Abstract
Purpose
CREB, MYCY1 and NAT10 are involved in cancer cell migration. However, the relationship between these three proteins and their role in laryngeal cancer cell migration remains unknown.
Methods
Transient gene transfection was performed in laryngeal cancer cells. Bioinformatics analysis was used to predict the binding of CREB to MYCT1 promoter. Binding of CREB to the promoter of MYCT1 was monitored by luciferase reporter assay and chromatin immuno-precipitation method in vitro and in vivo, respectively. Real-time RT-PCR and Western bolt were applied to detect gene transcription and translation levels, respectively. Laryngeal cancer cell migration was assayed by transwell chamber experiment.
Results
CREB protein expression was significantly up-regulated in laryngeal cancer tissues and associated with cancer differentiation, tumor stage, and lymphatic metastasis. CREB inhibits MYCT1 expression by direct binding to its promoter. Meanwhile, MYCT1 has a negative impact on the NAT10 gene expression. Furthermore, CREB promotes NAT10 expression via down-regulating the MYCT1 gene expression. In addition, contrary to MYCT1, CREB and NAT10 enhanced laryngeal cancer cell migration. MYCT1 and NAT10 significantly rescued the effects of CREB and MYCT1 on Hep2 cell migration, respectively.
Conclusion
CREB promotes laryngeal cancer cell migration via MYCT1/NAT10 axis, suggesting that CREB might be a potential prognostic marker in laryngeal cancer.
Keywords: laryngeal cancer, CREB, MYCT1, NAT10, migration
Introduction
Cancer is the second leading cause of death throughout the world.1 The prognosis of cancer patients is still poor because of the high frequency of cancer metastasis and recurrence.2,3 Studies have shown that cancer metastasis is related to abnormal gene expression and function in some signaling pathways.4–6 However, the molecular mechanisms of cancer metastasis remains vague. Thus, a better understanding of the molecular mechanisms responsible for cancer metastasis is crucial for identifying molecular targets to predict cancer prognosis.
Previous studies have revealed that CREB and NAT10 are associated with the migration of cancer cells. For example, Yang et al reported that CREB mediates liver cancer cell migration.7 Singh et al found that CREB plays a vital role in human breast cancer cell migration.8 CREB also participates in the epithelial-to-mesenchymal transition (EMT) of bladder cancer.9 Li et al discovered that CREB increases cancer cell migration.10 In colorectal cancer, the subcellular redistribution of NAT10 promotes cancer cell motility and NAT10 expression is correlated with invasive potential and poor clinical outcome, suggesting that NAT10 may be a useful prognostic marker and potential therapeutic target.11 In hepatocellular cancer, NAT10 mediates EMT and promotes metastasis.12
In our previous work, we found that MYCT1 over-expression could inhibit laryngeal cancer cell migration.13 However, the molecular mechanism of MYCT1 in laryngeal cancer cell migration is still unclear. In this study, we found a potential CRE binding site in the promoter of MYCT1 using bioinformatics analysis. Based on protein microarray detection, we discovered that NAT10 was down-regulated by MYCT1 in laryngeal cancer cells. Therefore, we speculate that CREB may participate in laryngeal cancer cell migration through MYCT1/NAT10 axis in laryngeal cancer.
Material and methods
Tissues and cell lines
Laryngeal cancer tissues and paired normal tissues were obtained from 35 patients in the Department of Otolaryngology, No 463 Hospital of PLA. All tissues were stored at -80°C immediately after being removed from the patients. All patients gave their written informed consent and the study was approved by the Ethics Committee of China Medical University. Human laryngeal cancer Hep2 cells were commercially purchased from the KeyGEN BioTECH Company (Nanjing, People’s Republic of China). Hep2 cells were cultured in RPMI 1640 (Thermo Fisher Scientific, Waltham, MA, USA) with 10% new-born calf serum (Hyclone, Logan, UT, USA) in a humidified atmosphere at 37°C in 5% CO2.
Bioinformatics prediction and plasmid construction
Two online software programs, TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) and JASPAR (http://jaspar.genereg.net/) were used to predict the possible transcription factors targeting MYCT1. Two fragments of MYCT1 5′-flanking sequence, -795/+12(P795) and -760/+12(P760), were cloned into pGL3-Basic vector (Promega Corporation, Fitchburg, WI, USA) by GenScript Company (Nanjing, People’s Republic of China). Complementary DNA fragments encoding human CREB, MYCT1, and NAT10 were constructed into pcDNA3.1 vector (GenScript Company).
Gene transfection
For transient transfection, Hep2 cells were transfected with the pcDNA3.1, pcDNA3.1-CREB, pcDNA3.1-MYCT1 or pcDNA3.1-NAT10 using Lipofectamine 3000 (Thermo Fisher Scientific). After 48 h of transfection, cells were harvested for further use.
RNA isolation and real-time polymerase chain reaction (PCR) detection
Total RNA was extracted from Hep2 cells using Trizol reagent (Takara, Dalian, People’s Republic of China). RNA concentration was measured by reading the absorbance at OD 260/280 nm. Real-time PCR detection was performed as previously described.14 All primer sequences used in the study are listed in Table 1. PCR conditions were as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s in ABI 7500 Real Time PCR system (Thermo Fisher Scientific). The transcription level of each gene was normalized by GAPDH as an internal control.
Table 1.
Primer sequences used in the study
| Name | Sequence |
|---|---|
| CREB-Forward primer | 5′-ACATTAGCCCAGGTATCTATGCCAG-3′ |
| CREB-Reverse primer | 5′-CCTGAATGACTCCATGGACTTGAAC-3′ |
| MYCT1-Forward primer | 5′-GCCAGAAAACTTTTGGGAGGA-3′ |
| MYCT1-Reverse primer | 5′-ATCCAGTTCTGTTGAGGCCG-3′ |
| NAT10-Forward primer | 5′-GGGATTGGCCTGCAGCATA-3′ |
| NAT10-Reverse primer | 5′-GGCTCCATGACCACATCCTT-3′ |
| GAPDH-Forward primer | 5′-TGGCACCCAGCACAATGAA-3′ |
| GAPDH-Reverse primer | 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′ |
| ChIP-Forward primer | 5′-ATAGTGGCCGAGCGCAGT-3′ |
| ChIP-Reverse primer | 5′-CACACCCGGCTAATTTTTGT-3′ |
Abbreviation: ChIP, chromatin immunoprecipitation.
Western blot analysis
Protein was isolated from tissues and cells were subjected to sonication in ice-cold radio immunoprecipitation assay lysis buffer. Denatured protein was separated on sodium dodecyl sulfate-polyacrylamide gel and transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). After blocking with 5% skim milk in tris buffered saline tween buffer for 2 h at room temperature, the samples were incubated with primary antibodies including CREB (Cell Signaling Technology, Danvers, MA, USA [9197]), MYCT1 (Abcam, Cambridge, UK [ab139945]), NAT10 (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA [sc-271770]) and GAPDH (Santa Cruz Biotechnology Inc. [sc-365062]), followed by incubation with appropriate peroxidase-coupled secondary antibodies. Proteins on PVDF membrane were visualized using an ECL kit (Thermo Fisher Scientific, Waltham, MA, USA). The relative integrated density values were calculated by ImageJ software and normalized with GAPDH.
Luciferase reporter assay
Hep2 cells were co-transfected with pcDNA3.1-CREB and MYCT1-promoter plasmid or Renilla. After transfection for 48 h, luciferase activities were analyzed using the Dual-Luciferase Reporter kit (Promega Corporation).
Chromatin immunoprecipitation (ChIP) assay
ChIP was performed using the ChIP express kit (Millipore). Briefly, Hep2 cells were fixed with 1% formaldehyde for 10 min. Immunoprecipitation was done with anti-CREB and anti-IgG antibodies. DNA was purified using spin columns and amplified by PCR. The primer sequences used in ChIP detection are also shown in Table 1.
Cell migration assay
Cell migration was assayed using a 24-well transwell chamber with a pore size of 8 µm (Corning Life Sciences, Corning, NY, USA). Cells were re-suspended in serum-free medium and then seeded on the top chamber. Medium with serum was added to the lower chamber as the chemoattractant. The cells on the lower chamber were fixed and then stained with Giemsa. The number of migrated cells was visualized and counted in randomly selected fields under a microscope.
Statistical analysis
All results are expressed as mean ± SD. Data were analyzed using Student’s t-test or one-way analysis of variance (ANOVA) by SPSS 19.0 (IBM Corporation, Armonk, NY, USA). P<0.05 was considered statistically significant.
Results
CREB expression in laryngeal cancer tissues
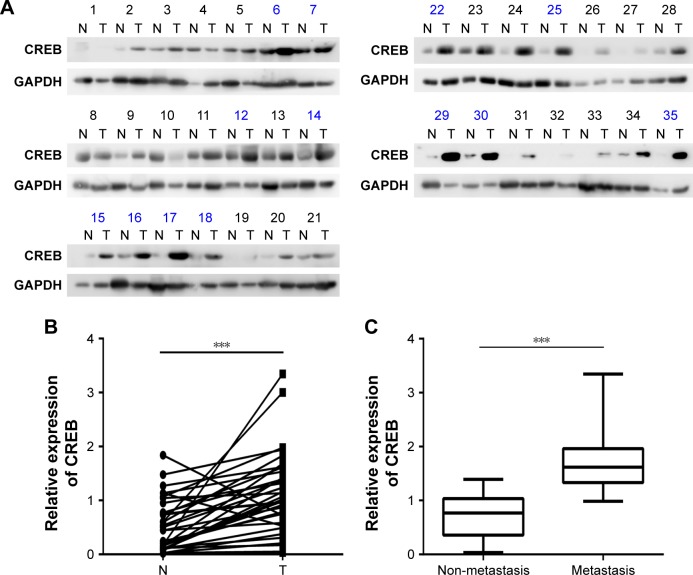
To explore the expression of CREB in laryngeal cancer tissues, Western blot was performed. The results showed that CREB was up-regulated in 28 of 35 (80%) laryngeal cancer tissues (Figure 1A). Statistically, CREB protein was significantly over-expressed in laryngeal cancer tissues in comparison with paired normal tissues (Figure 1B). Moreover, CREB protein expression was significantly higher in laryngeal cancer patients with lymphatic metastasis than in those without lymphatic metastasis (Figure 1C). In addition to lymphatic metastasis, we also found that CREB protein expression was significantly correlated with differentiation and tumor stage (Table 2), but not with age and gender (Table 2). These results suggested that CREB participates in the development of laryngeal cancer.
Figure 1.
Expression of CREB in LSCC tissues.
Notes: (A) CREB protein level in laryngeal cancer by Western blot. N, paired normal tissues; T, tumor tissues. Number in blue color indicates the tissue from the patient with metastasis. (B) Statistical analysis of CREB protein levels in laryngeal cancer. (C) Relative protein levels of CREB in cancer tissues of patients with metastasis and non-metastasis. ***P<0.001.
Abbreviation: LSCC, laryngeal squamous cell carcinoma.
Table 2.
Association of CREB expression with clinical characteristics in LSCC
| Clinicopathological characteristics | N | CREB expression (mean ± SD) |
P-value |
|---|---|---|---|
| Age (years) | 0.328 | ||
| <60 | 15 | 0.982±0.630 | |
| ≥60 | 20 | 1.232±0.810 | |
| Gender | 0.583 | ||
| Male | 28 | 1.161±0.790 | |
| Female | 7 | 0.986±0.506 | |
| Differentiation | <0.001 | ||
| Well differentiated | 11 | 0.811±0.491 | |
| Moderately and poorly differentiated | 24 | 1.813±0.491 | |
| TNM classification | <0.001 | ||
| I + II | 16 | 0.660±0.491 | |
| III + IV | 19 | 1.518±0.690 | |
| Lymph node metastasis | <0.001 | ||
| Positive | 13 | 1.797±0.681 | |
| Negative | 22 | 0.729±0.419 |
Note: One-way ANOVA was used to analyze the correlation between the CREB expression and clinical features of each patient.
Abbreviations: ANOVA, analysis of variance; LSCC, laryngeal squamous cell carcinoma.
CREB directly inhibits MYCT1 transcription
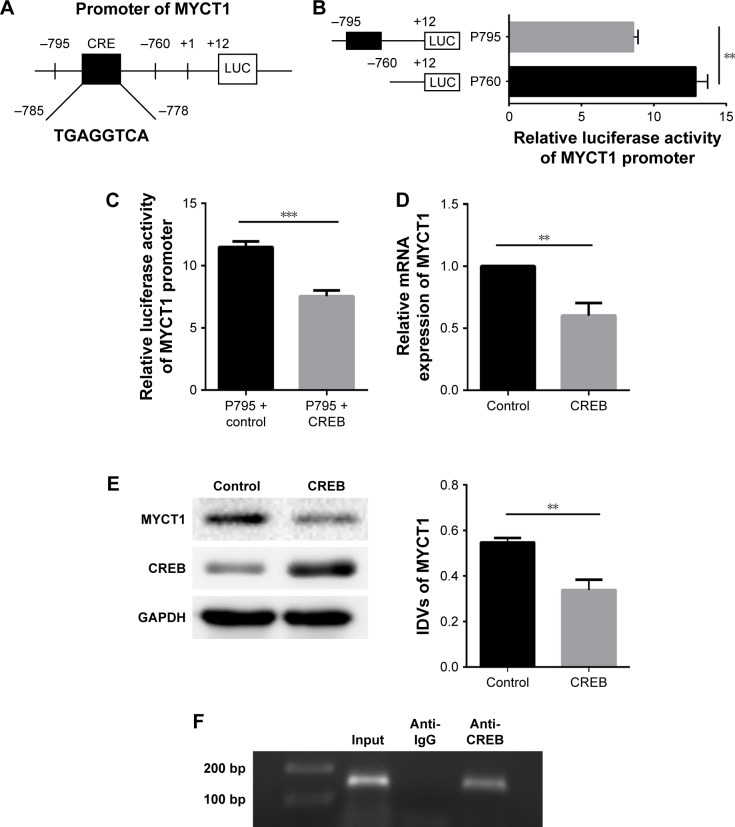
The results from TFSEARCH and JASPAR search programs revealed a CRE between -785 and -778 of MYCT1 promoter (Figure 2A), suggesting that MYCT1 is a potential target of CREB. We then constructed two expression vectors including -795/+12 (pGL3-P795) and -760/+12 (pGL3-P760) around the CRE region. Luciferase reporter gene assay results showed that the promoter activity of pGL3-P795 was significantly lower than that of pGL3-P760 (Figure 2B). Furthermore, the pGL3-P795 promoter activity was significantly decreased in CREB-transfected Hep2 cells compared to the control (Figure 2C). Both real-time PCR and Western blot results indicated that MYCT1 mRNA and protein levels in Hep2 cells transfected with CREB were significantly lower than those in the controls (Figure 2D and E). Chromatin immunoprecipitation detection results demonstrated that PCR product spanning the MYCT1 CRE sequence precipitated by anti-CREB primary antibody was obtained compared to the controls (Figure 2F). Thus, we conclude that CREB inhibits MYCT1 expression by direct binding to MYCT1 promoter.
Figure 2.
CREB inhibits MYCT1 transcription.
Notes: (A) Transcription factor CREB binding sites present in the MYCT1 promoter region. (B) MYCT1 promoter activity analysis. Hep2 cells were transfected with pGL3-P760 or pGL3-P795, followed by a luciferase reporter assay. Firefly luciferase activity was normalized to Renilla luciferase activity. (C) Effect of CREB on pGL3-P795 activity. pcDNA3.1 vector or pcDNA3.1-CREB was co-transfected with pGL3-P795 into Hep2 cells, followed by a luciferase reporter assay. (D) Effect of CREB on MYCT1 mRNA level. MYCT1 mRNA level was detected by qRT-PCR in Hep2 cells transfected with pcDNA3.1 or pcDNA3.1-CREB. (E) Effect of CREB on MYCT1 protein level. MYCT1 protein level was examined by Western blot in Hep2 cells transfected with pcDNA3.1 or pcDNA3.1-CREB. (F) Binding of CREB to MYCT1 in vivo. DNA fragments from Hep2 cells were amplified by PCR based on immunoprecipitation with anti-IgG or anti-CREB antibodies. **P<0.01; ***P<0.001.
Abbreviations: IDVs, integrated density values; qRT-PCR, quantitative reverse transcription-polymerase chain reaction.
CREB up-regulates NAT10 expression via repressing MYCT1 expression
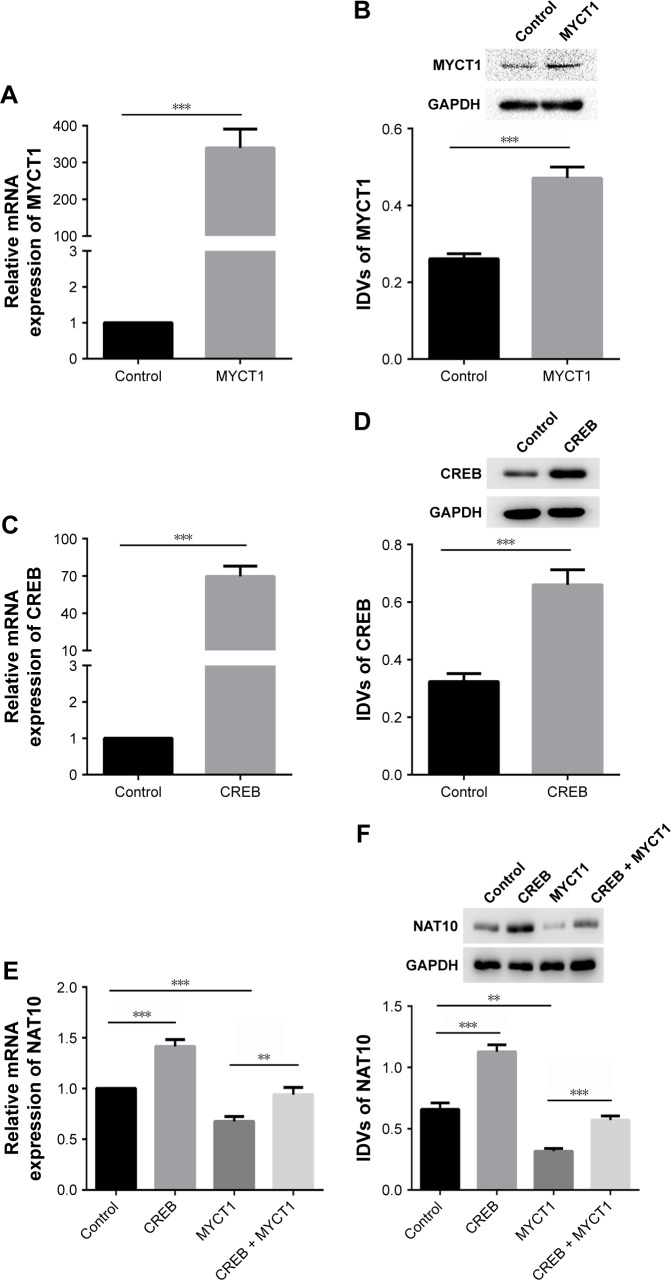
To further explore the downstream protein of MYCT1, a protein microarray was performed in two kinds of Hep2 cells transfected with pcDNA3.1-MYCT1 vector and pcDNA3.1-vector. The results showed that over 400 proteins were differentially expressed (Table S1). In particular, NAT10 was one of the most significantly down-regulated proteins in Hep2 cells with MYCT1 over-expression (Table S1). We found that MYCT1 could be down-regulated by CREB (Figure 2). Thus, we hypothesized that CREB may promote NAT10 expression by down-regulating MYCT1 gene expression. Compared to the controls, MYCT1 and CREB were significantly over-expressed both at mRNA and protein levels in Hep2 cells transfected with MYCT1 and CREB, indicating that transfection was successful (Figure 3A–D). We then detected the effects of MYCT1 and CREB on NAT10 expression in Hep2 cells. Results showed that MYCT1 and CREB significantly inhibited and enhanced the NAT10 gene expression in Hep2 cells compared to the controls (Figure 3E and F). Meantime, MYCT1 significantly rescued the effect of CREB on NAT10 expression both at mRNA and protein levels in Hep2 cells (Figure 3E and F). Taken together, these results indicate that CREB promotes NAT10 expression via down-regulating the MYCT1 gene expression.
Figure 3.
CREB up-regulates NAT10 expression via repressing MYCT1 expression.
Notes: (A) MYCT1 mRNA level in MYCY1-transfected Hep2 cells by real-time PCR method. (B) MYCT1 protein level in MYCY1-transfected Hep2 cells by Western blot detection. (C) CREB mRNA level in CREB-transfected Hep2 cells by real-time PCR method. (D) CREB protein level in CREB-transfected Hep2 cells by Western blot detection. (E) Effects of CREB and MYCT1 on NAT10 mRNA level in Hep2 cells by real-time PCR method. (F) Effects of CREB and MYCT1 on NAT10 protein level in Hep2 cells by Western blot detection. **P<0.01; ***P<0.001.
Abbreviation: IDVs, integrated density values.
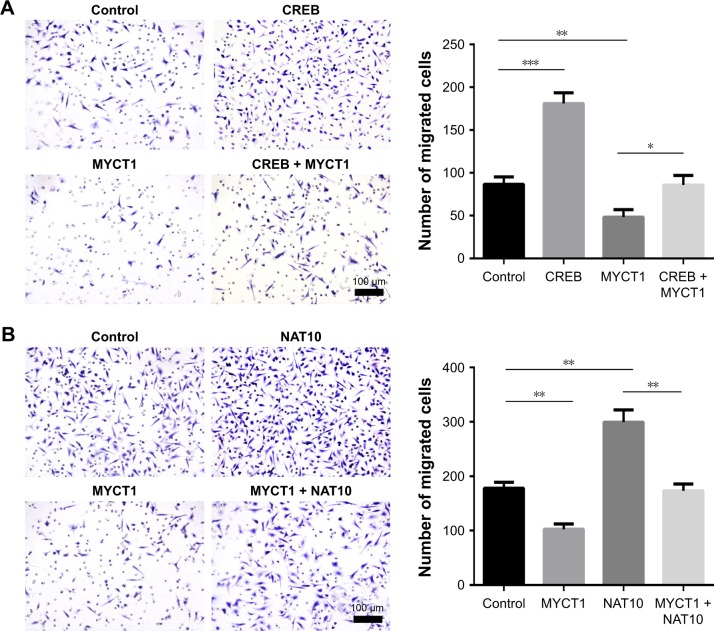
CREB promotes laryngeal cancer cell migration via MYCT1/NAT10 axis
We then investigated the roles of CREB, MYCT1, and NAT10 in laryngeal cancer cell migration. The results showed that numbers of migrated Hep2 cells transfected by CREB and MYCT1 were significantly increased and decreased compared to the controls, respectively, and MYCT1 significantly rescued the effect of CREB on Hep2 cell migration (Figure 4A). Similarly, NAT10 and MYCT1 significantly promoted and suppressed the Hep2 cell migration, respectively, and NAT10 significantly rescued the effect of MYCT1 on Hep2 cell migration (Figure 4B). Taken together, we conclude that CREB promotes laryngeal cancer cell migration via MYCT1/NAT10 axis.
Figure 4.
CREB promotes laryngeal cancer cell migration via MYCT1/NAT10 axis.
Notes: (A) Effects of CREB and MYCT1 on Hep2 cell migration. (B) Effects of MYCT1 and NAT10 on Hep2 cell migration. *P<0.05; **P<0.01; ***P<0.001.
Discussion
CREB transcription factor, a member of the CREB/ATF family, plays a key role in cell growth, proliferation, differentiation, apoptosis, and migration in human diseases.8,15–19 However, studies on CREB in laryngeal cancer are seldom reported. In our study, we demonstrated that CREB is highly expressed in the laryngeal cancer tissues and is positively correlated with cancer metastasis. Consistent with our results, CREB is over-expressed in different kinds of cancer such as gliomas, ovarian adenocarcinoma, liver cancer, and colorectal cancer.20–23
Based on the bioinformatics analysis, we found that MYCT1 is a potential target of CREB. Luciferase reporter results showed that CRE is a negative response element within MYCT1 promoter region. In Hep2 cells, CREB inhibited MYCT1 expression. ChIP assay results indicated that CREB directly binds the CRE sequence of MYCT1. These results suggest that CREB represses MYCT1 expression by directly binding to MYCT1 promoter. In our previous work, we found that MYC and YY1 directly promote and inhibit the transcription activities of MYCT1 in Hep2 cells.13,24 In addition to MYCT1, there are other targets of CREB. For example, CREB is a positive transcript factor by binding to the promoters of HULC and YAP in liver cancer.22,25 On the contrary, CREB is a negative regulator of CA9 and AP2-alpha in gastric cancer and melanoma.26,27
In the study, we discovered that MYCT1 suppresses NAT10 expression in Hep2 cells, further validating our protein microarray detection result. Moreover, CREB up-regulates NAT10 expression in Hep2 cells and the effect is rescued by MYCT1, suggesting that there exists a CREB/MYCT1/NAT10 axis in laryngeal cancer cells. We also found that both CREB and NAT10 enhanced laryngeal cancer cell migration contrary to MYCT1. Furthermore, MYCT1 and NAT10 could rescue the effects of CREB and MYCT1 on Hep2 cell migration. These results imply that CREB participates in Hep2 cell migration via MYCT1/NAT10 axis.
Several studies have reported that CREB is involved in some cancer metastasis through different signaling pathways. In colorectal cancer, miR-199a is associated with distant metastasis via activation of SIRT1 and inhibition of CREB/KISS1 signaling.28 In ovarian cancer, calcium dependent FAK/CREB/TNNC1 axis mediates the effect of MFAP5 on cancer metastatic potential.29
In this study, we first found that CREB protein expression is up-regulated and correlated with differentiation, tumor stage, and lymphatic metastasis in laryngeal cancer tissue. The novel CREB/MYCT1/NAT10 axis participates in laryngeal cancer cell migration. However, the mechanism of how MYCT1 regulates NAT10 expression is still unclear, which is a key problem to be solved in our future studies.
Conclusion
Taken together, our results indicate that CREB up-regulates NAT10 expression via inhibiting MYCT1 transcription leading to laryngeal cancer cell migration. CREB may serve as a critical target in prediction of laryngeal cancer prognosis.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81372876).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Zeng YJ, Zhang CD, Dai DQ. Impact of lymph node micrometastasis on gastric carcinoma prognosis: a meta-analysis. World J Gastroenterol. 2015;21(5):1628–1635. doi: 10.3748/wjg.v21.i5.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toiyama Y, Tanaka K, Kitajima T, et al. Serum angiopoietin-like protein 2 as a potential biomarker for diagnosis, early recurrence and prognosis in gastric cancer patients. Carcinogenesis. 2015;36(12):1474–1483. doi: 10.1093/carcin/bgv139. [DOI] [PubMed] [Google Scholar]
- 4.Gugnoni M, Sancisi V, Gandolfi G, et al. Cadherin-6 promotes EMT and cancer metastasis by restraining autophagy. Oncogene. 2017;36(5):667–677. doi: 10.1038/onc.2016.237. [DOI] [PubMed] [Google Scholar]
- 5.Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samaeekia R, Adorno-Cruz V, Bockhorn J, et al. miR-206 Inhibits Stemness and Metastasis of Breast Cancer by Targeting MKL1/IL11 Pathway. Clin Cancer Res. 2017;23(4):1091–1103. doi: 10.1158/1078-0432.CCR-16-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Z, Tsuchiya H, Zhang Y, Hartnett ME, Wang L. MicroRNA-433 inhibits liver cancer cell migration by repressing the protein expression and function of cAMP response element-binding protein. J Biol Chem. 2013;288(40):28893–28899. doi: 10.1074/jbc.M113.502682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh R, Shankar BS, Sainis KB. TGF-beta1-ROS-ATM-CREB signaling axis in macrophage mediated migration of human breast cancer MCF7 cells. Cell Signal. 2014;26(7):1604–1615. doi: 10.1016/j.cellsig.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 9.Xu X, Zhu Y, Liang Z, et al. c-Met and CREB1 are involved in miR-433- mediated inhibition of the epithelial-mesenchymal transition in bladder cancer by regulating Akt/GSK-3beta/Snail signaling. Cell Death Dis. 2016;7:e2088. doi: 10.1038/cddis.2015.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li D, Jin L, Alesi GN, et al. The prometastatic ribosomal S6 kinase 2-cAMP response element-binding protein (RSK2-CREB) signaling pathway up-regulates the actin-binding protein fascin-1 to promote tumor metastasis. J Biol Chem. 2013;288(45):32528–32538. doi: 10.1074/jbc.M113.500561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Hou W, Wang HL, et al. GSK-3beta-regulated N-acetyltransferase 10 is involved in colorectal cancer invasion. Clin Cancer Res. 2014;20(17):4717–4729. doi: 10.1158/1078-0432.CCR-13-3477. [DOI] [PubMed] [Google Scholar]
- 12.Ma R, Chen J, Jiang S, Lin S, Zhang X, Liang X. Up regulation of NAT10 promotes metastasis of hepatocellular carcinoma cells through epithelial-to-mesenchymal transition. Am J Transl Res. 2016;8(10):4215–4223. [PMC free article] [PubMed] [Google Scholar]
- 13.Fu S, Guo Y, Chen H, et al. MYCT1-TV, a novel MYCT1 transcript, is regulated by c-Myc and may participate in laryngeal carcinogenesis. PLoS One. 2011;6(10):e25648. doi: 10.1371/journal.pone.0025648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S, Sun YY, Zhang ZX, Li YH, Xu ZM, Fu WN. Transcriptional suppression of microRNA-27a contributes to laryngeal cancer differentiation via GSK-3beta-involved Wnt/beta-catenin pathway. Oncotarget. 2017;8(9):14708–14718. doi: 10.18632/oncotarget.14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Pietro R, di Giacomo V, Caravatta L, Sancilio S, Rana RA, Cataldi A. Cyclic nucleotide response element binding (CREB) protein activation is involved in K562 erythroleukemia cells differentiation. J Cell Biochem. 2007;100(4):1070–1079. doi: 10.1002/jcb.21106. [DOI] [PubMed] [Google Scholar]
- 16.Caravatta L, Sancilio S, di Giacomo V, Rana R, Cataldi A, Di Pietro R. PI3-K/Akt-dependent activation of cAMP-response element-binding (CREB) protein in Jurkat T leukemia cells treated with TRAIL. J Cell Physiol. 2008;214(1):192–200. doi: 10.1002/jcp.21186. [DOI] [PubMed] [Google Scholar]
- 17.Muranen T, Selfors LM, Hwang J, et al. ERK and p38 MAPK Activities Determine Sensitivity to PI3K/mTOR Inhibition via Regulation of MYC and YAP. Cancer Res. 2016;76(24):7168–7180. doi: 10.1158/0008-5472.CAN-16-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Illiano M, Nigro E, Sapio L, et al. Adiponectin down-regulates CREB and inhibits proliferation of A549 lung cancer cells. Pulm Pharmacol Ther. 2017;45:114–120. doi: 10.1016/j.pupt.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Knapp DJ, Hammond CA, Aghaeepour N, et al. Distinct signaling programs control human hematopoietic stem cell survival and proliferation. Blood. 2017;129(3):307–318. doi: 10.1182/blood-2016-09-740654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan X, Wang S, Zhu L, et al. cAMP response element-binding protein promotes gliomagenesis by modulating the expression of oncogenic microRNA-23a. Proc Natl Acad Sci U S A. 2012;109(39):15805–15810. doi: 10.1073/pnas.1207787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linnerth NM, Greenaway JB, Petrik JJ, Moorehead RA. cAMP response element-binding protein is expressed at high levels in human ovarian adenocarcinoma and regulates ovarian tumor cell proliferation. Int J Gynecol Cancer. 2008;18(6):1248–1257. doi: 10.1111/j.1525-1438.2007.01177.x. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Ma L, Weng W, et al. Mutual interaction between YAP and CREB promotes tumorigenesis in liver cancer. Hepatology. 2013;58(3):1011–1020. doi: 10.1002/hep.26420. [DOI] [PubMed] [Google Scholar]
- 23.Li P, Xue WJ, Feng Y, Mao QS. MicroRNA-205 functions as a tumor suppressor in colorectal cancer by targeting cAMP responsive element binding protein 1 (CREB1) Am J Transl Res. 2015;7(10):2053–2059. [PMC free article] [PubMed] [Google Scholar]
- 24.Qu SY, Sun YY, Li YH, Xu ZM, Fu WN. YY1 directly suppresses MYCT1 leading to laryngeal tumorigenesis and progress. Cancer Med. 2017;6(6):1389–1398. doi: 10.1002/cam4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G, Cheng Z, Liu F, Zhang H, Li J, Li F. CREB is a key negative regulator of carbonic anhydrase IX (CA9) in gastric cancer. Cell Signal. 2015;27(7):1369–1379. doi: 10.1016/j.cellsig.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 27.Melnikova VO, Dobroff AS, Zigler M, et al. CREB inhibits AP-2alpha expression to regulate the malignant phenotype of melanoma. PLoS One. 2010;5(8):e12452. doi: 10.1371/journal.pone.0012452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen ZL, Wang B, Jiang KW, et al. Downregulation of miR-199b is associated with distant metastasis in colorectal cancer via activation of SIRT1 and inhibition of CREB/KISS1 signaling. Oncotarget. 2016;7(23):35092–35105. doi: 10.18632/oncotarget.9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung CS, Yeung TL, Yip KP, et al. Calcium-dependent FAK/CREB/TNNC1 signalling mediates the effect of stromal MFAP5 on ovarian cancer metastatic potential. Nat Commun. 2014;5:5092. doi: 10.1038/ncomms6092. [DOI] [PMC free article] [PubMed] [Google Scholar]