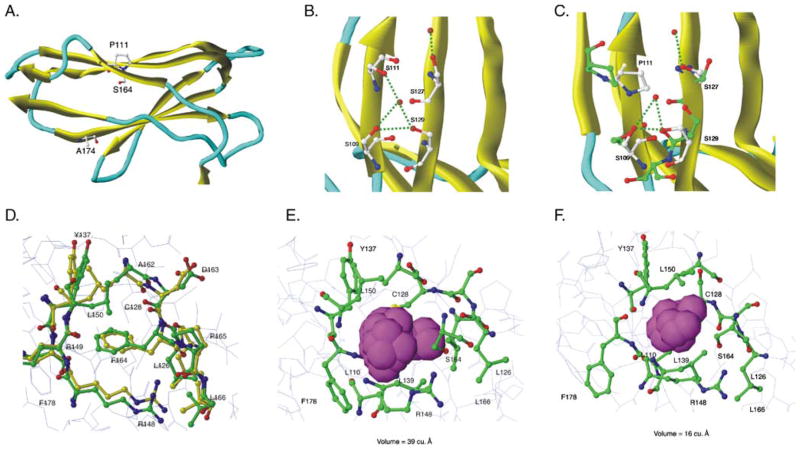
In order to understand the impact of polymorphism at residues 111, 164, and 174 in altering KIR2DS5 expression, molecular modeling of KIR2DS5 and its allelic products was performed. When the structure of KIR2DS5 was predicted based on the KIR2DS2 crystal structure (Saulquin et al. 2003), it was observed that residues 111 and 174 are on the external surface of the D2 domain while residue 164 is located within the domain (Fig. 5a; KIR2DS5 (*001)). The P/S polymorphism at residue 111 lies on a beta strand just C-terminal to the hinge loop region connecting the two domains. Inside the D2 domain, residue 164 lies on a beta strand and is surrounded by hydrophobic residues. Residue 174, which alters the level of KIR2DS5 on the cell surface, also lies on a beta strand and appears to interact with an adjacent strand.
Fig. 5.
Molecular modeling predicts that P111 and S164 cause extracellular domain instability. KIR2DS5 (*001 and *002) energy-minimized structures were generated from the previously-solved KIR2DS2 crystal structure of extracellular domains (PDB:1M4K). a The D2 domain of KIR2DS5 (*001) showing the positions of the three polymorphic amino acids that differ between the receptors encoded by KIR2DS5*001 (shown) and KIR2DS5*002 (S111, F164, T174). Residues 111 and 174 lie on the surface; residue 164 lies in the hydrophobic interior of the domain. b Initial predicted KIR2DS5 (*002) structure in the region of S111 showing a hydrogen bond network with nearby serines. MD simulations did not alter this network (data not shown). c Predicted KIR2DS5 (*001) structure in the region of P111 before (yellow) and after (green) MD simulations. The substitution of proline for serine at 111 alters the hydrogen bond network destabilizing the structure. d Initial predicted KIR2DS5 (*002) structure before and after MD simulations. Yellow represents carbon atoms before MD simulations and green, after simulations. Red represents oxygen and blue, nitrogen. F164 interacts with hydrophobic residues in the interior of the domain and these interactions are not altered following MD simulations. e A closer view of the 2DS5*001-encoded S164 region before MD simulations. The van der Waals spheres are represented by a space filling model with magenta representing the pocket size in the S164 region. The size of the pocket before MD simulations was 39 Å3. f A closer view of 2DS5 (*001) S164 region after MD simulations. Magenta van der Waals spheres represent the volume of the pocket after MD simulations; its size was 16 Å3
A network of hydrogen bonds either directly or indirectly involving water molecules links S111, S109, S129, and S127 (Fig. 5b). The rigid hydrogen bond structure involving S111 in KIR2DS5 (*002) is stable and not disturbed by molecular dynamics (MD) simulations. In contrast, substitution of proline at 111 as in KIR2DS5 (*001) disrupts this network, disrupting local folding and overall conformation (Fig. 5c). MD simulations of the F164 region in the surface-expressed KIR2DS5 (*002) demonstrated no significant change in the structure. Figure 5d shows the positioning of carbon atoms before (yellow) and after (green) simulations. F164 occupies a quarter of the pocket volume and makes hydrophobic contact with surrounding residues stabilizing the pocket. In contrast, MD simulations of the KIR2DS5 (*001) structure indicated a significant movement (>4.5 Å root mean squared deviation) of amino acids in the S164 region over 3 ns causing a compression of the pocket (Fig. 5e,f). The area measured in the S164 region before MD simulations was 39 Å3; it collapsed to 16 Å3 after MD simulations. This compression of the pocket is due to the hydrophobic collapse associated with the substitution of serine for phenylalanine at residue 164. In contrast, at residue 174, variation in the amino acid (A (*001) vs. T (*002)) is not predicted to alter domain structure.
Discussion
Based on an observation from our previous study (VandenBussche et al. 2006), we hypothesized that two of the 2D KIR stimulatory receptors, KIR2DS4 and KIR2DS5, which carry an alteration at a key amino acid required for KIR2DL2 folding (D47), would also be defective and fail to be expressed at the cell surface. Our initial flow cytometry analysis of NK and T cell lines transfected with KIR2DS4*001 or KIR2DS5*002 supplemented by gel electrophoresis of total KIR2DS protein to demonstrate mature isoforms suggested that these stimulatory receptors were expressed at the cell surface. Although the H47 did not abrogate extracellular expression of KIR2DS5, a mutation restoring D47 demonstrated that H47 did decrease the level of surface expression. In contrast, the N47 in KIR2DS4 compared to a mutant with D47 did not significantly alter the level on the cell surface. While our hypothesis was supported, the impact of H47 on the cell phenotype was less dramatic than expected. Our finding suggests, although KIR receptors exhibit ~94% sequence homology (Parham 2003), the effect of specific amino acid variations on the structure and expression of a receptor encoded by one locus may not be predictive of their impact on KIR receptors encoded by other loci.
This study also evaluated the impact of specific amino acid substitutions on cell surface expression of KIR2DS5. The amino acids studied differentiate the receptors encoded by KIR2DS5*002, chosen because of its predominance in individuals carrying the locus (Hou et al. 2008; Middleton et al. 2007b; Yawata et al. 2006), and KIR2DS5*001, chosen because it was an outlier in the phylogenetic analysis of KIR2DS5 allelic products. Haplotypes carrying KIR2DS5 usually carry other well-characterized stimulatory genes like KIR2DS1 or KIR2DS2 (Middleton et al. 2007b) so these may provide activating receptors for those individuals carrying this defective and possibly rare KIR2DS5 allele (Dohring et al. 1996; Vilches et al. 2000b).
Four amino acid differences distinguish KIR2DS5*001 from KIR2DS5*002 encoded proteins. One variation lies in the signal peptide, where a L/S interchange is found at residue 2 of a 21 amino acid long sequence. Variations in the amino acid sequence of the signal peptide have been shown to alter the level of extracellular expression of a variety of genes including the cytokine TGF-beta1 (Dunning et al. 2003) and CTLA4 (Anjos et al. 2002). Variation may affect translocation into the endoplasmic reticulum, signal peptide cleavage, or association with chaperones to impact folding or glycosylation. Site-directed mutagenesis was used to alter residue −20 in both KIR2DS5 allelic products; both mutants retained the expression properties of their wild type origins suggesting that signal peptide variation did not have any impact on expression. A similar amino acid substitution (A vs. S or T) at position 17 in the 20 amino acid long n-region of the signal peptide of CTLA4 impacts protein folding, glycosylation, and ultimately surface expression (Anjos et al. 2002); however, it is possible that the positioning of the KIR2DS5 polymorphism in the very short n-region of the signal peptide (four amino acids long, predicted using SignalP 3.0 at www.cbs.dtu.dk/services/SignalP/) is responsible for its lack of phenotype.
Alteration of either residue 111 or residue 164 in KIR2DS5 (*002) to the amino acid found in KIR2DS5 (*001) resulted in loss of cell surface expression as measured by flow cytometry and by the appearance of only the immature isoform upon gel electrophoresis of proteins encoded by mutated constructs. Reverse mutation of either residue in KIR2DS5 (*001) to the amino acid found in KIR2DS5 (*002) did not restore surface expression; mutation of both restored expression although not to wild type levels. Based on the crystal structure of KIR2DS2, residue 111 is located on the solvent accessible exterior. Using MD simulations, substitution of proline in the nonexpressed allelic product disrupts a hydrogen bond network that stabilizes the D2 domain structure in the expressed variant with S111. P111 appears to disrupt local folding and conformation resulting in intracellular sequestration. Another critical residue for cell surface expression, the amino acid at 164 is buried among hydrophobic core residues in the D2 domain. Alteration from F164 in the expressed version of KIR2DS5 to S164 leads to a hydrophobic collapse in the residue 164 region destabilizing the domain structure and resulting in only an immature intracellular isoform.
D2 domain residue 174 appeared to modulate the level of KIR2DS5 surface expression in the presence of S111 and F164. T174 was necessary to maintain wild type KIR2DS5 (*002) expression levels. In the crystal structure, residue 174 is on the exterior of the D2 domain in the angle formed between the D1 and D2 domains with no apparent consequences on KIR structure. However, it is possible that this position may impact intermolecular contacts of the KIR monomer with intracellular proteins such as endoplasmic reticulum or Golgi resident chaperones altering the rate of folding, glycosylation, and/or transport of the mature protein to the cell surface.
The response of a natural killer cell to a target is determined by the net sum of inhibitory and stimulatory signals received through KIR and other cell surface receptors. The majority of studies have focused on signals regulated by the presence or absence of specific KIR inhibitory receptors coupled with expression of their HLA ligands. In these cases, KIR haplotypic diversity coupled with allelic polymorphism of HLA, may determine susceptibility or resistance to disease (Bashirova et al. 2006). For example, an earlier study of the role of inhibitory receptors KIR2DL2 and KIR2DL3 in the resolution of hepatitis C infections suggested a correlation of the lower affinity KIR2DL3 receptor and its HLA-C1 ligand with viral clearance (Khakoo et al. 2004). In the case of autoimmune diseases like psoriatic arthritis, the lower inhibitory threshold in individuals missing ligands for inhibitory receptors KIR2DL1 and KIR2DL2/2DL3 appeared to result in a greater disease susceptibility (Martin et al. 2002). The activation threshold is also modulated by allelic diversity at an inhibitory KIR locus. For inhibitory receptor KIR3DL1, polymorphism controls the amount of the receptor on the cell surface as well as its ligand-binding affinity (Yawata et al. 2006).
Tipping the balance from inhibition to activation from the standpoint of the stimulatory KIR is less well studied, in part because the ligands are less well defined. Here, too, the presence or absence of specific loci in a diverse array of KIR haplotypes determines the occurrence of an activation signal for a particular stimulus. For example, in HIV-1 infection, KIR3DS1-expressing NK cells control viral replication, possibly by recognizing an HLA ligand bound to a viral or stress peptide (Alter et al. 2007). How allelic diversity at the stimulatory KIR loci impacts the strength of the stimulatory signal is not yet clear. Most stimulatory loci encode a single frequent allele (Hou et al. 2008; Yawata et al. 2006) so the impact of diversity may be limited to a small population of individuals. Allelic variation altering stimulatory KIR from cell membrane bound to secreted has been described for KIR2DS4 (Middleton et al. 2007a) but a role for secreted receptors has not yet been found. The KIR2DS5 variation described here is the first at a stimulatory locus to impact protein maturation and the level of surface expression. An understanding of the constraints of amino acid sequence variation on the folding, post-translational modification, and transport of stimulatory KIR to the NK cell surface should help elucidate the impact of the KIR polymorphism on function.
Acknowledgments
Funding from the Office of Naval Research N00014-05-1-0784 and N00014-06-1-0726 (C.K.H.) supported this research. The views expressed in this article are those of the authors and do not reflect the official policy of the Department of Navy, the Department of Defense, or the United States government. These studies were conducted using the Tissue Culture and Flow Cytometry and Cell Sorting Shared Resources of Lombardi Comprehensive Cancer Center supported by National Cancer Institute Cancer Center Support Grant CA51008. Computing time and staff support was in part provided by the Advanced Biomedical Computing Center at the National Cancer Institute (Frederick, MD). We thank Karen Creswell and Michelle Lombard for providing excellent technical assistance and Tiernan Mulrooney, William Frazier, and Phillip Posch for helpful discussions.
Abbreviations
- KIR
killer cell Ig-like receptor
- 2DL
two extracellular domains, long cytoplasmic tail
- 3DL
three extracellular domains, long cytoplasmic tail
- 2DS
two extracellular domains, short cytoplasmic tail
- MD
molecular dynamics
Contributor Information
Noriko K. Steiner, Department of Oncology, Lombardi Cancer Center, Georgetown University, Washington, DC 20057, USA
Sivanesan Dakshanamurthy, Department of Oncology, Lombardi Cancer Center, Georgetown University, Washington, DC 20057, USA.
Christopher J. VandenBussche, Department of Oncology, Lombardi Cancer Center, Georgetown University, Washington, DC 20057, USA. Tumor Biology Training Program, Lombardi Cancer Center, Washington, DC 20057, USA
Carolyn K. Hurley, Research Building Room E404, Georgetown University Medical Center, 3970 Reservoir Rd NW, Washington, DC 20057, USA
References
- Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjos S, Nguyen A, Ounissi-Benkalha H, Tessier MC, Polychronakos C. A common autoimmunity predisposing signal peptide variant of the cytotoxic T-lymphocyte antigen 4 results in inefficient glycosylation of the susceptibility allele. J Biol Chem. 2002;277:46478–46486. doi: 10.1074/jbc.M206894200. [DOI] [PubMed] [Google Scholar]
- Bashirova AA, Martin MP, McVicar DW, Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu Rev Genomics Hum Genet. 2006;7:277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular-dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- Campbell KS, Cella M, Carretero M, Lopez-Botet M, Colonna M. Signaling through human killer cell activating receptors triggers tyrosine phosphorylation of an associated protein complex. Eur J Immunol. 1998;28:599–609. doi: 10.1002/(SICI)1521-4141(199802)28:02<599::AID-IMMU599>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci USA. 1993;90:12000–12004. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T, York D, Pedersen L. Particle mesh Ewald—An N. Log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- Dohring C, Samaridis J, Colonna M. Alternatively spliced forms of human killer inhibitory receptors. Immunogenetics. 1996;44:227–230. doi: 10.1007/BF02602590. [DOI] [PubMed] [Google Scholar]
- Dunning AM, Ellis PD, McBride S, Kirschenlohr HL, Healey CS, Kemp PR, et al. A transforming growth factorbeta1 signal peptide variant increases secretion in vitro and is associated with increased incidence of invasive breast cancer. Cancer Res. 2003;63:2610–2615. [PubMed] [Google Scholar]
- Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory receptor KIR2DLI-HLA-Cw4 complex. Nat Immunol. 2001;2:452–460. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
- Fiser A, Do RKG, Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodridge JP, Lathbury LJ, Steiner NK, Shulse CN, Pullikotil P, Seidah NG, et al. Three common alleles of KIR2DL4 (CD158d) encode constitutively expressed, inducible and secreted receptors in NK cells. Eur J Immunol. 2007;37:199–211. doi: 10.1002/eji.200636316. [DOI] [PubMed] [Google Scholar]
- Hanson RN, Lee CY, Friel CJ, Dilis R, Hughes A, DeSombre ER. Synthesis and evaluation of 17alpha-20E-21-(4-substituted phenyl)-19-norpregna-1,3,5(10),20-tetraene-3,17beta-diols as probes for the estrogen receptor alpha hormone binding domain. J Med Chem. 2003;46:2865–2876. doi: 10.1021/jm0205806. [DOI] [PubMed] [Google Scholar]
- Hou L, Steiner NK, Chen M, Belle I, Kubit AL, Ng J, et al. Limited allelic diversity of stimulatory two domain killer immunoglobulin-like receptors. Hum Immunol. 2008;69:174–178. doi: 10.1016/j.humimm.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Hsu KC, Liu XR, Selvakumar A, Mickelson E, O’Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. 2002;169:5118–5129. doi: 10.4049/jimmunol.169.9.5118. [DOI] [PubMed] [Google Scholar]
- Katz G, Markel G, Mizrahi S, Arnon TI, Mandelboim O. Recognition of HLA-Cw4 but not HLA-Cw6 by the NK cell receptor killer cell Ig-like receptor two-domain short tail number 4. J Immunol. 2001;166:7260–7267. doi: 10.4049/jimmunol.166.12.7260. [DOI] [PubMed] [Google Scholar]
- Katz G, Gazit R, Arnon TI, Gonen-Gross T, Tarcic G, Markel G, et al. MHC class I-independent recognition of NK-activating receptor KIR2DS4. J Immunol. 2004;173:1819–1825. doi: 10.4049/jimmunol.173.3.1819. [DOI] [PubMed] [Google Scholar]
- Khakoo SI, Carrington M. KIR and disease: a model system or system of models? Immunol Rev. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck—a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Martin MP, Nelson G, Lee JH, Pellett F, Gao X, Wade J, et al. Cutting edge: susceptibility to psoriatic arthritis: influence of activating killer Ig-like receptor genes in the absence of specific HLA-C alleles. J Immunol. 2002;169:2818–2822. doi: 10.4049/jimmunol.169.6.2818. [DOI] [PubMed] [Google Scholar]
- Maxwell LD, Wallace A, Middleton D, Curran MD. A common KIR2DS4 deletion variant in the human that predicts a soluble KIR molecule analogous to the KIR1D molecule observed in the rhesus monkey. Tissue Antigens. 2002;60:254–258. doi: 10.1034/j.1399-0039.2002.600307.x. [DOI] [PubMed] [Google Scholar]
- Middleton D, Gonzalez A, Gilmore PM. Studies on the expression of the deleted KIR2DS4*003 gene product and distribution of KIR2DS4 deleted and nondeleted versions in different populations. Hum Immunol. 2007a;68:128–134. doi: 10.1016/j.humimm.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Middleton D, Meenagh A, Gourraud PA. KIR haplotype content at the allele level in 77 Northern Irish families. Immunogenetics. 2007b;59:145–158. doi: 10.1007/s00251-006-0181-7. [DOI] [PubMed] [Google Scholar]
- Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, et al. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- Pando MJ, Gardiner CM, Gleimer M, McQueen KL, Parham P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol. 2003;171:6640–6649. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- Parham P. Immunogenetics of killer-cell immunoglobulin-like receptors. Tissue Antigens. 2003;62:194–200. doi: 10.1034/j.1399-0039.2003.00126.x. [DOI] [PubMed] [Google Scholar]
- Parham P. Immunogenetics of killer cell immunoglobulin-like receptors. Mol Immunol. 2005a;42:459–462. doi: 10.1016/j.molimm.2004.07.027. [DOI] [PubMed] [Google Scholar]
- Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005b;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- Robinson J, Waller MJ, Stoehr P, Marsh SG. IPD—the Immuno Polymorphism Database. Nucleic Acids Res. 2005;33:D523–D526. doi: 10.1093/nar/gki032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulquin X, Gastinel LN, Vivier E. Crystal structure of the human natural killer cell activating receptor KIR2DS2 (CD158j) J Exp Med. 2003;197:933–938. doi: 10.1084/jem.20021624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CA, Laugier-Anfossi F, Vely F, Saulquin X, Riedmuller J, Tisserant A, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci USA. 2005;102:13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBussche CJ, Dakshanamurthy S, Posch PE, Hurley CK. A single polymorphism disrupts the killer Ig-like receptor 2DL2/ 2DL3 D1 domain. J Immunol. 2006;177:5347–5357. doi: 10.4049/jimmunol.177.8.5347. [DOI] [PubMed] [Google Scholar]
- Vilches C, Gardiner CM, Parham P. Gene structure and promoter variation of expressed and nonexpressed variants of the KIR2DL5 gene. J Immunol. 2000a;165:6416–6421. doi: 10.4049/jimmunol.165.11.6416. [DOI] [PubMed] [Google Scholar]
- Vilches C, Pando MJ, Rajalingam R, Gardiner CM, Parham P. Discovery of two novel variants of KIR2DS5 reveals this gene to be a common component of human KIR ‘B’ haplotypes. Tissue Antigens. 2000b;56:453–456. doi: 10.1034/j.1399-0039.2000.560510.x. [DOI] [PubMed] [Google Scholar]
- Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161:571–577. [PubMed] [Google Scholar]
- Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]