Abstract
Cellular senescence has been viewed as a tumor suppression mechanism and also as a contributor to individual aging. Widespread shortening of 3′ untranslated regions (3′ UTRs) in messenger RNAs (mRNAs) by alternative polyadenylation (APA) has recently been discovered in cancer cells. However, the role of APA in the process of cellular senescence remains elusive. Here, we found that hundreds of genes in senescent cells tended to use distal poly(A) (pA) sites, leading to a global lengthening of 3′ UTRs and reduced gene expression. Genes that harbor longer 3′ UTRs in senescent cells were enriched in senescence-related pathways. Rras2, a member of the Ras superfamily that participates in multiple signal transduction pathways, preferred longer 3′ UTR usage and exhibited decreased expression in senescent cells. Depletion of Rras2 promoted senescence, while rescue of Rras2 reversed senescence-associated phenotypes. Mechanistically, splicing factor TRA2B bound to a core “AGAA” motif located in the alternative 3′ UTR of Rras2, thereby reducing the RRAS2 protein level and causing senescence. Both proximal and distal poly(A) signals showed strong sequence conservation, highlighting the vital role of APA regulation during evolution. Our results revealed APA as a novel mechanism in regulating cellular senescence.
Cellular senescence was originally described as a process that limits the proliferation of cultured human cells. After extensive proliferation, senescence occurs because of telomere shortening and loss in the absence of endogenous telomerase activity (Hayflick and Moorhead 1961; Olovnikov 1996). In addition to telomere erosion, many stimuli and stresses can cause cellular senescence, including DNA double-strand breaks, strong mitogenic signals, oxidative stress, and ectopic expression of cyclin-dependent kinase inhibitors (CDKIs) (Xue et al. 2004; Rodier and Campisi 2011; Li et al. 2013). Numerous morphological and molecular markers of senescent cells have been identified in recent decades, which include a flattened and enlarged cell morphology, increased senescence-associated β-galactosidase (SA-β-gal) activity, reduced proliferation rate, and expression of tumor suppressors, cell cycle inhibitors, and DNA damage markers (Dimri et al. 1995; Busuttil et al. 2003; Lopez-Otin et al. 2013; Munoz-Espin and Serrano 2014). Cellular senescence is viewed as an important mechanism for preventing cancer (Campisi et al. 2001). It is also involved in normal embryonic development and tissue damage (Munoz-Espin et al. 2013; Storer et al. 2013; Munoz-Espin and Serrano 2014). Removing senescent cells expands the healthy lifespan of mice (Baker et al. 2016). These results demonstrate the significance of cellular senescence.
A number of studies have shown that dramatic changes in the transcriptome and/or proteome accompany the phenotypic alterations of senescent cells (Kim et al. 2013b; Mazin et al. 2013; Waldera-Lupa et al. 2014; Wei et al. 2015) and that the development of senescence-associated phenotypes can be regulated by stage-specific gene expression modules (Kim et al. 2013b). Therefore, understanding the regulation of gene expression and the corresponding regulatory networks is crucial to dissecting the mechanism of cellular senescence. Alternative polyadenylation (APA) is recognized as a crucial contributor to the regulation of mammalian gene expression (Di Giammartino et al. 2011; Elkon et al. 2013; Chen et al. 2017; Tian and Manley 2017). Cleavage and polyadenylation of nascent RNA is essential for maturation of the vast majority of eukaryotic mRNAs and determines the length of 3′ UTRs (Sachs 1990). The process requires several cis-acting RNA elements and dozens of trans-factors (Millevoi and Vagner 2010). The key cis-element is a six-nucleotide (nt) motif (termed the poly[A] signal or PAS), the canonical form of which is “AAUAAA.” The PAS determines recognition and cleavage by the 3′ end-processing machinery (Proudfoot and Brownlee 1976). The mammalian 3′ end-processing machinery contains several subcomplexes as well as additional accessory factors which mediate the precise processing of mRNA precursors (pre-mRNAs) together (Di Giammartino et al. 2011; Elkon et al. 2013).
With the increasing application of high-throughput sequencing technologies, genome-wide studies indicate that most eukaryotic mRNA genes have multiple polyadenylation (pA) sites (Tian et al. 2005; Wang et al. 2008; Zheng and Tian 2014). Alternative pA sites can reside in the 3′-most exon or upstream regions and can give rise to multiple mRNA transcripts that contain different coding sequences, 3′ UTRs, or both (Di Giammartino et al. 2011; Elkon et al. 2013). Importantly, both microRNAs (miRNAs) and RNA binding proteins (RBPs) targeting 3′ UTRs are able to regulate translational efficiency, degradation, and subcellular localization of mRNA or protein (Di Giammartino et al. 2011; Berkovits and Mayr 2015; Tian and Manley 2017). It is well known that APA plays important roles in a wide range of biological processes such as cell differentiation (Ji et al. 2009; Mangone et al. 2010; Hilgers et al. 2011; Li et al. 2012; Ulitsky et al. 2012; Fu et al. 2016; Hu et al. 2017), cell proliferation (Sandberg et al. 2008; Elkon et al. 2012; Hoffman et al. 2016), cell/tissue identity (Zhang et al. 2005; Derti et al. 2012; Smibert et al. 2012; Ni et al. 2013), and carcinogenesis (Mayr and Bartel 2009; Fu et al. 2011; Lin et al. 2012; Xia et al. 2014). However, whether APA is involved in senescence-associated gene expression and contributes to cellular senescence remains to be answered.
We therefore examined the potential role and possible mechanism of APA in cellular senescence by applying our polyadenylation sequencing (PA-seq) approach (Ni et al. 2013) in two cellular senescence models, the passage of mouse embryonic fibroblasts (MEFs) and aortic vascular smooth muscle cells of rats (rVSMCs) at different ages.
Results
Global lengthening of 3′ UTRs couples with decreased gene expression in senescent cells
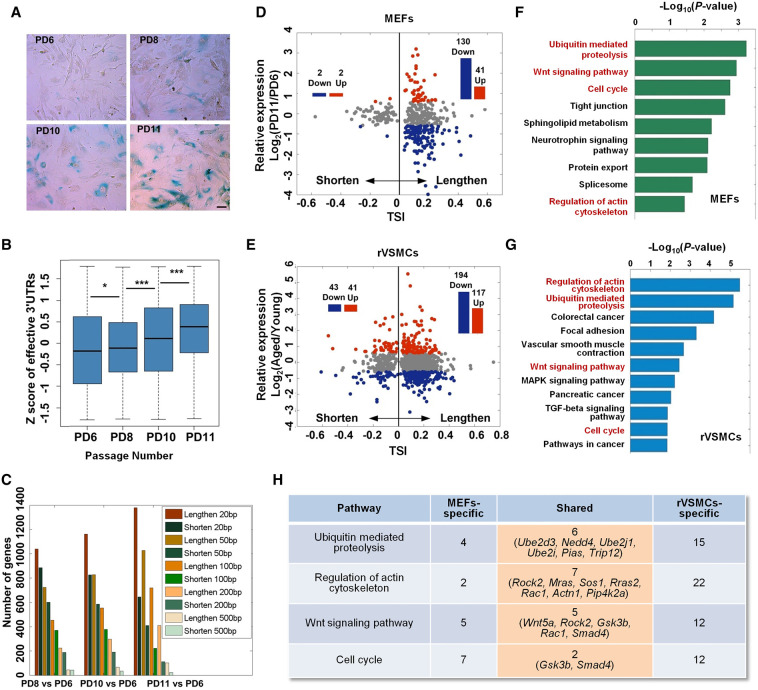
To determine whether APA plays a role during cellular senescence, we first used MEFs undergoing replicative senescence in vitro and having population doubling (PD) times of 6, 8, 10, and 11 passages (Fig. 1A; Supplemental Fig. S1; Dimri et al. 1995; Parrinello et al. 2003; Tian and Li 2014; Tigges et al. 2014). We applied RNA sequencing (RNA-seq) and PA-seq (Ni et al. 2013) to discern the relationship between gene expression and 3′ UTR length patterns in these cells. After confirming the reliability of called pA sites, such as genomic location distribution, poly(A) signal enrichment, and overlap with polyA_DB (Supplemental Figs. S2, S3; Supplemental Table S1), the effective 3′ UTR length (or weighted mean of 3′ UTR length) was used to estimate the relative trend of pA site usage and changes in 3′ UTR length according to our previous method, which took pA site location, the distance to a stop codon, and tag number into consideration (Ni et al. 2013). Effective 3′ UTRs showed a global lengthening trend during MEFs senescence (Fig. 1B), indicating a tendency to use the distal pA sites. To further evaluate the changes in 3′ UTR length at the level of individual genes, we compared effective 3′ UTR length in later passaged (PD8, PD10, and PD11) cells with that in the earlier passaged (PD6) cells using different cut-offs. Cells of later passages always had many more genes with a longer 3′ UTR compared with cells of the earlier passage (Fig. 1C). Moreover, the number of genes with a lengthened 3′ UTR gradually increased from PD8 to PD11, while the number of genes with a shortened 3′ UTR continuously decreased (Fig. 1C). The same trend of global 3′ UTR lengthening was further confirmed by adopting a different methodology, the relative usage of distal polyadenylation sites (RUD) index (Ji et al. 2011), using separate RNA-seq data from the same MEFs used in PA-seq (Supplemental Figs. S4, S5; Supplemental Table S2). Biological replicates of PD11 MEFs had more genes with a longer 3′ UTR compared with those of PD6 MEFs (Supplemental Fig. S6; Supplemental Table S3). Together, these results demonstrate the global lengthening of 3′ UTRs in senescent MEFs.
Figure 1.
Global lengthening of 3′ UTRs in senescent cells. (A) SA-β-gal staining for MEFs of PD6, PD8, PD10, and PD11 passages. (B) Box plot for Z-score transformed effective 3′ UTRs across PD6, PD8, PD10, and PD11 MEFs. (C) Number of genes with lengthened and shortened effective 3′ UTRs. (D) Scatter plot between TSI and expression changes for genes with significantly longer or shorter 3′ UTRs in senescent MEFs (PD11 vs. PD6). TSI denotes tandem UTR isoform switch index (see Methods). The red and blue histograms represent up- and down-regulation (1.5-fold) in senescent cells, respectively. (E) Scatter plot between TSI and expression changes for genes with significantly longer or shorter 3′ UTRs in rVSMCs (old vs. young), similar to that in D. (F,G) Pathways significantly enriched (P < 0.01, Fisher's exact test) for genes tended to use distal pA sites (TSI > 0 in D and E) in senescent MEFs (F) and VSMCs derived from old rats (G). The shared senescence-related pathways are marked in red text. (H) Overlap of genes in the shared pathways between senescent MEFs and rat VSMCs. The number of genes is shown in each grid and shared gene names are in brackets.
To determine whether cell cycle affects the length of 3′ UTRs during senescence, PD6 MEF cells were subjected to serum starvation to drive them into G0 phase (Gustincich and Schneider 1993). The global pattern of 3′ UTR length in G0 cells was most similar to that in PD6 compared with other passages (Supplemental Figs. S5A, S7; Supplemental Tables S1, S2), implying that the global lengthening of 3′ UTRs resulted from senescence rather than from cell cycle alterations.
A longer 3′ UTR region could provide more opportunities for regulation by miRNAs and/or RBPs, which would influence mRNA and/or protein abundance at the post-transcriptional level (Mayr and Bartel 2009; Zheng and Tian 2014). In line with this hypothesis, a global decline in gene expression was observed for genes preferring distal pA sites during senescence (Supplemental Fig. S8A,C–E). In contrast, control genes with a single pA site or favoring proximal pA sites during senescence did not show such a trend (Supplemental Fig. S8B,F,G). Meanwhile, changes in APA and gene expression at the individual gene level were also analyzed by comparing PA-seq data from PD11 and PD6 MEFs using TSI (tandem 3′ UTR isoform switch index), a higher value of which indicates 3′ UTR lengthening (Fu et al. 2011; Li et al. 2012). Genes with a significantly lengthened 3′ UTR outnumbered those containing a shortened one (Fig. 1C,D; Supplemental Fig. S9). Within 3′ UTR lengthened genes, more genes showed reduced expression than elevated expression (P < 5.6 × 10−12, binomial test) (Fig. 1D, right half), while no such difference was observed for genes with shorter 3′ UTRs (Fig. 1D, left half). These data suggest that global 3′ UTR lengthening is associated with decreased gene expression during cellular senescence.
To explore whether APA-induced global lengthening of 3′ UTRs occurred in other senescence systems, we performed PA-seq in VSMCs derived from young and aged rats (Supplemental Table S4). The results showed that senescent rVSMCs and MEFs had a similar ratio between numbers of genes with longer and shorter 3′ UTRs (Supplemental Fig. S9). A significant overlap of genes with longer 3′ UTR usage was observed in these two senescence models (Supplemental Figs. S10, S11). The observation that more genes with lengthened 3′ UTRs tended to be down-regulated was also verified in rVSMCs (P < 1.5 × 10−5, binomial test) (Fig. 1E). Together, the results indicate that APA-mediated 3′ UTR lengthening is involved in gene expression regulation in multiple cellular senescence systems.
Genes preferred distal pA sites in senescent cells enrich in senescence-associated pathways
To further understand the correlation between pA site selection and cellular senescence, we performed pathway analysis on genes preferring distal pA sites in PD11 compared with PD6 MEFs. Genes containing a significantly longer 3′ UTR in PD11 compared with PD6 MEFs were enriched in pathways that are highly pertinent to cellular senescence (Fig. 1F), as were the genes whose 3′ UTRs progressively lengthened in four different passages of MEFs (PD6, PD8, PD10, and PD11) (Supplemental Fig. S12). We also found similar enrichment in senescence-associated pathways between old and young rVSMCs (Fig. 1G). Notably, four of the shared pathways in senescent MEFs and rVSMCs, including ubiquitin-mediated proteolysis, the Wnt signaling pathway, cell cycle, and regulation of the actin cytoskeleton (Fig. 1F,G), are linked to cellular senescence (Amberg et al. 2012; Chandler and Peters 2013; Deschenes-Simard et al. 2014; Hofmann et al. 2014). Further examination of the genes preferring distal pA sites in mouse and rat revealed that these two species also possessed common genes undergoing APA regulation (Fig. 1H), which may serve as good candidates to study the function of 3′ UTR lengthening in cellular senescence.
APA-induced longer 3′ UTR of Rras2 reduces protein production and promotes cellular senescence
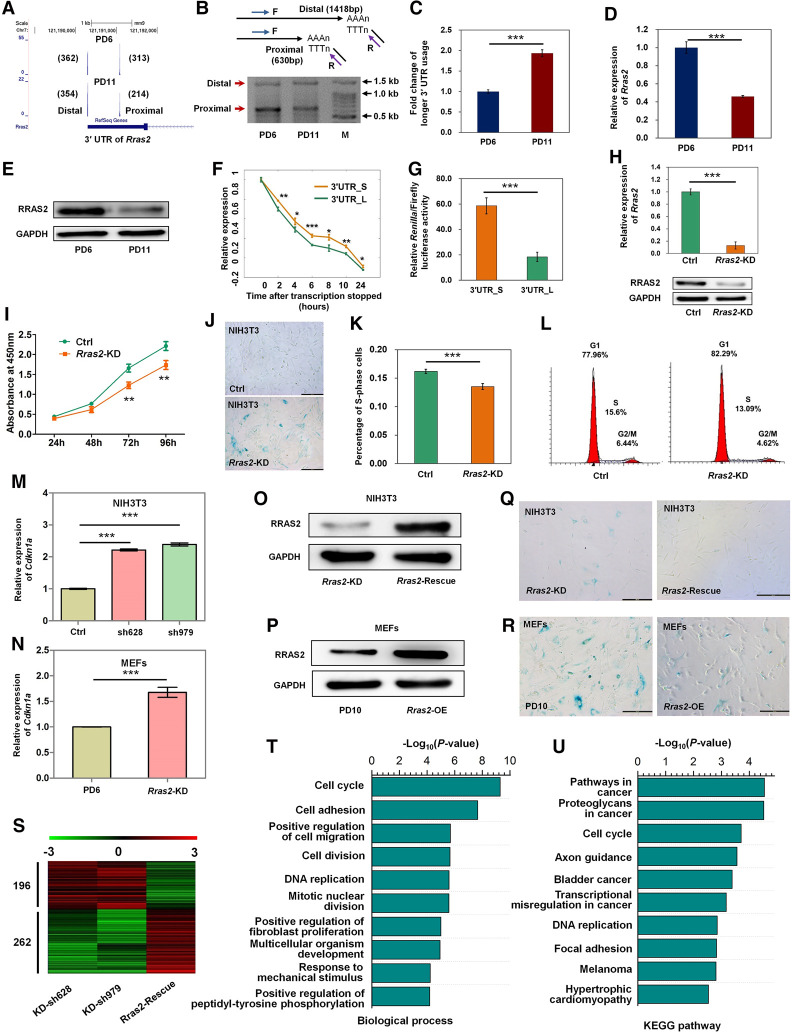
To identify candidate genes that can affect cellular senescence through alternate pA site usage, we applied the following criteria: (1) favoring distal pA site usage both in senescent mouse and rat cells; (2) belonging to the shared four senescence-associated pathways (Fig. 1H); and (3) exhibiting decreased expression during senescence. Based on these criteria, we focused on Rras2. The tendency of Rras2 to use distal pA sites in senescent cells was further confirmed. The UCSC Genome Browser displayed a higher usage of distal pA tags in both senescent MEFs (Fig. 2A) and rVSMCs (Supplemental Fig. S13). 3′ Rapid Amplification of cDNA Ends (3′ RACE) showed a reduced amplicon intensity of the proximal pA site in PD11 MEFs compared with PD6 MEFs (Fig. 2B). Real-time reverse transcription polymerase chain reaction (qRT-PCR) demonstrated a higher usage of longer 3′ UTRs in senescent MEFs (Fig. 2C). Rras2 mRNA and protein levels were both down-regulated in senescent MEFs (Fig. 2D,E), consistent with the idea that longer 3′ UTRs tended to display a decreased mRNA abundance. To investigate how APA-induced increase in Rras2 3′ UTR length contributes to gene expression changes, we evaluated the RNA degradation rate of Rras2 transcripts containing long and short 3′ UTRs. After blocking transcription followed by qRT-PCR, we found that Rras2 transcripts with a longer 3′ UTR were less stable than those with a shorter 3′ UTR (Fig. 2F). We then inserted the shorter 3′ UTR (represented as 3′ UTR_S) and the longer 3′ UTR (3′ UTR_L) with a mutated proximal PAS of Rras2 into a dual-luciferase reporter system, respectively. The reporter gene containing 3′ UTR_L exhibited significantly reduced luciferase activities compared with that containing 3′ UTR_S (Fig. 2G), implying that the APA-induced longer 3′ UTR of Rras2 down-regulated RRAS2 protein abundance.
Figure 2.
Down-regulation of Rras2 through 3′ UTR lengthening promotes cellular senescence in mouse cells. (A) PA-seq track of proximal and distal pA site usage in PD6 and PD11 MEFs. Raw tag numbers reflected by both width and height of each pA peak are indicated in parentheses. (B) Changes of distal and proximal pA site usage between PD6 and PD11 MEF cells detected by 3′ RACE. The 3′ RACE strategy is shown in the upper panel. F and R indicate forward and reverse primers, respectively. PCR product sizes for distal and proximal pA sites are shown in parentheses. (C) The Rras2 gene has increased usage of the distal pA site in MEF PD11 compared to PD6 detected by qRT-PCR. (D,E) qRT-PCR (D) and Western blot (E) of Rras2 expression in PD6 and PD11 MEFs. Gapdh serves as the internal control. (F) Stability comparison of Rras2 isoforms with 3′ UTR_S and 3′ UTR_L in mouse fibroblast cell line NIH3T3. (G) Luciferase activity from a reporter system containing 3′ UTR_S or 3′ UTR_L. (H) qRT-PCR (upper panel) and Western blot (lower panel) validation of Rras2 knockdown in NIH3T3 cells. (I) Proliferation rate evaluation of NIH3T3 cells without (Ctrl) and with Rras2 knockdown (Rras2-KD, sh979) by Cell Counting Kit-8 (CCK-8). (J) SA-β-gal staining in NIH3T3 cells without (Ctrl) and with Rras2 knockdown (Rras2-KD). (K) FACS analysis of four replicate samples showed a lower percentage of Rras2-KD NIH3T3 cells in S phase compared with control cells. (L) Percentage of cells in S, G1, and G2/M phase in one replicate. (M,N) Rras2 depletion promoted Cdkn1a expression in mouse NIH3T3 cells (M) and MEFs (N) evaluated by qRT-PCR. (O,P) Western blot validations of Rras2 rescue in Rras2-KD NIH3T3 cells (O) and Rras2-overexpression (OE) in senescent MEFs (P). GAPDH serves as the internal control. (Q) Higher SA-β-gal staining caused by reduced Rras2 expression was rescued by overexpression of Rras2 (Rras2-OE) in mouse NIH3T3 cells. (R) Rras2-OE reversed senescence in primary MEF cells as evaluated by SA-β-gal staining assay. (S) Heat map of log2-transformed expression ratio of differentially expressed genes shared by Rras2-KD and Rras2-Rescue cells compared with the control NIH3T3 cells. Red and green denotes increased and decreased expression, respectively. The numbers of down- and up-regulated genes are shown on the left. (T,U) Enrichment of all differentially expressed genes shown in S for biological process (T) and the KEGG pathway (U), as determined by DAVID. (***) P < 0.001, (**) P < 0.01, (*) P < 0.05, two-tailed t-test.
To determine whether decreased levels of RRAS2 trigger senescence-associated phenotypes, we depleted RRAS2 with two short hairpin RNAs (shRNA-mediated knockdown; Rras2-KD) in mouse NIH3T3 cells (Fig. 2H; Supplemental Fig. S14A) and observed delayed cell proliferation and increased SA-β-gal staining (Fig. 2I,J; Supplemental Fig. S14B,C). Cell cycle analysis showed that RRAS2-depletion reduced the percentage of S phase cells (Fig. 2K,L). Cdkn1a, which encodes cyclin-dependent kinase inhibitor 1A (P21), showed increased expression upon Rras2-KD (Fig. 2M). Moreover, Rras2-KD in primary MEFs has similar effects to those in NIH3T3 cells (Fig. 2N; Supplemental Fig. S15). In addition, reintroduction of RRAS2 (Rras2-Rescue) into Rras2-KD cells rescued the expression level of RRAS2 (Fig. 2O) and gave rise to reduced SA-β-gal signals (Fig. 2Q). Through RNA-seq profiling of Rras2-KD and Rras2-Rescue NIH3T3 cells, we found that altered RRAS2 expression caused expression changes in a variety of genes (Fig. 2S). Gene Ontology (GO) and pathway analysis revealed that those genes showing opposite expression trends were enriched in senescence-relevant biological processes, including cell cycle, cell adhesion, and DNA replication (Fig. 2T,U; Supplemental Table S5). More importantly, overexpression of RRAS2 (Fig. 2P) was able to reverse the SA-β-gal staining (Fig. 2R) and rescue Cdkn1a expression levels in senescent MEFs (Supplemental Fig. S16). Taken together, these data indicate that RRAS2 plays a crucial role in delaying cellular senescence.
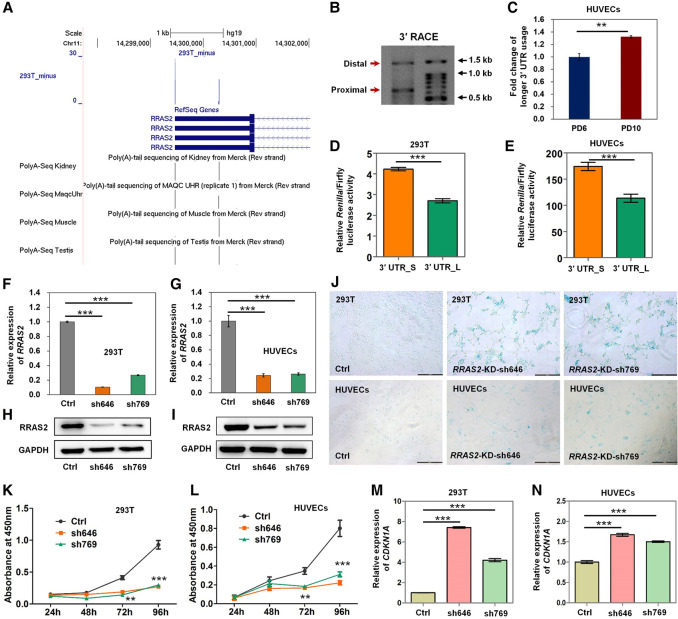
To address whether APA in Rras2 is evolutionarily conserved between rodents and human, we verified the existence of its two pA sites in both human embryonic kidney (HEK) 293T cells and human umbilical vein endothelial cells (HUVECs) (Kim et al. 2007; Muck et al. 2008; Cardus et al. 2013; Zhang et al. 2014). Our PA-seq data from 293T cells (Ni et al. 2013) and public PolyA-seq data from human tissues confirmed the existence of both pA sites in human RRAS2 (Fig. 3A). The two pA sites were validated by 3′ RACE (Fig. 3B), and their PCR products were cloned and sequenced by the Sanger method to confirm the existence of poly(A) or poly(T) sequence at the end of the amplicon (Supplemental Fig. S17). Of note, senescent HUVEC cells also favored the distal pA site (Fig. 3C), which generated less protein (Fig. 3D,E). Knockdown of RRAS2 (Fig. 3F–I) increased SA-β-gal activity (Fig. 3J), slowed cell proliferation rate (Fig. 3K,L), and induced CDKN1A expression in both 293T cells and HUVECs (Fig. 3M,N). These findings indicate an evolutionarily conserved mechanism that longer 3′ UTR of Rras2 causes reduced protein production and then results in senescence in both rodents and human cells.
Figure 3.
Decreased RRAS2 protein level via 3′ UTR lengthening causes senescence in human cells. (A) PA-seq track of 293T cells and PolyA-seq track of four representative human samples for RRAS2 in the UCSC Genome Browser. (B) 3′ RACE products separated by agarose gel electrophoresis to confirm distal and proximal pA sites in human cells. Right lane, molecular weight marker. (C) RRAS2 has a higher usage of the distal pA site in HUVECs at PD10 than at PD6, as determined by qRT-PCR. (D,E) Luciferase activity from a reporter containing the short 3′ UTR compared with that from the reporter containing 3′ UTR_L in 293T cells (D) and HUVECs (E). (F,G) Validation of RRAS2 knockdown by two different shRNAs (sh646 and sh769) by qRT-PCR in 293T cells (F) and HUVECs (G). (H,I) Western blot confirmation of RRAS2 knockdown in 293T cells (H) and HUVECs (I). GAPDH served as the loading control. (J) SA-β-gal staining for 293T cells (upper panels) and HUVECs (lower panels) without (Ctrl) and with RRAS2 knockdown (RRAS2-KD). Scale bar, 200 µm. (K,L) CCK-8 analysis of 293T cell (K) and HUVEC (L) proliferation without (Ctrl) and with RRAS2 knockdown. (M,N) Increased CDKN1A expression upon RRAS2 knockdown in 293T cells (M) and HUVECs (N), as determined by qRT-PCR. (***) P < 0.001, (**) P < 0.01, two-tailed t-test.
Splicing factor TRA2B represses RRAS2 protein level through binding to its alternative 3′ UTR and contributes to cellular senescence
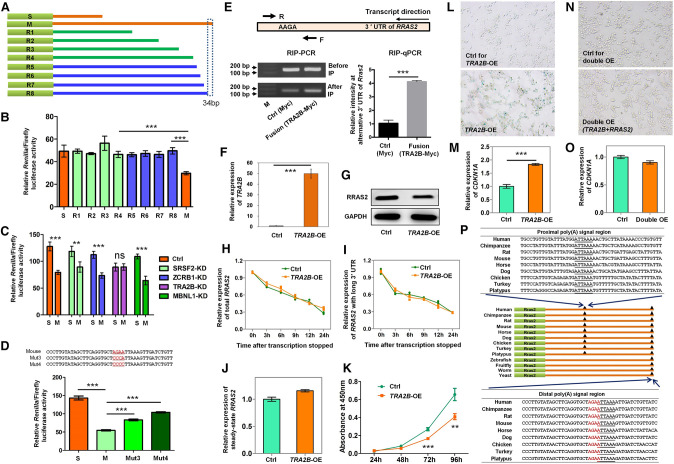
We reasoned that cis-elements recognized by either miRNAs or RBPs in the alternative 3′ UTR of Rras2 contribute to decreased RRAS2 protein production. To this end, we divided the 3′ UTR of Rras2 into four regions by deleting sequences of different lengths (shown as R1–R4 in Fig. 4A) and inserted them separately into a luciferase reporter, which was then transfected into mouse cells. Subsequent luciferase assays demonstrated significantly reduced luciferase activity from the construct containing a long 3′ UTR (resulting from mutated proximal PAS, labeled “M”) compared with the short 3′ UTR (labeled “S”) and the R1–R4 constructs (Fig. 4B). Thus, the sequence of last 148 base pairs (bp) located in the alternative 3′ UTR of Rras2 contains key motifs or elements essential for protein production.
Figure 4.
Binding of TRA2B to the alternative 3′ UTR of Rras2 causes cellular senescence via reduced RRAS2 expression. (A) Schematic diagram of the truncation strategy to identify key cis regulatory elements. M denotes full length 3′ UTR with mutated proximal poly(A) signal. S represents 3′ UTR using the proximal pA site. R1–R4 refer to four truncated 3′ UTR fragments constructed for the first round screen. R5–R8 denote four additional constructs for the second-round screen. (B) Luciferase assays for all constructs indicated in A. (C) Luciferase assay screen for key trans-acting factors in candidate RBP knockdown cells. (ns) Not significant. (D) Luciferase assay in mouse cells transfected with constructs containing mutations introduced in the potential region containing the key cis-element. “AGAA” is the core binding motif of TRA2B (Grellscheid et al. 2011). (E) TRA2B binding to the alternative 3′ UTR of Rras2 evaluated by RIP-PCR and RIP-qPCR. The PCR primer pair (F and R, forward and reverse, respectively) was designed to span the “AGAA” motif. TRA2B was fused to Myc protein to serve as the ectopically expressed fusion protein. A vector expressing Myc protein served as the internal control (Ctrl). Anti-Myc was used to pull down RNA from MEF cells either expressing Ctrl (Myc) or Fusion (TRA2B-Myc) protein. The same amount of RNA was applied for RIP-PCR (left panel) and RIP-qPCR (right panel). (F) qRT-PCR validation of TRA2B overexpression (TRA2B-OE) in human 293T cells. (G) Decreased RRAS2 protein level upon TRA2B overexpression detected by Western blot in 293T cells. (H,I) RRAS2 RNA degradation rate was assayed for total isoforms (H) and isoform with longer 3′ UTRs (I) in TRA2B-OE and control human cells by qRT-PCR. (J) Steady-state mRNA level of RRAS2 was quantified in TRA2B-OE and control human cells by qRT-PCR. (K) Overexpression of TRA2B slows down cell proliferation as detected by the CCK-8 assay. (L,M) TRA2B-OE caused increased SA-β-gal staining (L) and CDKN1A expression (M) in 293T cells. (N,O) Overexpression of RRAS2 in TRA2B up-regulated human cells reversed SA-β-gal staining (N) and CDKN1A expression (O). (P) Sequence alignment near proximal and distal pA sites of Rras2 in representative species. The PAS is underlined while the TRA2B core binding motif “AGAA” is highlighted in red text. (***) P < 0.001, (**) P < 0.01, two-tailed t-test.
To further narrow the range of key elements, four more reporter constructs harboring the truncated 3′ UTRs (presented as R5–R8 in Fig. 4A) were generated. The construct lacking a 34-bp element showed significantly reduced luciferase activity, indicating that this element is critical for down-regulation of protein production (Fig. 4A,B). No miRNA binding sites were predicted in this 34-bp region; therefore, we searched RBPmap (Paz et al. 2014) for potential RBPs that might recognize this element and identified four RBPs (SPSF2, ZCRB1, TRA2B, and MBNL1). Following individual knockdown of the four RBPs, we measured the luciferase activities of S and M constructs (Fig. 4C). Depletion of TRA2B, a known splicing factor involved in mRNA processing, cell proliferation, and migration (Yang et al. 2015), eliminated the difference in luciferase activities between the S and M fragments-containing reporters (Fig. 4C), suggesting that TRA2B played an important role in down-regulating protein production. These data implied that the 34-bp element in the 3′ UTR of Rras2 is required for TRA2B-mediated down-regulation of RRAS2.
The “AGAA” element may serve as the core sequence of the TRA2B binding motif (Fig. 4D; Grellscheid et al. 2011). Considering that “AGAA” shared 1 nt with the downstream PAS “ATTAAA” in the alternative 3′ UTR of Rras2, we mutated the first 3 nt to avoid disruption of the PAS (“AGAA” to “CCCA”) (designated Mut3 in Fig. 4D). All 4 nt were also mutated (“AGAA” to “CCCC”) (denoted Mut4 in Fig. 4D) to validate the effects that are dependent on the whole core sequence. According to luciferase reporter assays, the Mut3 construct partially restored luciferase activity compared with that for M, while Mut4 showed a higher capability of restoring the repressed luciferase activity than Mut3 (Fig. 4D). To explore whether TRA2B directly binds to the alternative 3′ UTR of Rras2, we carried out RNA immunoprecipitation coupled with both regular and quantitative PCR (RIP-PCR and RIP-qPCR) assays. The results showed an enrichment of TRA2B-binding signal (Fig. 4E), suggesting direct binding of TRA2B to the alternative 3′ UTR of Rras2. To further evaluate the role of “AGAA,” we moved this motif to two upstream positions in the alternative 3′ UTR of Rras2. The “AGAA” motif at a more proximal position did not repress protein production (Supplemental Fig. S18), implying that the “AGAA” motif in the alternative 3′ UTR of the Rras2 gene was required but not sufficient to repress protein levels. Together, these data suggest that the “AGAA” motif within the alternative 3′ UTR of Rras2 is essential for TRA2B binding and repression of RRAS2 protein production.
To further confirm the contribution of TRA2B-RRAS2 regulation to cellular senescence, we overexpressed TRA2B in 293T cells, which show a higher usage of the distal pA site in RRAS2 and can serve as an appropriate cell model to perform the test, given that TRA2B bound to the alternative 3′ UTR of Rras2 (Fig. 3A). As expected, ectopic expression of TRA2B led to decreased RRAS2 protein level (Fig. 4F,G). Notably, neither RNA stability (Fig. 4H,I) nor mRNA steady-state levels of RRAS2 (Fig. 4J) showed considerable changes upon TRA2B overexpression, suggesting that the reduced RRAS2 protein level was caused by repressed translation through binding of TRA2B to the alternative 3′ UTR. Up-regulation of TRA2B led to a reduced proliferation rate (Fig. 4K), higher SA-β-gal staining level, and increased CDKN1A expression (Fig. 4L,M). Recovery of RRAS2 expression by additional overexpression of RRAS2 in TRA2B-overexpressed cells attenuated the CDKN1A expression and SA-β-gal staining to basal levels (Fig. 4N,O). Taken together, these data demonstrated that binding of TRA2B to the alternative 3′ UTR of Rras2 results in decreased RRAS2 expression, which in turn accelerated cellular senescence.
Key elements involved in APA regulation of Rras2 are evolutionarily conserved
Since Rras2 prefers the distal pA site in senescent mouse, rat, and human cells (Figs. 2A–C, 3C; Supplemental Fig. S13), it would be interesting to have an evolutionary view on related Rras2 sequences. Comparative genomic analysis revealed that the 3′ UTR of Rras2 was extremely conserved in representative species from galliformes to primates (Supplemental Fig. S19), implying that regulation of 3′ UTR length by APA is of evolutionary significance. Notably, all listed animals contained a canonical PAS (AA/TTAAA) near the proximal pA site (Fig. 4P). This strongly indicated that APA-regulated Rras2 expression plays a crucial role during evolution. The sequence adjacent to the distal pA site within the 3′ UTR of Rras2 also contained the canonical PAS “ATTAAA” and the TRA2B binding motif “AGAA,” which were conserved from birds to mammals (Fig. 4P), indicating that a strong selection pressure is likely to have driven the molecular evolution for TRA2B-mediated regulation of APA in Rras2.
Discussion
In this study, we discovered that APA-mediated 3′ UTR lengthening played a role in cellular senescence. As exemplified by Rras2 (Fig. 5), we extended the functional importance of APA to the aging field and provided a novel perspective for understanding the mechanism underlying cellular senescence. However, upstream factors controlling 3′ UTR lengthening during cellular senescence need to be further explored. Given that certain core factors in the 3′ processing machinery were known to play a role in APA regulation (Takagaki et al. 1996; Zheng and Tian 2014), we therefore surveyed the gene expression of 24 important polyadenylation trans-factors. Most of these factors underwent a trend of decreased expression during MEFs senescence (Supplemental Fig. S20), consistent with recent findings that up-regulation of polyadenylation factors was associated with 3′ UTR shortening (Mayr and Bartel 2009; Xia et al. 2014). These results provided a possible mechanistic explanation for 3′ UTR lengthening during cellular senescence and deserved further investigation.
Figure 5.
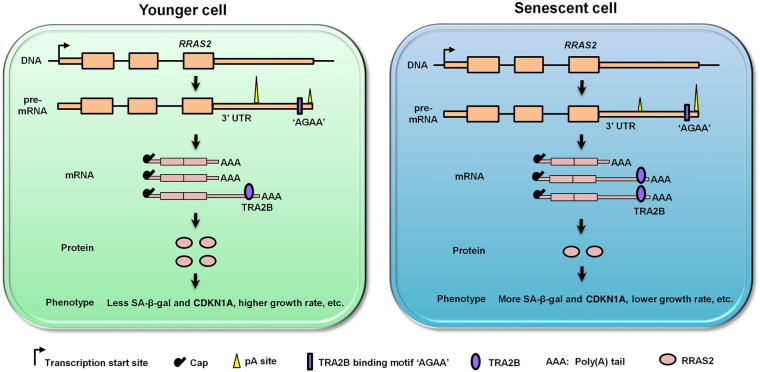
Working model for APA as a new mechanism in regulating cellular senescence. Genes, such as RRAS2, favor distal pA site usage during senescence, leading to 3′ UTR lengthening. Splicing factor TRA2B bound to its core cis-element “AGAA” located in the alternative 3′ UTR of RRAS2 and repressed RRAS2 protein production, thereby leading to senescence-associated phenotypes.
Genes preferring distal pA sites showed a global lengthening of the 3′ UTR and a trend of decreased mRNA levels in both senescent MEFs and rVSMCs (Fig. 1), supplementing the observation that genes favoring proximal pA sites tended to have increased mRNA levels in cancer cells (Mayr and Bartel 2009; Xia et al. 2014). We also found that many genes displayed opposite pA site usage preference in senescent cells compared with cancer cells. Eighty-two and 166 genes that favored proximal pA sites in seven tumor types were prone to use distal pA sites in senescent MEFs and rVSMCs, respectively (Supplemental Fig. S21). In addition, 35 genes preferring shorter 3′ UTRs in multiple cancer cells tended to use longer 3′ UTRs in both senescent MEFs and rVSMCs (Supplemental Fig. S21). These findings supported a model that interaction between condition-specific trans-acting factors and dynamic changes in 3′ UTR length determined by APA could contribute to opposite biological processes, such as cellular senescence and tumor development.
There are two major categories of cellular senescence, developmentally programmed senescence and stress-induced premature senescence (SIPS) (Munoz-Espin and Serrano 2014). Here, we showed that MEFs in replicative senescence underwent global 3′ UTR lengthening. VSMCs derived from old rats were likely to undergo a combination of replicative senescence and varieties of stress-induced senescence. Whether SIPS itself will induce global 3′ UTR lengthening needs to be further determined. Thus, more senescence models are required to fully understand the prevalence and functional relevance of 3′ UTR lengthening. In conclusion, our results provide evidence that APA contributes to the regulation of gene expression during cell senescence in multiple species, implying APA-regulated gene expression may be evolutionarily conserved.
Methods
Cell isolation, cultivation, and total RNA extraction
Primary MEFs were isolated from embryos of a 12.5- to 14-d pregnant C57BL/6 mouse according to a previously described method (Todaro and Green 1963). Details were described in Supplemental Methods. Total RNA was isolated using TRIzol reagent (Invitrogen for both MEFs and rVSMCs from rats of different ages (2 wk and 2 yr).
SA-β-gal staining, cell proliferating rate assay, and cell cycle analysis
SA-β-gal activity (Dimri et al. 1995) was monitored in mouse (MEFs, NIH3T3) and human (293T, HUVECs) cells using a senescence detection kit (BioVision, cat. no. K320-250). Mouse (NIH3T3) and human (293T, HUVECs) cells with or without candidate gene knockdown were cultured and assayed for cell proliferation using a Cell Counting Kit-8 (CCK-8) according to the vendor's instructions (Dojindo). Details of SA-β-gal staining and cell cycle analysis are described in Supplemental Methods.
PA-seq and RNA-seq library construction, mapping, and peak calling
The PA-seq libraries were constructed according to our established protocol (Ni et al. 2013). The dUTP-based strand-specific RNA-seq libraries were constructed by following a previously described protocol (Parkhomchuk et al. 2009). Both types of libraries were sequenced using an Illumina HiSeq2000 platform in a paired-end 2 × 101-bp manner. Processed raw data were aligned to the mouse genome (version mm9) or rat genome (version rn5) using TopHat2 (Kim et al. 2013a). Detailed peak calling of pA sites and expression analysis were included in Supplemental Methods. All called pA clusters in MEFs are listed in Supplemental Table S6.
Comparison of APA profiles between MEFs of different passages
The tandem 3′ UTR isoform switch index values were calculated according to a published approach (Fu et al. 2011; Li et al. 2012). Genes with expression fold-change ≥ 1.5 and FPKM ≥ 1 in both samples are shown in Figure 1, D and E. The genes with a P-value cut-off of 0.05 (corresponding to a false discovery rate [FDR], estimated by the Benjamini–Hochberg method with R software [R Core Team 2015]) were considered to have significantly changed APA among the different passages. Specifically, an FDR ≤ 0.05 with a positive TSI (Li et al. 2012) implies a lengthening 3′ UTR across the different passages of cellular senescence; an FDR ≤ 0.05 with a negative TSI implies a shortening 3′ UTR. All APA genes with detailed information, including expression value, effective 3′ UTR length, and RUD index (Ji et al. 2011), are included in Supplemental Table S7. Comparison of lengthening or shortening of 3′ UTRs is included in Supplemental Table S8.
Pathway enrichment analysis
The Database for Annotation, Visualization, and Integrated Discovery (DAVID) (Huang da et al. 2009) was used for the pathway enrichment analysis, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was selected.
Western blotting and qRT-PCR
Primary antibodies (RRAS2, 1:1000, cat. no. ab209078, Abcam; GAPDH, 1:2000–3000, cat. no. sc-32233, Santa Cruz Biotechnology; CDKN2A, 1:500–1000, cat. no. sc-1207, Santa Cruz Biotechnology) targeting proteins of interest were used for the Western blot. A detailed description is included in Supplemental Methods. Total RNA used for expression quantification was reverse-transcribed into cDNA using random primers, and then mRNA levels were quantified by qRT-PCR and normalized to that of Gapdh (Roche LightCycler). For pA site usage quantification, RNAs were reverse-transcribed with oligo(dT) primer, followed by PCR with two pairs of primers (proximal and distal) targeting different regions of the cDNAs. Specifically, the region targeted by the proximal pair is common to both APA isoforms and the region targeted by the distal pair is unique to the longer isoform. qPCR signals from the proximal and distal pairs of primers were compared to indicate the relative expression of the two isoforms. All primer sequence information is listed in Supplemental Table S9.
RNA stability, RNA immunoprecipitation, and luciferase assay
For evaluation of RNA stability, cells were treated with actinomycin D (5 µg/mL) for 0, 2, 4, 6, 8, 10, and 24 h. After the indicated time, total RNAs were isolated and analyzed by qRT-PCR. Primer sequences are shown in Supplemental Table S9. RNA immunoprecipitation (RIP) was performed according to a previously published protocol with minor modification (Zhao et al. 2008). The Dual-Luciferase Reporter 1000 Assay System (Promega) was carried out to evaluate the impact of protein production for different lengths of 3′ UTRs. Detailed information of RIP-PCR and the luciferase assay are described in Supplemental Methods.
Data access
The raw PA-seq and RNA-seq data from this study have been submitted to the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra/) under accession number SRP065821.
Supplementary Material
Acknowledgments
We thank Dr. Haijian Wang for providing the psiCHECK-2 Vector, Professor Hongyan Wang for luciferase assay instrument support, and Professors Li Jin and Jun Zhu for insightful suggestions regarding bioinformatic analyses. This work was supported by the National Basic Research Program of China (973 program: 2013CB530700 and 2015CB943000), National Natural Science Foundation of China (31771336, 31471192, 31521003, and 31471205), and the 111 Project of China (B13016). We thank Genergy Biotech (Shanghai) Co., Ltd. for the deep sequencing service. We thank Jeremy Allen, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for language polishing of this manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.224451.117.
Freely available online through the Genome Research Open Access option.
References
- Amberg D, Leadsham JE, Kotiadis V, Gourlay CW. 2012. Cellular ageing and the actin cytoskeleton. Subcell Biochem 57: 331–352. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, Zhong J, Saltness RA, Jeganathan KB, Verzosa GC, Pezeshki A, et al. 2016. Naturally occurring p16Ink4a-positive cells shorten healthy lifespan. Nature 530: 184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovits BD, Mayr C. 2015. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 522: 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busuttil RA, Rubio M, Dolle ME, Campisi J, Vijg J. 2003. Oxygen accelerates the accumulation of mutations during the senescence and immortalization of murine cells in culture. Aging Cell 2: 287–294. [DOI] [PubMed] [Google Scholar]
- Campisi J, Kim SH, Lim CS, Rubio M. 2001. Cellular senescence, cancer and aging: the telomere connection. Exp Gerontol 36: 1619–1637. [DOI] [PubMed] [Google Scholar]
- Cardus A, Uryga AK, Walters G, Erusalimsky JD. 2013. SIRT6 protects human endothelial cells from DNA damage, telomere dysfunction, and senescence. Cardiovasc Res 97: 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler H, Peters G. 2013. Stressing the cell cycle in senescence and aging. Curr Opin Cell Biol 25: 765–771. [DOI] [PubMed] [Google Scholar]
- Chen W, Jia Q, Song Y, Fu H, Wei G, Ni T. 2017. Alternative polyadenylation: methods, findings, and impacts. Genomics Proteomics Bioinformatics 15: 287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. 2012. A quantitative atlas of polyadenylation in five mammals. Genome Res 22: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschenes-Simard X, Lessard F, Gaumont-Leclerc MF, Bardeesy N, Ferbeyre G. 2014. Cellular senescence and protein degradation: breaking down cancer. Cell Cycle 13: 1840–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL. 2011. Mechanisms and consequences of alternative polyadenylation. Mol Cell 43: 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci 92: 9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Drost J, van Haaften G, Jenal M, Schrier M, Oude Vrielink JA, Agami R. 2012. E2F mediates enhanced alternative polyadenylation in proliferation. Genome Biol 13: R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. 2013. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 14: 496–506. [DOI] [PubMed] [Google Scholar]
- Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, Xu A. 2011. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res 21: 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Yang D, Su W, Ma L, Shen Y, Ji G, Ye X, Wu X, Li QQ. 2016. Genome-wide dynamics of alternative polyadenylation in rice. Genome Res 26: 1753–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grellscheid S, Dalgliesh C, Storbeck M, Best A, Liu Y, Jakubik M, Mende Y, Ehrmann I, Curk T, Rossbach K, et al. 2011. Identification of evolutionarily conserved exons as regulated targets for the splicing activator Tra2β in development. PLoS Genet 7: e1002390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustincich S, Schneider C. 1993. Serum deprivation response gene is induced by serum starvation but not by contact inhibition. Cell Growth Differ 4: 753–760. [PubMed] [Google Scholar]
- Hayflick L, Moorhead PS. 1961. The serial cultivation of human diploid cell strains. Exp Cell Res 25: 585–621. [DOI] [PubMed] [Google Scholar]
- Hilgers V, Perry MW, Hendrix D, Stark A, Levine M, Haley B. 2011. Neural-specific elongation of 3′ UTRs during Drosophila development. Proc Natl Acad Sci 108: 15864–15869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman Y, Bublik DR, Ugalde AP, Elkon R, Biniashvili T, Agami R, Oren M, Pilpel Y. 2016. 3′UTR shortening potentiates microRNA-based repression of pro-differentiation genes in proliferating human cells. PLoS Genet 12: e1005879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann JW, McBryan T, Adams PD, Sedivy JM. 2014. The effects of aging on the expression of Wnt pathway genes in mouse tissues. Age (Dordr) 36: 9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Li S, Park JY, Boppana S, Ni T, Li M, Zhu J, Tian B, Xie Z, Xiang M. 2017. Dynamic landscape of alternative polyadenylation during retinal development. Cell Mol Life Sci 74: 1721–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci 106: 7028–7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Luo W, Li W, Hoque M, Pan Z, Zhao Y, Tian B. 2011. Transcriptional activity regulates alternative cleavage and polyadenylation. Mol Syst Biol 7: 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Kim MS, Seu YB, Chung HY, Kim JH, Kim JR. 2007. Regulation of replicative senescence by insulin-like growth factor-binding protein 3 in human umbilical vein endothelial cells. Aging Cell 6: 535–545. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013a. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Byun HO, Jee BA, Cho H, Seo YH, Kim YS, Park MH, Chung HY, Woo HG, Yoon G. 2013b. Implications of time-series gene expression profiles of replicative senescence. Aging Cell 12: 622–634. [DOI] [PubMed] [Google Scholar]
- Li Y, Sun Y, Fu Y, Li M, Huang G, Zhang C, Liang J, Huang S, Shen G, Yuan S, et al. 2012. Dynamic landscape of tandem 3′ UTRs during zebrafish development. Genome Res 22: 1899–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhang Y, Fu J, Han L, Xue L, Lv C, Wang P, Li G, Tong T. 2013. FOXA1 mediates p16INK4a activation during cellular senescence. EMBO J 32: 858–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Li Z, Ozsolak F, Kim SW, Arango-Argoty G, Liu TT, Tenenbaum SA, Bailey T, Monaghan AP, Milos PM, et al. 2012. An in-depth map of polyadenylation sites in cancer. Nucleic Acids Res 40: 8460–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. 2013. The hallmarks of aging. Cell 153: 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. 2010. The landscape of C. elegans 3′UTRs. Science 329: 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin P, Xiong J, Liu X, Yan Z, Zhang X, Li M, He L, Somel M, Yuan Y, Phoebe Chen YP, et al. 2013. Widespread splicing changes in human brain development and aging. Mol Syst Biol 9: 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Vagner S. 2010. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res 38: 2757–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muck C, Micutkova L, Zwerschke W, Jansen-Durr P. 2008. Role of insulin-like growth factor binding protein-3 in human umbilical vein endothelial cell senescence. Rejuvenation Res 11: 449–453. [DOI] [PubMed] [Google Scholar]
- Munoz-Espin D, Serrano M. 2014. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol 15: 482–496. [DOI] [PubMed] [Google Scholar]
- Munoz-Espin D, Canamero M, Maraver A, Gomez-Lopez G, Contreras J, Murillo-Cuesta S, Rodriguez-Baeza A, Varela-Nieto I, Ruberte J, Collado M, et al. 2013. Programmed cell senescence during mammalian embryonic development. Cell 155: 1104–1118. [DOI] [PubMed] [Google Scholar]
- Ni T, Yang Y, Hafez D, Yang W, Kiesewetter K, Wakabayashi Y, Ohler U, Peng W, Zhu J. 2013. Distinct polyadenylation landscapes of diverse human tissues revealed by a modified PA-seq strategy. BMC Genomics 14: 615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov AM. 1996. Telomeres, telomerase, and aging: origin of the theory. Exp Gerontol 31: 443–448. [DOI] [PubMed] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A. 2009. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res 37: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. 2003. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol 5: 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz I, Kosti I, Ares M Jr, Cline M, Mandel-Gutfreund Y. 2014. RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res 42: W361–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, Brownlee GG. 1976. 3′ non-coding region sequences in eukaryotic messenger RNA. Nature 263: 211–214. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- Rodier F, Campisi J. 2011. Four faces of cellular senescence. J Cell Biol 192: 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. 1990. The role of poly(A) in the translation and stability of mRNA. Curr Opin Cell Biol 2: 1092–1098. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads BD, Carlson J, Brown JB, et al. 2012. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep 1: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storer M, Mas A, Robert-Moreno A, Pecoraro M, Ortells MC, Di Giacomo V, Yosef R, Pilpel N, Krizhanovsky V, Sharpe J, et al. 2013. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell 155: 1119–1130. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL. 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87: 941–952. [DOI] [PubMed] [Google Scholar]
- Tian XL, Li Y. 2014. Endothelial cell senescence and age-related vascular diseases. J Genet Genomics 41: 485–495. [DOI] [PubMed] [Google Scholar]
- Tian B, Manley JL. 2017. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 18: 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33: 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigges J, Krutmann J, Fritsche E, Haendeler J, Schaal H, Fischer JW, Kalfalah F, Reinke H, Reifenberger G, Stuhler K, et al. 2014. The hallmarks of fibroblast ageing. Mech Ageing Dev 138: 26–44. [DOI] [PubMed] [Google Scholar]
- Todaro GJ, Green H. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol 17: 299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Subtelny AO, Koppstein D, Bell GW, Sive H, Bartel DP. 2012. Extensive alternative polyadenylation during zebrafish development. Genome Res 22: 2054–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldera-Lupa DM, Kalfalah F, Florea AM, Sass S, Kruse F, Rieder V, Tigges J, Fritsche E, Krutmann J, Busch H, et al. 2014. Proteome-wide analysis reveals an age-associated cellular phenotype of in situ aged human fibroblasts. Aging (Albany NY) 6: 856–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei YN, Hu HY, Xie GC, Fu N, Ning ZB, Zeng R, Khaitovich P. 2015. Transcript and protein expression decoupling reveals RNA binding proteins and miRNAs as potential modulators of human aging. Genome Biol 16: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, Li W. 2014. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat Commun 5: 5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Wu J, Zheng W, Wang P, Li J, Zhang Z, Tong T. 2004. Sp1 is involved in the transcriptional activation of p16INK4 by p21Waf1 in HeLa cells. FEBS Lett 564: 199–204. [DOI] [PubMed] [Google Scholar]
- Yang L, Tao T, Wang Y, Bao Z, He X, Cui G. 2015. Knocking down the expression of TRA2β inhibits the proliferation and migration of human glioma cells. Pathol Res Pract 211: 731–739. [DOI] [PubMed] [Google Scholar]
- Zhang H, Lee JY, Tian B. 2005. Biased alternative polyadenylation in human tissues. Genome Biol 6: R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhen YZ, Lin YJ, Liu J, Wei J, Xu R, Hu G. 2014. KNDC1 knockdown protects human umbilical vein endothelial cells from senescence. Mol Med Rep 10: 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Tian B. 2014. RNA-binding proteins in regulation of alternative cleavage and polyadenylation. Adv Exp Med Biol 825: 97–127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.