Abstract
Background
Ribosomes, the organelles responsible for the translation of mRNA, are comprised of four rRNAs and ~ 80 ribosomal proteins (RPs). Although canonically assumed to be maintained in equivalent proportions, some RPs have been shown to possess differential expression across tissue types. Dysregulation of RP expression occurs in a variety of human diseases, notably in many cancers, and altered expression of some RPs correlates with different tumor phenotypes and patient survival. Little work has been done, however, to characterize overall patterns of RP transcript (RPT) expression in human cancers.
Methods
To investigate the impact of global RPT expression patterns on tumor phenotypes, we analyzed RPT expression of ~ 10,000 human tumors and over 700 normal tissues from The Cancer Genome Atlas (TCGA) using t-distributed stochastic neighbor embedding (t-SNE). Clusters of tumors identified by t-SNE were then analyzed with chi-squared and t-tests to compare phenotypic data, ANOVA to compare individual RPT expression, and Kaplan-Meier curves to assess survival differences.
Results
Normal tissues and cancers possess distinct and readily discernible RPT expression patterns that are independent of their absolute levels of expression. In tumors, RPT patterning is distinct from that of normal tissues, identifies heretofore unrecognized tumor subtypes, and in many cases correlates with molecular, pathological, and clinical features, including survival.
Conclusions
RPT expression patterns are both tissue-specific and tumor-specific. These could be used as a powerful and novel method of tumor classification, offering a potential clinical tool for prognosis and therapeutic stratification.
Electronic supplementary material
The online version of this article (10.1186/s12885-018-4178-z) contains supplementary material, which is available to authorized users.
Keywords: Diamond-Blackfan anemia, Shwachman-diamond syndrome, t-SNE, Ribosomopathy, 5q- syndrome
Background
Eukaryotic ribosomes are among the most highly evolutionarily conserved organelles, comprised of four ribosomal RNAs (rRNAs) and approximately 80 ribosomal proteins (RPs). Responsible for translating mRNA into proteins, ribosomes were long believed to be nonspecific “molecular machines” with unvarying structures and function in different biological contexts. Recent evidence has shown, however, that some individual RPs are expressed in tissue-specific patterns and can differentially contribute to ribosome composition, affect rRNA processing, and regulate translation [1]. Despite the complexity of RP assembly in ribosomes, early studies of ribosome function revealed that the catalytic activity responsible for peptide bond formation might depend only on the presence of rRNAs and a small number of core RPs [2]. This finding, in conjunction with the observation that some RPs are expressed in a tissue-specific manner, has led to speculation that one purpose for the evolutionary emergence of RPs may have been to confer translational specificity and adaptability [1, 3].
An increasing body of evidence continues to show that RPs do, in fact, have important roles in imbuing ribosomes with mRNA translational specificity. During embryonic development, RPs are expressed at different levels across tissue types, and loss of RPs due to mutation or targeted knockdown produces specific developmental abnormalities in plants, invertebrates, and vertebrates. The tissue-specific patterning that occurs as a consequence of individual RP loss suggests that some RPs serve to guide the translation of specific subsets of transcripts in order to influence cellular development. While the mechanism(s) by which RPs confer translation specificity are not entirely known, one may involve the alteration of ribosome affinity for transcripts with specific cis-regulatory elements, including internal ribosome entry sites (IRES) elements and upstream open reading frames (uORFs) [1].
RPs also participate in a variety of extra-ribosomal functions. In normal contexts, ribosome assembly from individual rRNAs and RPs is a tightly regulated process, with unassembled RPs undergoing rapid degradation. Disruption of ribosomal biogenesis by any number of extracellular or intracellular stimuli induces ribosomal stress, leading to an accumulation of unincorporated RPs. In some cases, these free RPs may participate in a variety of extra-ribosomal functions, including the regulation of cell cycle progression, immune signaling, and cellular development. Many free RPs bind to and inhibit MDM2, a potentially oncogenic E3 ubiquitin ligase that interacts with and promotes the degradation of the TP53 tumor suppressor. The resulting stabilization of TP53 triggers cellular senescence or apoptosis in response to the inciting ribosomal stress. Additional extra-ribosomal functions of RPs are numerous, and have been recently reviewed [4, 5].
Given their role in regulating gene translation, cellular differentiation, and organismal development, it is perhaps unsurprising that altered RP expression has been implicated in human pathology. Indeed, an entire class of diseases has been shown to be associated with haploinsufficient expression or mutation in individual RPs. These so-called “ribosomopathies,” including Diamond-Blackfan Anemia (DBA) and Shwachman-Diamond Syndrome (SDS), are characterized by early onset bone marrow failure, variable developmental abnormalities and a life-long cancer predisposition that commonly involves non-hematopoietic tissues [6, 7]. The loss of proper RP stoichiometry and ensuing ribosomal stress result in increased ribosome-free RPs, which bind to MDM2 and impair its ubiquitin-mediated degradation of TP53 [6, 8–10]. The resulting TP53 stability is believed to underlie the bone marrow failure that affects the erythroid or myeloid lineages in DBA and SDS, respectively. The developmental abnormalities of the ribosomopathies are variable and associate with specific RP loss or mutation. For example, RPL5 loss in DBA is specifically associated with cleft palate and other craniofacial abnormalities whereas RPL11 loss is associated with isolated thumb malformations [11].
Ribosomopathy-like properties have also been observed in various cancers. We have recently shown that RP transcripts (RPTs) were dysregulated in two murine models of hepatoblastoma and hepatocellular carcinoma (HCC) in a tumor-specific manner and in patterns unrelated to tumor growth rates [12]. These murine tumors also displayed abnormal rRNA processing and increased binding of free RPs to MDM2, reminiscent of the aforementioned inherited ribosomopathies.
Perturbations of several individual RPs have been found in numerous human cancers, including those of the breast, pancreas, bladder, brain and many other tissues [13–25]. Mutations and deletions of RP-encoding genes have also been found in endometrial cancer, colorectal cancer, glioma, and various hematopoietic malignancies [26–28]. Indeed, the Chr. 5q- abnormality associated with myelodysplastic syndrome and the accompanying haploinsufficiency of RPS14 is considered one of the prototype “acquired” ribosomopathies that are often classified together with DBA, SDS and other inherited ribosomopathies [6]. Although many free RPs can induce cellular senescence during ribosomal stress via the MDM2-TP53 pathway, not all RPs possess such tumor suppressor functions. RPS3A overexpression, for example, actually transforms NIH3T3 mouse fibroblasts and induces tumor formation in nude mice [29].
A recent attempt to summarize the heterogeneity of RPT expression in human cancers was limited to describing expression differences of single RPTs among cancer cohorts, without accounting for larger patterns of variation that might better distinguish tumors from one another [3]. RPT expression patterns were, however, examined in normal tissues using the dimensionality-reduction technique Principal Component Analysis (PCA) in the aforementioned study. These results provided hints of cell-specific patterning in the hematopoietic tissues examined, but not all cell types clustered into obviously distinct groups.
In the current work, we leverage a machine learning technique known as t-distributed stochastic neighbor embedding (t-SNE) to identify distinct patterns of global RPT expression across both normal human tissues and cancers. Like PCA, t-SNE is a dimensionality reduction technique used to visualize patterns in a data set [30]. With either technique, patterns shared between data points are represented with clustering. However, t-SNE differs from PCA in that it performs particularly well with highly dimensional data and is able to distinguish non-linear relationships and patterns. With this technique, we show that virtually all normal tissues and tumors can be reliably distinguished from one another based on their signature RPT expression patterns. Tumors differ from normal tissues, but retain sufficient remnants of normal tissue patterning to allow for their origin to be easily discerned. Finally, we show that a number of cancers possess subtypes of RPT expression patterns that correlate in readily understandable ways with molecular markers, various tumor phenotypes, and survival.
Methods
Accessing ribosomal protein transcript expression data
RNA-seq whole-transcriptome expression data for 9844 tumors and 716 normal tissues from The Cancer Genome Atlas (TCGA) was accessed using the University of California Santa Cruz (UCSC) Xenabrowser [31]. Only primary tumors were included for analysis, apart from the melanoma (SKCM) cohort, as the vast majority of tumors with sequencing data in this cohort were metastatic (78%). The total number of samples analyzed in each cohort can be found in Additional file 1: Table S1. For each of the 30 cancer cohorts, RNA-seq data was selected according to the label “gene expression RNAseq (polyA+ IlluminaHiSeq).” “IlluminaGA” RNA-seq expression data was used for the cohort Uterine Corpus Endometrial Carcinoma (UCEC), as this group of data had more samples than the “IlluminaHiSeq” group. For all cancer cohorts, expression data for 80 cytoplasmic RP genes were extracted and base-two exponentiated, as the raw RPKM (Reads Per Kilobase per Million mapped reads) expression data was stored log-transformed. The sum of total RPKM counts for all RP genes was calculated for each sample, and relative expression of each RP gene in a sample was calculated by dividing the RPKM gene expression by this summed expression.
Visualizing ribosomal protein transcript expression
Principal component analyses and t-SNE analyses of RPT relative expression in normal tissues and tumor samples were performed using TensorFlow r1.0 and Tensorboard [32]. t-SNE analyses were performed at a learning rate (epsilon) of 10 with 5000 iterations or until the visualization stabilized. t-SNE was initially performed in two dimensions for all analyses; data sets that could not be cleanly visualized with two dimensions, particularly those with a large number of samples, were visualized with three-dimensional t-SNE. Multiple analyses were performed with perplexity settings varying between 6 and 15 for all individual cohort analyses and 10–30 for all grouped cohort analyses, with final perplexity settings for each analysis chosen to maximize cluster distinctions. Clusters of at least 10 samples which distinctly separated visually from other samples were named and samples from these clusters were identified. 3D area maps of RPT relative expression were generated using Microsoft Excel, with each sample listed across the x-axis, RPTs listed across the z-axis, and relative expression of each RPT across the y-axis.
Comparing t-SNE clusters
Relative expression of RPTs were compared between t-SNE clusters with Analyses of Variance (ANOVA) using R version 3.3.2 [33]. ANOVA p-values were log10-transformed and used to generate Volcano plots comparing expression patterns between clusters. Volcano plots were graphed with Graphpad Prism 7 (GraphPad Software, Inc., La Jolla, CA).
Clinical and survival data for each TCGA cancer cohort were accessed again using the UCSC Xenabrowser under the data heading “Phenotypes.” For each cohort, survival curves of tumors in each t-SNE cluster were compared with Mantel-Haenszel (log-rank) and Gehan-Breslow-Wilcoxon methods using Graphpad Prism 7. Categorical clinical variables were compared between clusters of tumors with chi-squared tests. Continuous variables which were normally distributed were compared with t-tests assuming heteroskedasticity, and non-normally-distributed variables were compared with Wilcoxon sign-rank tests. All statistical tests were two-tailed.
Co-regulated RPTs
Certain groups of RPTs possessed recurring, highly-significant differences between multiple t-SNE clusters, including RPL3, RPL8, RPS4X, and RPL13. For each TCGA cohort with a cluster that possessed significantly different relative expression of one of these transcripts, relative expression of all other RPTs was compared between the identified cluster and other tumors in the same cohort. Co-regulated transcripts were defined as those with consistent differences in relative expression when comparing clusters of interest to other tumors from the same cohort (Table 1). For example, five TCGA cohorts had a t-SNE cluster with significant relative overexpression of RPL8 and RPL30. When comparing relative expression of other RPTs between these clusters and other tumors from the same cohorts, all five clusters with high RPL8 and RPL30 also displayed, on average, lower relative expression of RPL10 and higher relative expression of RPL7.
Table 1.
Recurring patterns of RPT expression in tumors across cancer cohorts
| Primary Difference | Co-regulated RPTs | Observations | ||
|---|---|---|---|---|
| Lower expression | Higher Expression | Cohort | Cluster | |
| Low RPL3 | RPL5, RPL10, RPL17, RPL22, RPL26, RPS23 | RPL28, RPL35, RPL36, RPL38, RPLP2 | THCAa | 1 |
| GBMLGG | 3, 4 | |||
| LIHC | 2, 3 | |||
| KIRC | 3 | |||
| THYM | 2 | |||
| PRAD | 2, 3 | |||
| PAAD | 1 | |||
| PCPG | 1 | |||
| DLBC | 2 | |||
| High RPL8 and RPL30 | RPL10, RPL13A, RPL19, RPL21, RPL34, RPL37A, RPL4, UBA52, RPL5, RPS11, RPS12, RPS13, RPS17, RPS27A, RPS8, RPS9 | RPL36A, RPL7, RPS20 | BRCA | 3 |
| LIHC | 3 | |||
| PRAD | 2 | |||
| LUNG | 1 | |||
| SKCM | 2 | |||
| HNSC | 2 | |||
| High RPS4X | RPL13, RPL23, RPL37, RPL8, RPLP1, RPS16, RPS2 | RPS3A, RPL17 | PRAD | 1 |
| KIRC | 1 | |||
| THCA | 1, 2 | |||
| STAD | 2 | |||
| LAML | 1 | |||
| CESC | 2 | |||
| High RPL13 | RPL10, RPL10A, RPL26, RPL3, RPL35A, RPL41, RPL5, RPL7, RPL7A, RPS12, RPS13, RPS14, RPS15A, RPS18, RPS23, RPS27, RPS3, RPS3A, RPS4X, RPS5 | RPL12, RPL13, RPL18, RPL24, RPL27A, RPL28, RPL32, RPL35, RPL36, RPL37, RPL38, RPL39, RPLP1, RPLP2, RPS15, RPS17, RPS19, RPS20, RPS24, FAU, RPS4Y1, RPS9 | PRAD | 3 |
| UCEC | 3 | |||
| KIRC | 3 | |||
Four recurring patterns of expression distinguishing tumor clusters from one another were observed in multiple clusters across cancer cohorts, as shown in Figs. 2 and 3a. For each pattern, the most significant and defining RPT expression difference is listed under “primary difference.” Other significant RPT expression differences associated with these patterns are listed under “co-regulated RPTs.” “Low” and “high” expression is defined relative to other tumors within a cancer cohort. Clusters with the described pattern are listed under “observations”
aWhile tumors in this cluster had relatively lower expression of RPL3, other RPTs were not co-regulated in the same manner
RP gene copy number variations (CNVs)
CNV data for TCGA tumors was accessed using the UCSC Xenabrowser under the data heading “copy number (gistic2_thresholded).” Positive values were classified as amplifications, and negative values were classified as deletions. The frequency of amplifications and deletions in RP genes were compared between clusters of tumors in each TCGA cohort using chi-squared tests and adjusted for 5% false discovery rate. Within each cancer cohort, clusters of tumors with significantly greater incidence of a CNV compared to other tumor clusters, and which possessed > 90% incidence of this copy number variation, were included in Table 2.
Table 2.
RP gene copy number alterations associated with t-SNE clusters
| Genes | Type | Location | Cohort | Cluster | Frequency |
|---|---|---|---|---|---|
| RPL19, RPL23 | Amplification | 17q12 | BRCA | 1 | 98.9%, 83.1% |
| RPL8, RPL30 | Amplification | 8q22 | BRCA | 3 | 100%, 97.5% |
| LIHC | 3 | 98.8%, 98.8% | |||
| PRAD | 2 | 95.1%, 97.5% | |||
| LUNG | 1 | 100%, 94.3% | |||
| SKCM | 2 | 94.2%, 88.5% | |||
| HNSC | 2 | 93.2%, 85.3% | |||
| RPS3 | Amplification | 11q13 | BRCA | 2 | 100% |
| LUNG | 5 | 90.5% | |||
| RPS16 | Amplification | 19q13 | LUNG | 6 | 100% |
| RPL13A, RPL18, RPL28, RPS5, RPS9, RPS11, RPS16, RPS19 | Deletion | 19q13 | GBMLGG | 5 | 98.8% - 99.4% |
| RPL11, RPL22, RPL5, RPS8 | Deletion | 1p | GBMLGG | 5 | 97.5% |
| RPS24 | Deletion | 10q22 | GBMLGG | 3 | 90.5% |
Some tumor clusters were significantly associated with greater incidence of copy number alterations than other tumors from the same cancer cohorts (α < 0.01); clusters with > 90% of tumors possessing a given copy number alteration are included in this table. Cancer cohorts with no tumor clusters strongly associating with a RP copy number alteration were excluded from this table
Classification models
Using RPT relative expression in tumors and normal tissues, classification models were created using both logistic regression and feed-forward, fully-connected artificial neural networks (ANNs) [34]. LR models were used for binary classifiers and developed with Stata SE 14 (StataCorp LP, College Station, TX) with c-statistics, sensitivity, and specificity reported in Additional file 1: Table S2. ANN models were generated for classifiers with multiple outcomes (e.g. tissue of origin models) and binary classifiers with a LR model that failed to converge.
ANN models were created and tested using TensorFlow with graphics processing unit (GPU) acceleration on a Titan X Pascal (NVIDIA, Inc. Santa Clara, CA). To reduce bias, samples were balanced for both training and testing by cancer cohort such that each training and test set had the same number of samples from each cohort. 60% of data sets were used for training and 10% for validation and hyper-parameter tuning. Hyper-parameter sweeps were used to test all possible combinations of the following: learning rate (0.001, 0.002, 0.005, 0.01), batch size (100, 500, none), dropout rate (0.9, 0.95, 1), hidden layer structure (both one and two layers with sizes varying between 0 and 200 in increments of 25), and L2 regularization rate (0.00001, 0.0001, 0.001). All ANNs utilized ReLU activation functions. Neural network training performance was monitored with Tensorboard and stopped once validation accuracy had plateaued. The remaining 30% of data comprised a separate test set, which was used to test the final model’s classification accuracy once the hyper-parameters were chosen and the model trained. Performance of ANN models on the separate test sets were reported as classification accuracies in Additional file 1: Table S2.
Results
t-SNE identifies tissue- and tumor-specific RPT expression
RNA-seq expression data for 9844 tumors (30 cancer types) and 716 matched normal tissues were obtained from The Cancer Genome Atlas (TCGA) [35]. Relative expression of RPTs was calculated for all samples and first analyzed using PCA. To a modest degree, normal tissue samples could be distinguished by their RPT expression patterns, though many tissue types demonstrated considerable overlap (Fig. 1a and Additional file 1: Figure S1A). Patterns of RPT expression in tumors were even more heterogeneous, and most cancer cohorts did not cluster discretely (Fig. 1b).
Fig. 1.
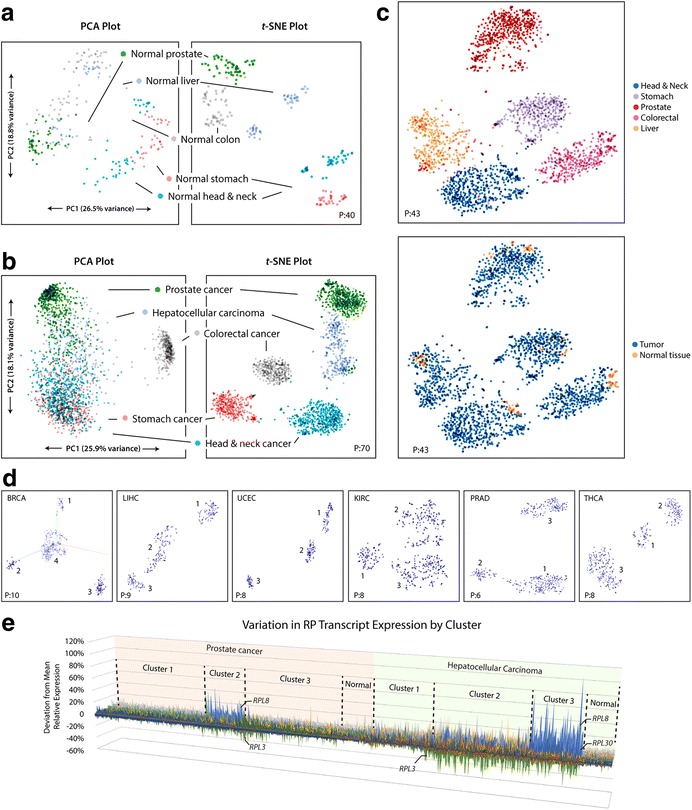
t-SNE better identifies clusters of RPT expression than PCA. a. Relative expression of RPTs in normal tissues from five cohorts was analyzed with PCA. In both methods, clustering occurs when samples possess similar underlying patterns of variation. t-SNE provides more distinct clusters that better associate with tissue of origin, indicating that normal tissues have distinct patterns of RPT expression. Axes are not labeled with t-SNE, as points are not mapped linearly and axes are not directly interpretable. b. Similar analyses in tumors. PCA clusters are poorly defined and do not correlate strongly with tumor type. t-SNE clusters are distinct and strongly associate with cancer type, indicating that tumors possess unique patterns of RPT expression based on their tissue of origin. c. Combined t-SNE analysis of RPT expression in normal tissue and tumor samples. Normal tissues and tumors cluster together but can be distinguished from one another, indicating that the latter retain a pattern of RPT expression resembling that of the normal tissue from which they originated. d. Many single cancer cohorts demonstrate sub-clustering by t-SNE. Clustering of six cohorts are provided as examples here. The number of clusters found in each cohort is listed in Additional file 1: Table S1. e. 3D area map of RPT relative expression in tumors from two cancer cohorts, sorted by cluster. The x-axis represents individual tumors, the z-axis represents individual RPTs, and the y-axis represents deviation from the mean relative expression. Cluster 2 of prostate cancer and Cluster 3 of HCC are both comprised of tumors with high relative expression of RPL8 and low RPL3. See Additional file 1: Figure S1, S2, and S5 for additional t-SNE plots of tumors and normal tissues. Perplexity settings for t-SNE analyses are designated in each plot by “P:”. For all analyses, learning rate (epsilon) = 10 and iterations = 5000
Samples were then analyzed with t-SNE, which more clearly identified clusters of variation due to its ability to identify non-linear relationships among RPTs (Fig. 1a and b and Additional file 1: Figure S1B). Clustering of normal tissue samples correlated nearly perfectly with tissue type. Tumors also demonstrated clustering that strongly associated with tissue type, with 20 cohorts segregating into largely distinct and non-overlapping groups. When both normal tissues and tumors were analyzed together with t-SNE, samples also generally grouped into large clusters according to tissue type. Normal tissues, however, localized into smaller sub-clusters distinct from tumors (Fig. 1c and Additional file 1: Figure S2). Thus, while samples nearly always possessed RPT expression patterns specific to their tissue type, normal tissues and tumors could be readily distinguished from one another.
Five cancer cohorts, including cholangiocarcinoma (CHOL), lung (LUNG), bladder (BLCA), cervical (CESC), and uterine carcinosarcoma (UCS), were comprised of tumors that lacked tissue-specific RPT expression profiles and did not group into distinct clusters. These tumors displayed significant overlap with each other as well as with tumors from the remaining five cohorts – liver HCC (LIHC), colorectal (COADREAD), mesothelioma (MESO), pancreatic (PAAD), and skin cutaneous melanoma (SKCM) – which otherwise clustered distinctly from one another (Additional file 1: Figure S3). Additionally, two clusters of tumors were found that did not associate with tissue of origin (Additional file 1: Figure S4). The first contained 143 tumors from 15 cohorts, 98% of which had amplification and relative up-regulation of RPL19, RPL23, and ERBB2 (Her2/Neu). The second contained 77 tumors from 12 cohorts with no clearly discernable or unifying RPT expression pattern.
t-SNE identifies sub-types of RPT expression within cancer types
Analyzed individually, 19 of 30 cancer types demonstrated sub-clustering of RPT expression with t-SNE (Fig. 1d, Additional file 1: Figure S5, and Additional file 1: Table S1). Graphing RPT relative expression by cluster using a 3D area map illustrated the different patterns of expression detected by t-SNE (Fig. 1e). In some cases, these clusters differed from one another in the expression pattern of numerous RPTs, as seen with Clusters 1 and 3 of prostate cancer. In other cases, expression patterns appeared to be dominated by the differential relative expression of one or two RPTs, as seen with prostate cancer Cluster 2 and HCC Cluster 3, both of which possess tumors that overexpress RPL8 and under-express RPL3 (Fig. 1e). While all clusters were distinct from normal tissues (Fig. 1c and Additional file 1: Figure S2), some were more similar to normal tissues than others, such as prostate cancer Cluster 1 and HCC Cluster 1 (Fig. 1e).
Classification models
While t-SNE analyses are useful for visualization and pattern discovery, they do not alone provide a direct means for classification of future samples. Thus, with the knowledge that RPTs have both tissue- and tumor-specific expression patterns, we constructed various tumor classifier models based on these patterns. The constructed models consisted of both artificial neural network (ANN) and logistic regression [35] classifiers, and are listed in Additional file 1: Table S2. An ANN model classified tumors by RPT content according to their tissue of origin on a separate test set with 93% accuracy. Similarly, a LR model distinguished tumors from normal tissues with > 98% accuracy. Other LR models could distinguish glioblastoma multiforme tumors from other brain cancers with 100% accuracy and were able to stratify both uterine and kidney clear cell tumors according to prognostic group with > 95% accuracy.
Characterizing tumor clusters identified by t-SNE
In order to quantify the differences in RPT expression that exist between clusters of tumors identified by t-SNE, RPT relative expression was compared among clusters of tumors with Analysis of Variance (ANOVA) and graphed with volcano plots (Figs. 2 and 3a). Small but highly significant differences in the expression of dozens of RPTs occurred in nearly every tumor cluster (P as low as 10− 220). As was the case with prostate cancer and HCC, these expression patterns were often dominated by particularly significant differences in expression of one or two RPTs, most commonly RPL3, RPS4X, RPL8, RPL30, and RPL13. Other tumor clusters, notably those involving the uterus, brain, and lung, possessed more complex differences involving larger numbers of RPTs (Figs. 2 and 3a).
Fig. 2.
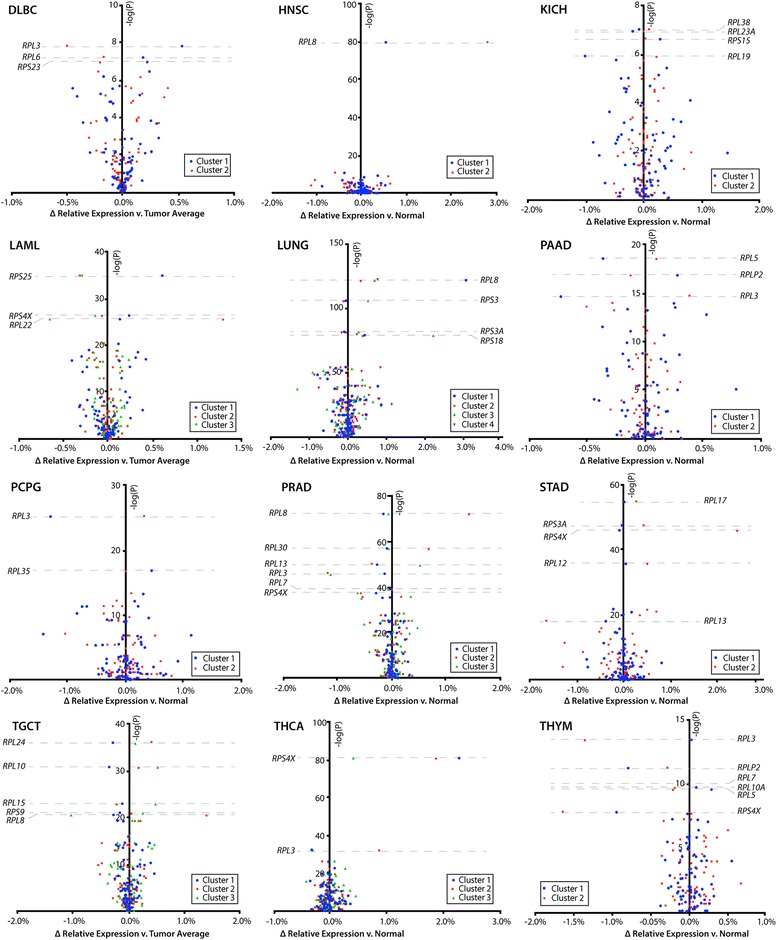
Volcano plots of relative RPT expression in tumor clusters in twelve cancer cohorts. Relative expression of RPTs was compared between tumor clusters in each included cancer cohort with ANOVA tests. The negative log of the ANOVA P-value for each RPT is displayed on the y-axis and the difference in relative expression across tumor clusters is displayed on the x-axis. RPTs near the top of the graphs are most significantly differentially expressed between tumor clusters. Note that nearly every RPT in virtually all cancer cohorts falls above –log(P) of 2, corresponding to P < 0.01 and indicating that tumor clusters have significantly distinct expression of virtually all RPTs. For each cohort, the number of samples in each cluster are shown under the label “n”. Additional volcano plots of seven other cancer cohorts are continued in Fig. 3
Fig. 3.
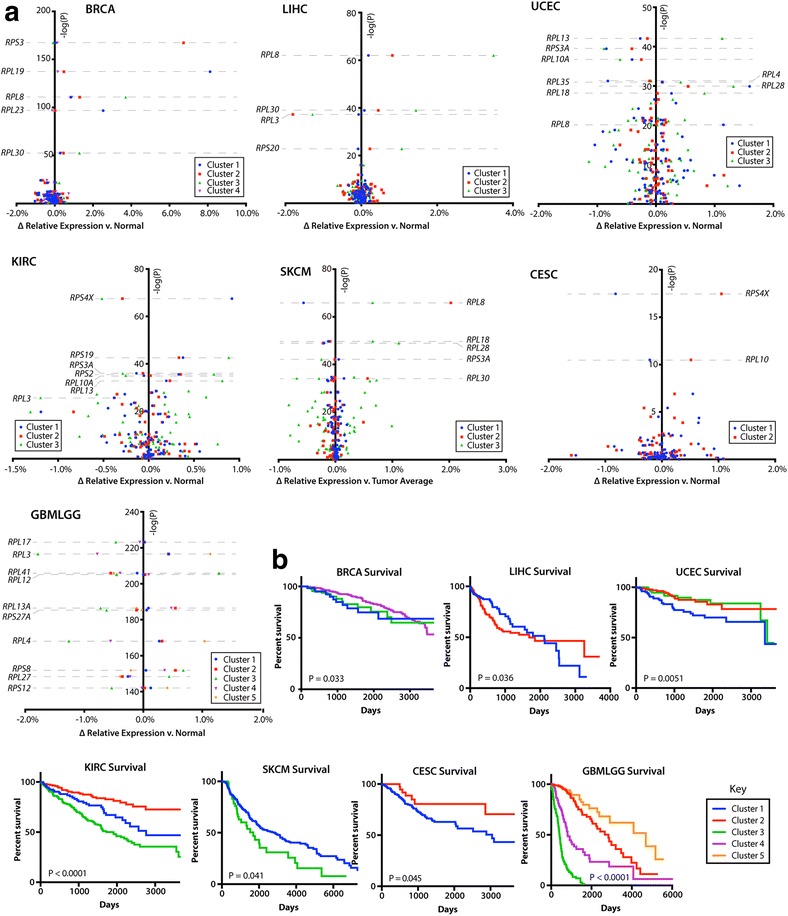
Volcano plots of relative RPT expression in tumor clusters associated with survival. a. Volcano plots comparing RPT relative expression between tumor clusters were generated, as in Fig. 2, for the remaining seven cancer cohorts which possessed tumor sub-clustering by t-SNE. Note that for the sake of clarity, clusters 5 and 6 are excluded from the LUNG cohort plot. These clusters correlated near perfectly with amplification and highly significant up-regulation of RPS3 and RPS16, respectively (Table 2). b. Patient survival by t-SNE cluster. Of the 19 cancer cohorts with sub-clustering of RPT expression patterns by t-SNE, seven possessed clusters that correlated with survival. Significance was determined with log-rank and Wilcoxon rank sum tests where appropriate, using all survival data available, including any data points beyond what are displayed in the survival curves
Several recurrent alterations in RPT expression were found among the 19 cancer cohorts with sub-clustering (Table 1). Nine of these cancer clusters, arising from thyroid, brain, liver, kidney clear cell, thymoma, prostate, pancreatic, pheochromocytoma and paraganglioma, and B-cell lymphoma, contained tumors with low relative expression of RPL3. These clusters also shared expression patterns with other RPTs, including the relative down-regulation of RPL5 and up-regulation of RPL36 and RPL38. Excluding thyroid cancers, all other tumor clusters with low RPL3 also shared 11 other similarly co-regulated RPTs. Additionally, six cancer cohorts – prostate, breast, liver, lung, melanoma, and head and neck – contained tumor clusters distinguished by overexpression of RPL8, RPL30 and RPS20, with shared expression patterns of 19 other RPTs. Relative up-regulation of RPS4X occurred in tumors from six cohorts, all of which showed similar co-expression patterns of nine other RPTs. Finally, tumor clusters overexpressing RPL13 were found in prostate, uterine and kidney clear cell carcinoma and shared similar patterns of expression of 42 other RPTs (Figs. 2 and 3a and Table 1).
In some cases, RP gene copy number variations (CNVs) were associated with and could explain the observed clustering (Table 2). Notably, the aforementioned RPL8/RPL30 overexpression pattern strongly correlated with co-amplification of a region on 8q22–24 containing genes encoding these two RPs as well as the Myc oncoprotein and PVT1, a long-non-coding RNA (lncRNA) with oncogenic properties [36, 37]. Similarly, an amplicon containing RPL19, RPL23, and ERBB2 (Her2/Neu) was amplified in 99% of the breast cancers in Cluster 1. Some tumor clusters associated with specific CNVs to a lesser degree. For example, 48% of tumors in kidney clear cell carcinoma Cluster 3 possessed deletions of RPL12, RPL35, and RPL7A on 9q33–34. Similarly, half of brain cancers in Cluster 1 possessed a 1p/19q13 co-deletion, compared to nearly 100% of tumors in Cluster 5 with this deletion (Table 2). Other tumor clusters in various cancer cohorts had differences in overall CNV frequencies. In testicular cancer, 39 RP genes were amplified at different frequencies among the three clusters. Endometrial cancer Cluster 1 and HCC Cluster 2 had more CNVs overall, but no RP gene was amplified or deleted with a frequency of greater than 65% in any given tumor cluster.
Many tumor clusters – each representing a distinct RPT expression pattern - significantly associated with various clinical parameters, molecular markers, and tumor phenotypes (Table 3). This was particularly true for brain, testicular, thyroid, lung, and endometrial cancers. Tumor clusters in HCC and head and neck cancers strongly correlated with etiologically-linked infections. For example, chronic hepatitis B infection was two-fold more common in HCC patients with Cluster 2 tumors compared to other HCC patients. Similarly, chronic HPV infection was 4.7-fold more frequent in head and neck cancer patients with Cluster 1 tumors compared to other patients in this cohort. Patient gender also associated with tumor clustering to varying but significant degrees in kidney clear cell carcinoma and AML. Notably, these clusters also associated with differential relative expression of the X-chromosome encoded RPS4X. Other clinical markers and tumor phenotypes significantly associated with tumor clustering can be found in Table 3.
Table 3.
Tumor phenotypes and clinical parameters associated with t-SNE clustering
| Feature | Cluster | Comparison | P-value |
|---|---|---|---|
| Breast cancer | |||
| Her2/Neu status | Cluster 1–88.2% | Other tumors – 7.7% | < 0.0001 |
| Hepatocellular carcinoma | |||
| Hepatitis B | Cluster 2–38.4% | Other tumors – 18.6% | < 0.0001 |
| Endometrial carcinoma | |||
| Type II (serous) type | Cluster 1–32.4% | Other tumors – 5.2% | < 0.0001 |
| Locoregional disease or recurrence | Cluster 1–8.6% | Other tumors – 2.2% | 0.004 |
| Lung cancer | |||
| Adenocarcinoma | Cluster 2–80.0% | Other tumors – 37.8% | < 0.0001 |
| Squamous cell carcinoma | Cluster 4–73.6% | Other tumors – 34.2% | < 0.0001 |
| Kidney clear cell carcinoma | |||
| Gender | Cluster 1–92.9% female | Other tumors – 29.0% female | |
| Longest dimension | Cluster 2–1.5 cm | Other tumors – 1.8 cm | < 0.0001 |
| Histologic grade 3 or 4 | Cluster 3–69.0% | Other tumors – 41.5% | < 0.0001 |
| Pathologic stage III or IV | Cluster 3–58.5% | Other tumors – 25.5% | < 0.0001 |
| Thyroid carcinoma | |||
| Gender | Cluster 3–35.4% male | Other tumors – 11.3% male | < 0.0001 |
| Tall cell subtype | Cluster 1–17.2% | Other tumors – 5.03% | 0.0001 |
| Follicular subtype | Cluster 3–27.7% | Other tumors – 10.5% | < 0.0001 |
| Acute myeloid leukemia | |||
| Gender | Cluster 1–100% female Cluster 3–81.5% male |
Cluster 2–53.6% female | < 0.0001 |
| Complex (> 3 distinct) cytogenetic abnormalities | Cluster 3–21.0% | Other tumors – 5.5% | 0.0025 |
| Testicular cancer | |||
| Seminoma subtype | Cluster 1–100% | Other tumors – 14.5% | < 0.0001 |
| Embryonal carcinoma subtype | Cluster 2–94.6% | Other tumors – 20.6% | < 0.0001 |
| Teratoma subtype | Cluster 3–84.0% | Other tumors – 14.7% | < 0.0001 |
| Head and neck cancer | |||
| HPV infection | Cluster 1–46.1% | Other tumors – 9.8% | 0.0001 |
| Skin cutaneous melanoma | |||
| Breslow depth | Cluster 1–4.8 cm | Other tumors – 7.6 cm | 0.0083 |
| Brain cancer | |||
| Glioblastoma | Cluster 3–100% | Other tumors – 0% | < 0.0001 |
| Astrocytoma low-grade glioma | Cluster 2–53.0% Cluster 4–62.7% |
Other tumors – 2.5% | < 0.0001 |
| Non-astrocytoma low-grade glioma | Cluster 1–86.7% Cluster 5–97.0% |
Other tumors – 29.1% | < 0.0001 |
Tumor phenotypes and clinical markers were compared between tumor clusters using chi-squared tests, with significance defined as α < 0.01. “Other tumors” are comprised of all tumors from the same cancer cohort not falling into the given cluster. Data were obtained using the UCSC Xenabrowser, under the data heading “Phenotypes.” Cancer cohorts with no tumor clusters significantly associating with any clinical parameter were excluded from this table
Tumor clusters were often predictive of survival, including some clusters that did not significantly associate with any other known tumor subtype (Fig. 3b). For example, Clusters 2 and 4 of the brain cancer cohort, which could not otherwise be distinguished by any known clinical parameter or tumor subtype, possessed vastly different survival patterns. Other cancer cohorts with significant survival differences among clusters included breast, liver, endometrial, kidney clear cell, melanoma, and cervical cancers.
Discussion
By investigating expression patterns of individual RPTs and utilizing more traditional and less powerful linear forms of dimensionality reduction such as PCA, previous studies have found modest evidence of tissue-specific patterning of RPT expression in some normal tissues and even less evidence in malignant tumors [3]. The failure to reproducibly identify recurrent and convincing patterning is presumably due to the complex regulation of RPT expression and the fact that many of the RPT relationships are non-linear. As shown here, however, the machine learning algorithm t-SNE provides a more elegant and robust dimensionality reduction that better highlights the distinct underlying patterns of RPT expression in both tumors and the normal tissues from which they originate.
Consistent with the more restricted and tentative conclusions of previous findings, our results using t-SNE clearly demonstrate that RPT expression patterns are not only tissue-specific but provide the ability to define tissue and tumor differences with a heretofore unachievable degree of resolution. The small cluster of 77 neoplasms that did not associate with their respective tissue clusters (Additional file 1: Figure S4) may represent either a subset of tumors that have lost control of their underlying tissue-specific expression patterns or that originated from a minority subpopulation of normal cells whose RPT expression is not representative of the remainder of the tissue.
In addition to their tissue-specific patterning, virtually all tumors showed perturbations of RPT expression that readily allowed them to be distinguished from the normal tissues from which they originated. For some cancers, the tumor-specific patterning of RPT expression was relatively homogeneous and could not otherwise be subcategorized. Most cohorts, however, were comprised of subgroups of tumors with distinct RPT expression patterns, all of which nonetheless remained distinguishable from normal tissue. The fact that many of these patterns correlated with molecular and clinical features implicates RPT expression patterns in tumor biology.
Aside from potentially altering translation, the notion that altered RP expression might influence the behaviors of both normal tissues and tumors is not new. In the ribosomopathies, the binding of any one of about a dozen RPs to MDM2 with subsequent stabilization of TP53 is thought to underlie the bone marrow failure that accompanies these disorders [6, 9, 10]. It has been proposed that subsequent circumvention of this TP53-mediated senescence by mutation and/or dysregulation of the p19ARF/MDM2/TP53 pathway is responsible for the propensity for eventual neoplastic progression [38]. In cancers, the binding of free RPs to MDM2 has been shown to mediate the response to ribosomal-stress-inducing chemotherapeutics such as actinomycin D and 5-fluorouracil [20, 39, 40].
Individual RPs have also been associated with specific tumor phenotypes. For example, RPL3 expression is a determinant of chemotherapy response in certain lung and colon cancers. RPL3 also associates with the high-risk neuroblastoma subtype and may have a role in the acquisition of lung cancer multidrug resistance [19–21]. Breast cancers with elevated expression of RPL19 are more sensitive to apoptosis-promoting drugs that induce endoplasmic reticulum stress [13]. RPS11 and RPS20 have been proposed as prognostic markers in glioblastoma [16] and the down-regulation of RPL10 correlates with altered treatment response to dimethylaminoparthenolide (DMAPT) in pancreatic cancer [22].
Our results also significantly extend the findings of previous studies by demonstrating that, in the vast majority of cancers, subsets of RPTs are expressed coordinately and have additional interpretive power when examined in the context of global RPT expression patterning. This suggests that further insights into the roles RPTs have in tumor development may be revealed by evaluating RPT relative expression. For example, the regulation of chemotherapy response by RPL3 described above may be found to occur in other cancer types once the expression of RPL3 relative to other RPTs has been accounted for. The apparent crucial role of RPT patterning in tumors may explain why a previous study found conflicting results when examining the expression of individual RPs in tumors [14].
Our results suggest a more ubiquitous role for RPL3 in regulating tumor phenotypes, beyond that already described in colorectal carcinoma, lung cancers, and neuroblastoma [19–21]. Of the recurring RPT expression patterns discovered by t-SNE, the pattern associated with RPL3 down-regulation occurred most frequently and involved tumors from nine cancer cohorts. Many clusters of tumors with down-regulated RPL3, including HCC, kidney clear cell cancer, and brain cancer, possessed inferior survival. The fact that relative down-regulation of RPL3 occurred in these tumor clusters with predictable expression of 11 other RPTs suggests that RPL3 may be acting in concert with these other identified RPs to exert its effects.
Other recurrent RPT expression patterns across cancer cohorts involved RPS4X, RPL13, RPL8 and RPL30 (Table 1). Altered RPS4X expression, found in six cancer cohorts, associated with unique expression of nine other RPTs, strongly suggesting an underlying coordinated expression, the mechanism of which remains to be identified. As with RPL3, deregulated RPS4X has been previously associated with various tumors and tumor phenotypes, including subgroups of colorectal carcinoma, a myelodysplasia risk signature and poor prognosis in bladder cancer [15, 18, 41]. Interestingly, some of our tumor clusters with altered RPS4X expression were comprised of a greater proportion of females than males (Table 1 and Table 3), perhaps reflecting RPS4X’s residence on the X chromosome. Although the cause of perturbed RPS4X expression in these tumor clusters is unknown, altered methylation patterns on chromosome X have been described in different subsets of cancers [42, 43] and could be responsible for the expression patterns detected by t-SNE.
Unlike RPL3 and RPS4X, RPL13’s role in tumor development is less clear. RPL13 activation has been described in a subset of gastrointestinal malignancies and correlated with greater proliferative capacity and attenuated chemoresistance [44], but further evidence for a role of RPL13 in tumor development is lacking. Furthermore, clinical correlations of the prostate, uterine and kidney cancer t-SNE clusters described here with relative overexpression of RPL13 were inconsistent. Uterine cancers with high relative RPL13 expression tended to correlate with favorable survival, whereas prostate cancers with high RPL13 showed no differences in prognosis or clinical features. In contrast, kidney clear cell carcinomas with high RPL13 expression tended to be of higher pathologic grade and were associated with significantly poorer survival (Tables 1 and 3, and Fig. 3b). The fact that these clusters shared similar patterning of 42 other RPTs suggests that the inciting factors responsible for higher RPL13 expression are not only shared by these tumors but coordinately regulate a common subset of RPTs, with different biological outcomes likely reflecting other tissue-specific factors.
In some cases, RPT expression patterns could be accounted for in part by CNVs, as exemplified by the recurrent RPL8 and RPL30 overexpression pattern (Tables 1 and 2). Virtually all tumors with this expression pattern possessed co-amplification of a region on 8q22–24 that includes RPL8, RPL30, and the oncogenes MYC and PVT1. Amplification of this region has been previously described in breast cancers and correlates with chemoresistance and metastasis [36, 37, 45–47]. Our results indicate that this amplification and the ensuing overexpression of RPL8 and RPL30 also occurs in subsets of melanoma, liver, prostate, lung, and head and neck cancers. CNVs of RPL19 and RPL23 in breast cancer (Table 2) likely occur due to their co-amplification with ERBB2 on 17q12. Overexpression of RPL19 has previously been described in a subset of breast cancers [13]. The small cluster of 144 tumors that did not group according to tissue of origin (Additional file 1: Figure S4), comprised of tumors from 15 cohorts, also shared amplification of this region on 17q12, indicating that this CNV is not restricted to breast cancers and ultimately affects global RPT expression patterning. Amplification of a region on 11q13 that contains RPS3, occurring in a cluster of breast cancers and HCCs, has been previously described in both cancers and is thought to confer unfavorable prognosis due to amplification of the adjacent oncogene EMS1 [48, 49]. The co-deletion of 19q13 along with 1p, which together includes 12 RP genes, has been described in low-grade gliomas and confers a favorable prognosis [50, 51].
The co-overexpression RPS25 and RPS4X detected in one cluster of AML (Fig. 2) has been previously identified as contributing to the poor risk signature in myelodysplastic syndrome [41]. This also associated with significant differential expression of 37 RPTs, which is consistent with our finding that RPS25 and RPS4X overexpression occur within the context of a larger and coordinated pattern of RPT expression. The RPS25 and RPS4X overexpressing AML cases likely possess a similar molecular alteration to those with the poor risk signature in MDS.
Collectively, our findings provide strong evidence to support the notion that RPT regulation by both tumors and normal tissues is complex, ordered, and highly coordinated. Although the means by which altered RPT patterns influence the pathogenesis and/or behavior of tumors remain incompletely understood, several non-mutually exclusive mechanisms can be envisioned. First, changes in RP levels may influence overall ribosome composition, thereby affecting their affinity for certain classes of transcripts and/or the efficiency with which they are translated. One such class of transcripts may be those with IRES elements, cis-regulatory sequences found in the 5′-untranslated regions of more than 10% of cellular mRNAs. IRES elements are found with particularly high frequency in transcripts encoding proteins involved in cell cycle control and various types of stress responses. Efficient translation of these IRES-containing transcripts has been shown to depend on specific RPs, notably RPS25, RPS19 and RPL11 [52–54]. Changes in ribosome affinity for IRES elements have been shown to reduce translation of tumor suppressors such as p27 and TP53 and to promote cancer development [55].
RPs may also influence cancer development via extra-ribosomal pathways. In addition to their stabilization of TP53 mediated by binding to and inactivating MDM2, specific RPs have been shown to inactivate Myc; to inhibit the Myc target Lin28B; to activate NF-κB, cyclins, and cyclin-dependent kinases and to regulate a variety of other tumorigenic functions and immunogenic pathways [4, 5].
In addition to providing evidence that tumors may use RPs to direct tumor phenotypes, our findings have allowed us to leverage the tissue- and tumor-specificity of RPT expression to generate highly sensitive and specific models that allow for precise tumor identification and sub-classification (Additional file 1: Table S2). Clinically, these might be useful for determining the tissue of origin of undifferentiated tumors and for predicting long-term behaviors in otherwise homogeneous cancers such as kidney clear cell carcinoma and those of the central nervous system (Fig. 3b). With more samples and further refinement to ANN structures, future iterations of these models will likely have even greater discriminatory power.
A limitation of using data from TCGA is the fact that transcript expression does not always correlate with protein expression, particularly in cancers [56–58]. Thus, it is difficult to predict how the different tissue-specific RPT expression patterns we identified correlate with actual protein expression in these cancers and/or with the numerous post-translational modifications that can alter RP behaviors [59, 60]. As this is a cross-sectional study, we also recognize that causality cannot be inferred, and it remains unknown whether altered RPT expression is an early or late event in tumorigenesis despite its predictive value. Furthermore, while RPT expression patterns appear to have significant predictive value in the large dataset we have analyzed, further cross-validation with additional transcriptional data in both primary tumors and metastatic lesions will be important in confirming potential clinical utility. Finally, additional molecular analyses of the identified t-SNE clusters with whole-transcriptome sequencing data, pathway analysis, whole-genome DNA mutation data, and DNA methylation patterning may offer additional insights into the biological mechanisms that link altered RPT expression with tumor phenotypes.
Conclusions
In summary, machine learning-based approaches have allowed us to show unequivocally that RPTs are expressed in distinct patterns across tissue types. This tissue-specificity persists in tumors, yet normal tissues and tumors can be readily distinguished from one another with high degrees of accuracy and confidence. Many cancers can be further sub-categorized into heretofore unrecognized, yet clinically important, subtypes based only upon RPT expression patterns. Several patterns of RPT expression recur across cancer types, suggesting common underlying modes of transcriptional regulation. Our results indicate that the expression of RPTs in tumors is biologically coordinated, clinically meaningful, and can be leveraged to create potential clinical tools for tumor classification and therapeutic stratification.
Additional file
Supplementary Information. Contains Figure S1-S5, Table S1-S2, and their respective legends. (DOCX 1621 kb)
Acknowledgements
We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan X Pascal GPU used for this research.
Funding
This work was supported by NIH grant RO1 CA174713 and by The Hyundai Hope on Wheels Scholar Grant to EVP. JMD was supported by the Clinical Scientist Training Program at the University of Pittsburgh School of Medicine. None of the funding bodies had a role in the interpretation of data or in the writing of the manuscript.
Availability of data and materials
This work is in part based on data generated from TCGA Research Network (http://cancergenome.nih.gov) [35]. Clinical annotation files and gene expression data were downloaded from UCSC Xenabrowser (https://xenabrowser.net) [31]. Source code for python scripts using Tensorflow to generate t-SNE plots are available from https://zenodo.org/record/1037563 (DOI: 10.5281/zenodo.1037563).
Abbreviations
- ANOVA
Analysis of variance
- CNV
Copy number variation
- DBA
Diamond-blackfan anemia
- IRES
Internal ribosome entry site
- LR
Logistic regression
- MDS
Myelodysplastic syndrome
- RP
Ribosomal protein
- RPKM
Reads-per-kilobase-million
- RPT
Ribosomal protein transcript
- TCGA
The cancer genome atlas
- t-SNE
t-distributed stochastic neighbor embedding
- uORF
Upstream open reading frame
Authors’ contributions
JMD conceived the study, supervised research, developed software, and performed analyses. APD performed analyses and interpreted data. EVP interpreted data. All authors wrote, read, and approved the final manuscript.
Ethics approval and consent to participate
No ethics approval was required for this work. All utilized public omics data sets were generated by others who obtained ethical approval.
Consent for publication
Not applicable.
Competing interests
Authors JMD and EVP are currently applying for a patent relating to the content of this manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12885-018-4178-z) contains supplementary material, which is available to authorized users.
References
- 1.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noller HF, Hoffarth V, Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 3.Guimaraes JC, Zavolan M. Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 2016;17(1):236. doi: 10.1186/s13059-016-1104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou X, Liao WJ, Liao JM, Liao P, Lu H. Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol. 2015;7:92–104. doi: 10.1093/jmcb/mjv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruggero D, Shimamura A. Marrow failure: a window into ribosome biology. Blood. 2014;124:2784–2792. doi: 10.1182/blood-2014-04-526301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yelick PC, Trainor PA. Ribosomopathies: global process, tissue specific defects. Rare Dis. 2015;3:e1025185. doi: 10.1080/21675511.2015.1025185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russo A, Russo G. Ribosomal proteins control or bypass p53 during nucleolar stress. Int J Mol Sci. 2017;18(1):140. doi: 10.3390/ijms18010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shenoy N, Kessel R, Bhagat TD, Bhattacharyya S, Yu Y, McMahon C, Verma A. Alterations in the ribosomal machinery in cancer and hematologic disorders. J Hematol Oncol. 2012;5:32. doi: 10.1186/1756-8722-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boultwood J, Pellagatti A, Wainscoat JS. Haploinsufficiency of ribosomal proteins and p53 activation in anemia: diamond-Blackfan anemia and the 5q- syndrome. Adv Biol Regul. 2012;52:196–203. doi: 10.1016/j.advenzreg.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Gazda HT, Sheen MR, Vlachos A, Choesmel V, O'Donohue MF, Schneider H, Darras N, Hasman C, Sieff CA, Newburger PE, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulkarni S, Dolezal JM, Wang H, Jackson L, Lu J, Frodey BP, Dosunmu-Ogunbi A, Li Y, Fromherz M, Kang A, et al. Ribosomopathy-like properties of murine and human cancers. PLoS One. 2017;12:e0182705. doi: 10.1371/journal.pone.0182705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong M, Kim H, Kim I. Ribosomal protein L19 overexpression activates the unfolded protein response and sensitizes MCF7 breast cancer cells to endoplasmic reticulum stress-induced cell death. Biochem Biophys Res Commun. 2014;450:673–678. doi: 10.1016/j.bbrc.2014.06.036. [DOI] [PubMed] [Google Scholar]
- 14.Lai MD, Xu J. Ribosomal proteins and colorectal cancer. Curr Genomics. 2007;8:43–49. doi: 10.2174/138920207780076938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung Y, Lee S, Choi HS, Kim SN, Lee E, Shin Y, Seo J, Kim B, Jung Y, Kim WK, et al. Clinical validation of colorectal cancer biomarkers identified from bioinformatics analysis of public expression data. Clin Cancer Res. 2011;17:700–709. doi: 10.1158/1078-0432.CCR-10-1300. [DOI] [PubMed] [Google Scholar]
- 16.Yong WH, Shabihkhani M, Telesca D, Yang S, Tso JL, Menjivar JC, Wei B, Lucey GM, Mareninov S, Chen Z, et al. Ribosomal proteins RPS11 and RPS20, two stress-response markers of glioblastoma stem cells, are novel predictors of poor prognosis in glioblastoma patients. PLoS One. 2015;10:e0141334. doi: 10.1371/journal.pone.0141334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Artero-Castro A, Castellvi J, Garcia A, Hernandez J, Ramon y Cajal S, Lleonart ME. Expression of the ribosomal proteins Rplp0, Rplp1, and Rplp2 in gynecologic tumors. Hum Pathol. 2011;42:194–203. doi: 10.1016/j.humpath.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 18.Paquet ER, Hovington H, Brisson H, Lacombe C, Larue H, Tetu B, Lacombe L, Fradet Y, Lebel M. Low level of the X-linked ribosomal protein S4 in human urothelial carcinomas is associated with a poor prognosis. Biomark Med. 2015;9:187–197. doi: 10.2217/bmm.14.115. [DOI] [PubMed] [Google Scholar]
- 19.Russo A, Saide A, Smaldone S, Faraonio R, Russo G. Role of uL3 in multidrug resistance in p53-mutated lung cancer cells. Int J Mol Sci. 2017;18(3). 10.3390/ijms18030547. [DOI] [PMC free article] [PubMed]
- 20.Russo A, Saide A, Cagliani R, Cantile M, Botti G, Russo G. rpL3 promotes the apoptosis of p53 mutated lung cancer cells by down-regulating CBS and NFκB upon 5-FU treatment. Sci Rep. 2016;6:38369. doi: 10.1038/srep38369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan FH, Pandian V, Ramraj S, Natarajan M, Aravindan S, Herman TS, Aravindan N. Acquired genetic alterations in tumor cells dictate the development of high-risk neuroblastoma and clinical outcomes. BMC Cancer. 2015;15:514. doi: 10.1186/s12885-015-1463-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi C, Wang Y, Guo Y, Chen Y, Liu N. Cooperative down-regulation of ribosomal protein L10 and NF-kappaB signaling pathway is responsible for the anti-proliferative effects by DMAPT in pancreatic cancer cells. Oncotarget. 2017;8:35009–35018. doi: 10.18632/oncotarget.16557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan H, Li J, Jia Y, Wu J, Yuan L, Li M, Wei J, Xu B. Silencing of ribosomal protein L34 (RPL34) inhibits the proliferation and invasion of esophageal cancer cells. Oncol Res. 2017;25(7):1061–1068. doi: 10.3727/096504016X14830466773541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kardos GR, Dai MS, Robertson GP. Growth inhibitory effects of large subunit ribosomal proteins in melanoma. Pigment Cell Melanoma Res. 2014;27:801–812. doi: 10.1111/pcmr.12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sim EU, Chan SL, Ng KL, Lee CW, Narayanan K. Human ribosomal proteins RPeL27, RPeL43, and RPeL41 are upregulated in nasopharyngeal carcinoma cell lines. Dis Markers. 2016;2016:5179594. doi: 10.1155/2016/5179594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ajore R, Raiser D, McConkey M, Joud M, Boidol B, Mar B, Saksena G, Weinstock DM, Armstrong S, Ellis SR, et al. Deletion of ribosomal protein genes is a common vulnerability in human cancer, especially in concert with TP53 mutations. EMBO Mol Med. 2017;9:498–507. doi: 10.15252/emmm.201606660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goudarzi KM, Lindstrom MS. Role of ribosomal protein mutations in tumor development (review) Int J Oncol. 2016;48:1313–1324. doi: 10.3892/ijo.2016.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fancello L, Kampen KR, Hofman IJ, Verbeeck J, De Keersmaecker K. The ribosomal protein gene RPL5 is a haploinsufficient tumor suppressor in multiple cancer types. Oncotarget. 2017;8:14462–14478. doi: 10.18632/oncotarget.14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naora H, Takai I, Adachi M, Naora H. Altered cellular responses by varying expression of a ribosomal protein gene: sequential coordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J Cell Biol. 1998;141:741–753. doi: 10.1083/jcb.141.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Maaten LJPH, G. E. Visualizing high-dimensional data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 31.University of California Santa Cruz. Xenabrowser. https://xenabrowser.net/. Accessed 31 July 2017.
- 32.TensorFlow. https://www.tensorflow.org. Accessed 31 July 2017.
- 33.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing 2008. http://www.R-project.org.
- 34.Dreiseitl S, Ohno-Machado L. Logistic regression and artificial neural network classification models: a methodology review. J Biomed Inform. 2002;35:352–359. doi: 10.1016/S1532-0464(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarver AL, Murray CD, Temiz NA, Tseng YY, Bagchi A. MYC and PVT1 synergize to regulate RSPO1 levels in breast cancer. Cell Cycle. 2016;15:881–885. doi: 10.1080/15384101.2016.1149660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tseng YY, Bagchi A. The PVT1-MYC duet in cancer. Mol Cell Oncol. 2015;2:e974467. doi: 10.4161/23723556.2014.974467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Keersmaecker K. Ribosomopathies and the paradox of cellular hypo- to hyperproliferation. Blood. 2015;125:1377–1382. doi: 10.1182/blood-2014-10-569616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esposito D, Crescenzi E, Sagar V, Loreni F, Russo A, Russo G. Human rpL3 plays a crucial role in cell response to nucleolar stress induced by 5-FU and L-OHP. Oncotarget. 2014;5:11737–11751. doi: 10.18632/oncotarget.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun XX, Dai MS, Lu H. 5-fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J Biol Chem. 2007;282:8052–8059. doi: 10.1074/jbc.M610621200. [DOI] [PubMed] [Google Scholar]
- 41.Sridhar K, Ross DT, Tibshirani R, Butte AJ, Greenberg PL. Relationship of differential gene expression profiles in CD34(+) myelodysplastic syndrome marrow cells to disease subtype and progression. Blood. 2009;114:4847–4858. doi: 10.1182/blood-2009-08-236422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaligné R, Popova T, Mendoza-Parra MA, Saleem MAM, Gentien D, Ban K, Piolot T, Leroy O, Mariani O, Gronemeyer H, et al. The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer. Genome Res. 2015;25:488–503. doi: 10.1101/gr.185926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spatz A, Borg C, Feunteun J. X-chromosome genetics and human cancer. Nat Rev Cancer. 2004;4:617–629. doi: 10.1038/nrc1413. [DOI] [PubMed] [Google Scholar]
- 44.Kobayashi T, Sasaki Y, Oshima Y, Yamamoto H, Mita H, Suzuki H, Toyota M, Tokino T, Itoh F, Imai K, Shinomura Y. Activation of the ribosomal protein L13 gene in human gastrointestinal cancer. Int J Mol Med. 2006;18:161–170. [PubMed] [Google Scholar]
- 45.Hu G, Chong RA, Yang Q, Wei Y, Blanco MA, Li F, Reiss M, Au JS, Haffty BG, Kang Y. MTDH activation by 8q22 genomic gain promotes Chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parris TZ, Kovacs A, Hajizadeh S, Nemes S, Semaan M, Levin M, Karlsson P, Helou K. Frequent MYC coamplification and DNA hypomethylation of multiple genes on 8q in 8p11-p12-amplified breast carcinomas. Oncogene. 2014;3:e95. doi: 10.1038/oncsis.2014.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taghavi A, Akbari ME, Hashemi-Bahremani M, Nafissi N, Khalilnezhad A, Poorhosseini SM, Hashemi-Gorji F, Yassaee VR. Gene expression profiling of the 8q22-24 position in human breast cancer: TSPYL5, MTDH, ATAD2 and CCNE2 genes are implicated in oncogenesis, while WISP1 and EXT1 genes may predict a risk of metastasis. Oncol Lett. 2016;12:3845–3855. doi: 10.3892/ol.2016.5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ormandy CJ, Musgrove EA, Hui R, Daly RJ, Sutherland RL. Cyclin D1, EMS1 and 11q13 amplification in breast cancer. Breast Cancer Res Treat. 2003;78:323–335. doi: 10.1023/A:1023033708204. [DOI] [PubMed] [Google Scholar]
- 49.Yuan BZ, Zhou X, Zimonjic DB, Durkin ME, Popescu NC. Amplification and overexpression of the EMS 1 oncogene, a possible prognostic marker, in human hepatocellular carcinoma. J Mol Diagn. 2003;5:48–53. doi: 10.1016/S1525-1578(10)60451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barbashina V, Salazar P, Holland EC, Rosenblum MK, Ladanyi M. Allelic losses at 1p36 and 19q13 in gliomas: correlation with histologic classification, definition of a 150-kb minimal deleted region on 1p36, and evaluation of CAMTA1 as a candidate tumor suppressor gene. Clin Cancer Res. 2005;11:1119–1128. [PubMed] [Google Scholar]
- 51.Vogazianou AP, Chan R, Bäcklund LM, Pearson DM, Liu L, Langford CF, Gregory SG, Collins VP, Ichimura K. Distinct patterns of 1p and 19q alterations identify subtypes of human gliomas that have different prognoses() Neuro-Oncology. 2010;12:664–678. doi: 10.1093/neuonc/nop075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horos R, Ijspeert H, Pospisilova D, Sendtner R, Andrieu-Soler C, Taskesen E, Nieradka A, Cmejla R, Sendtner M, Touw IP, von Lindern M. Ribosomal deficiencies in diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood. 2012;119:262–272. doi: 10.1182/blood-2011-06-358200. [DOI] [PubMed] [Google Scholar]
- 53.Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23:2753–2764. doi: 10.1101/gad.1832209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muhs M, Yamamoto H, Ismer J, Takaku H, Nashimoto M, Uchiumi T, Nakashima N, Mielke T, Hildebrand PW, Nierhaus KH, Spahn CM. Structural basis for the binding of IRES RNAs to the head of the ribosomal 40S subunit. Nucleic Acids Res. 2011;39:5264–5275. doi: 10.1093/nar/gkr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bellodi C, Krasnykh O, Haynes N, Theodoropoulou M, Peng G, Montanaro L, Ruggero D. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 2010;70:6026–6035. doi: 10.1158/0008-5472.CAN-09-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen G, Gharib TG, Huang CC, Taylor JM, Misek DE, Kardia SL, Giordano TJ, Iannettoni MD, Orringer MB, Hanash SM, Beer DG. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002;1:304–313. doi: 10.1074/mcp.M200008-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Koussounadis A, Langdon SP, Um IH, Harrison DJ, Smith VA. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci Rep. 2015;5:10775. doi: 10.1038/srep10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, et al. Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol Cell Proteomics. 2004;3:960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- 59.Sauert M, Temmel H, Moll I. Heterogeneity of the translational machinery: variations on a common theme. vol. 114. France: Biochimie; 2015. p. 39–47. [DOI] [PMC free article] [PubMed]
- 60.Simsek D, Barna M. An emerging role for the ribosome as a nexus for post-translational modifications. Curr Opin Cell Biol. 2017;45:92–101. doi: 10.1016/j.ceb.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information. Contains Figure S1-S5, Table S1-S2, and their respective legends. (DOCX 1621 kb)
Data Availability Statement
This work is in part based on data generated from TCGA Research Network (http://cancergenome.nih.gov) [35]. Clinical annotation files and gene expression data were downloaded from UCSC Xenabrowser (https://xenabrowser.net) [31]. Source code for python scripts using Tensorflow to generate t-SNE plots are available from https://zenodo.org/record/1037563 (DOI: 10.5281/zenodo.1037563).


