Abstract
Bovine serum albumin (BSA) adsorbed on amorphous silicon dioxide (SiO2) nanoparticles was studied as a function of pH across the range of 2 to 8. Aggregation, surface charge, surface coverage, and protein structure were investigated over this entire pH range. SiO2 nanoparticle aggregation is found to depend upon pH and differs in the presence of adsorbed BSA. For SiO2 nanoparticles truncated with hydroxyl groups, the largest aggregates were observed at pH 3, close to the isoelectric point of SiO2 nanoparticles, whereas for SiO2 nanoparticles with adsorbed BSA, the aggregate size was the greatest at pH 3.7, close to the isoelectric point of the BSA-SiO2 complex. Surface coverage of BSA was also the greatest at the isoelectric point of the BSA-SiO2 complex with a value of ca. 3 ± 1 × 1011 molecules cm−2. Furthermore, the secondary protein structure was modified when compared to the solution phase at all pH values, but the most significant differences were seen at pH 7.4 and below. It is concluded that protein–nanoparticle interactions vary with solution pH, which may have implications for nanoparticles in different biological fluids (e.g., blood, stomach, and lungs).
I. INTRODUCTION
Nanotechnology is a rapidly growing field, thereby increasing concerns over potential human exposure to and environmental accumulation of nanoparticles. Oxide nanoparticles, including silicon dioxide (SiO2), are widely used in industrial applications, consumer products, and the biomedical field.1–3 Commercially available nanoparticles approved for food and pharmaceutical use, Cab-O-Sil HS-5, have been chosen for these studies to span the range of potential applications. The potential of these nanoparticles to enter the human body increases with each new application and use. Furthermore, the disposal of these particles includes disposing into water bodies. Therefore, it is critical that nanoparticle interactions with biological systems and biological components are studied in aqueous environments.
Oxide nanoparticles, including metal oxide nanoparticles, have been widely studied for protein–nanoparticle interactions.4 Certain metal oxide nanoparticles, such as zinc oxide, can readily dissociate in aqueous media leaching metal ions which can cause toxicity.5–7 SiO2 nanoparticles are of interest because they are widely accepted as nontoxic8 and therefore are becoming increasingly utilized.9 As a result, their potential for accumulation and transport in aqueous environments is high.
Nanoparticles that enter the human body are immediately coated with a protein corona, a dynamic entity comprised of biological components that encounter the nanoparticle including mainly proteins as well as some salts and other molecules.10,11 The protein corona then interacts directly with cells and tissues in the body and determines the fate of nanoparticles.10–12 Therefore, studying the nature of the protein corona becomes crucial in determining the implications of internalized nanoparticles toward human health. For these studies, we have used a model protein, bovine serum albumin (BSA), to investigate the effects of pH on the corona.
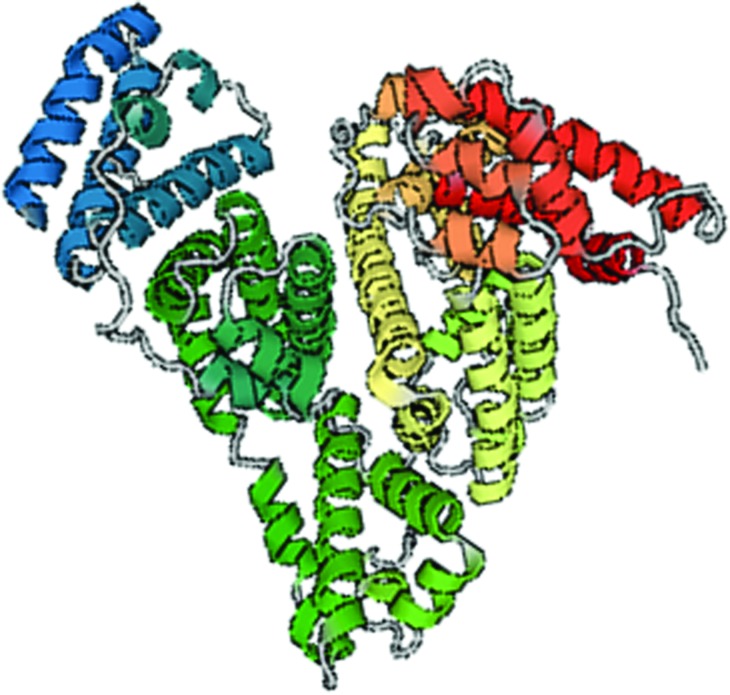
Serum albumins are important for human health studies because they are the most abundant protein in the blood, making up slightly more than half of the total protein content.13,14 Furthermore, BSA is 98% similar to the human analog while being more widely studied.15,16 The pH-dependent structural changes of both BSA and its human analog human serum albumin have been well characterized.17–22 BSA exists in three known forms, depending upon pH. The normal (N) form exists above pH 4.5, at physiological conditions, and has a compact, triangular, or heart shape (Fig. 1).23 As the pH lowers toward more acidic conditions, the BSA molecule extends its structure, becoming more linear.15,16 The extended (E) form of BSA exists below pH 4.0.15,16 Between the N- and E-forms of BSA, there exists the fast (F) form (pH 4.0–4.5); in this region, BSA quickly transitions from the N-form to the E-form.15,16 Further, the secondary structure of BSA has been studied using circular dichroism spectroscopy and attenuated total reflectance Fourier transform infrared (ATR-FTIR) spectroscopy. Most commonly, the alpha-helix content reported for BSA in aqueous media is 66%–68%.24–26 However, it has been shown that under acidic conditions and/or upon adsorption, the alpha helical structure of BSA decreases.13,27
Fig. 1.
Structure of BSA in its native form, “N-form,” as appears at physiological pH (PDB ID 4F5S).
The adsorption of proteins, including BSA, on nanoparticle surfaces has been investigated in terms of impacts on nanoparticle aggregation, protein coverage, and protein structure.27–33 Investigations on the effects of pH have been conducted and shown that protein adsorption is the greatest at or near the isoelectric point,34–37 and there is not necessarily a pH-dependent trend for protein adsorption to nanoparticle surfaces away from the isoelectric point.37 Aggarwal et al. conducted a study on pure silica and zirconia nanoparticle surfaces and a mixture of nanoparticle surfaces and found that the composite surface better retains the native protein structure upon adsorption.38
ATR-FTIR spectroscopy is particularly useful for real-time in situ analysis of protein adsorption on metal oxide nanoparticles.30,39–42 This technique probes the interfacial region between a nanoparticle thin film and the protein due to the evanescent wave produced from each reflection as well as the transparency of the metal oxide nanoparticle layer in the infrared region.39,42 Proteins exhibit characteristic absorption bands due to vibrational modes of the protein backbone. In particular, three amide regions exist, the amide I region which extends from 1600 to 1700 cm−1 is mainly due to C=O stretching; the amide II and III regions are due (mainly) to the out-of-phase and in-phase coupled motions of the C-N stretching and the N-H bending, respectively.13,43,44 The amide I and amide III regions can be used to determine secondary structure information; however, the amide III band has a much lower intensity than amide I, and therefore, the amide I peak is often used for secondary structure analyses.33,45
The amide I band has well characterized component bands of different frequencies corresponding to secondary protein structural motifs such as alpha helices.13,24,28,40,43,44,46–48 Additionally, the amide I peak will shift to lower wavenumbers when there are an increasing number of hydrogen bonds between the protein and the surface.49 The frequencies of these absorbance bands may shift as a function of pH, and additional bands arise due to protonation/deprotonation of acidic and basic sites within the amino acid residues that make up the protein.41,42 pH-dependent studies of proteins on nanoparticle surfaces have indicated that the isoelectric point of the protein corresponds to the greatest adsorption onto the nanoparticle surface, independent of the point of zero charge for the nanoparticle substrate.34–37 However, other studies indicate that the isoelectric point of the substrate determines maximum protein adsorption.48,50 Quantification of protein adsorption has been done in the past using ATR-FTIR spectroscopy in an equilibrium state; however, here we use thermogravimetric analysis (TGA) to determine the number of protein molecules which adsorb to a nanoparticle surface.37,40 Previous studies of BSA adsorption have focused primarily on secondary structure analysis rather than protein surface coverage.13,31,33,51,52
In this study, we combine ATR-FTIR spectroscopy and TGA to obtain the time- and pH-dependent spectra of BSA on SiO2 and quantify the adsorption and the structure of BSA on the surface. Protein adsorption, protein coverage, and protein structure have not been investigated previously in an integrated study. We also have measured nanoparticle aggregation and surface charge using dynamic light scattering (DLS) to determine the isoelectric points of our materials and the tendency toward aggregation of these nanoparticles in aqueous environments. The results from these experiments provide an important step in better understanding protein adsorption on nanoparticle surfaces and the resulting protein corona as a function of solution pH.
II. EXPERIMENT
A. Nanoparticle characterization
The hydrophilic fumed SiO2, Cab-O-Sil HS-5®, was obtained from the Cabot Corporation and used with no further processing. Transmission electron microscopy (TEM, JEOL JEM-1230) was used to determine the size and size distribution of individual nanoparticles. A 100 μg/ml solution of SiO2 nanoparticles in isopropanol was prepared and sonicated using a probe sonicator for 10 min at an amplitude of 40% in 20 s on and 10 s off intervals. A single drop was added to a Formvar-coated copper TEM grid (Ted-Pella, Inc.) and allowed to dry overnight before imaging. The particle size was determined using imagej software. Brunauer-Emmett-Teller (BET, Quantachrome BET Nova 4200e) analysis was used to determine the surface area of the nanoparticles. SiO2 nanoparticles were allowed to degas at 300 °C overnight before running seven-point N2-BET analysis.
B. Hydrodynamic diameter and surface charge
DLS (Beckman-Coulter Delsa Nano C) was used to determine the hydrodynamic diameter of particles in solution as a function of pH. Using the same instrument, the zeta potential was also measured on the same samples to determine the surface charge as a function of pH. To determine the impact of protein adsorption on the nanoparticle size and zeta potential, 5 mg of SiO2 was mixed with 5 ml of 1 mg/ml BSA (>96% purity, Sigma-Aldrich). Nanoparticles with or without protein were suspended in pH-adjusted solution. Pure Optima® water was adjusted to the pH of interest using 1 M HCl or 0.8 M NaOH. Suspensions were sonicated in a bath sonicator for 10 min and then stored overnight to equilibrate the protein coating protein coating. Immediately before measuring and to prevent nanoparticle settling, suspensions were again sonicated for 10 min in a bath sonicator. Measurements in the presence and the absence of BSA were compared. Samples were run in triplicate, using the method of cumulants to determine the diameter and its standard deviation.
C. Surface coverage
TGA (Pyris 1 TGA, Perkin-Elmer) was used to quantify BSA adsorption on SiO2 nanoparticles. A mass of 4 mg of SiO2 was mixed with 1 ml of 1 mg/ml BSA solution. Samples were sonicated for 20 min before incubation at 4 °C for 24 h. The incubation time and temperature were used to obtain a uniform coating and prevent bacterial growth, respectively. Samples were washed by centrifugation at 1000 g for 20 min. The supernatant was removed, and 1 ml of pure water was added. The nanoparticles were resuspended in water by mixing via pipetting. Samples were dried at room temperature for two days. For TGA measurements, samples were heated at a rate of 5 °C/min from room temperature to 700 °C under a flow of air. The mass loss below 100 °C was assumed to be due to water, and the mass loss above 100 °C was assumed to be due to protein.40 The amount of BSA molecules per surface area of SiO2 was calculated. This calculation was done using the specific surface area measured by BET analysis, the initial mass of the nanoparticles alone and the mass of the desorbed protein, according to Lehman et al.40
D. Protein adsorption measurements using vibrational spectroscopy
Real-time in situ analysis of BSA adsorption on SiO2 nanoparticles was investigated using ATR-FTIR (Thermo-Fisher) spectroscopy. A suspension of 8 mg of SiO2 nanoparticles per 1 ml of Optima® water was prepared and sonicated for 5 min. This uniform suspension was deposited into an even layer on the amorphous material transmitting infrared radiation crystal element (PIKE Technologies) and allowed to dry overnight under a stream of dry air purge. A horizontal flow cell was attached to the unit, and pure Optima® water was run through the flow cell at approximately 1 ml/min to remove any loosely bound nanoparticles from the crystal element. A 1 mg/ml solution of BSA was prepared in pH-adjusted water and run through the flow cell for 90 min with spectra collected every 10 min. Then, pure Optima® water was run through the flow cell for 60 min with spectra collected every 10 min to observe desorption behavior.
E. Protein secondary structure determination
ATR-FTIR spectra of BSA adsorption onto SiO2 nanoparticles over time were further processed for direct comparisons between conditions. First, the spectrum of water on SiO2 nanoparticles was subtracted from each time point to achieve a linear baseline at 1800 cm−1, which helps ensure that the amide peaks are due to protein adsorption and not water. Then, spectra were curve fitted to a Gaussian–Lorentzian shape using five component peaks in the amide I region, determined from the second derivative spectra of the solution phase BSA and constrained by previous literature values.13,24,46 Curve fitted spectra were further modified to have a linear baseline at zero in the amide I region and a peak maximum of exactly 1 absorbance unit in the amide I region.
III. RESULTS AND DISCUSSION
A. Nanoparticle characterization
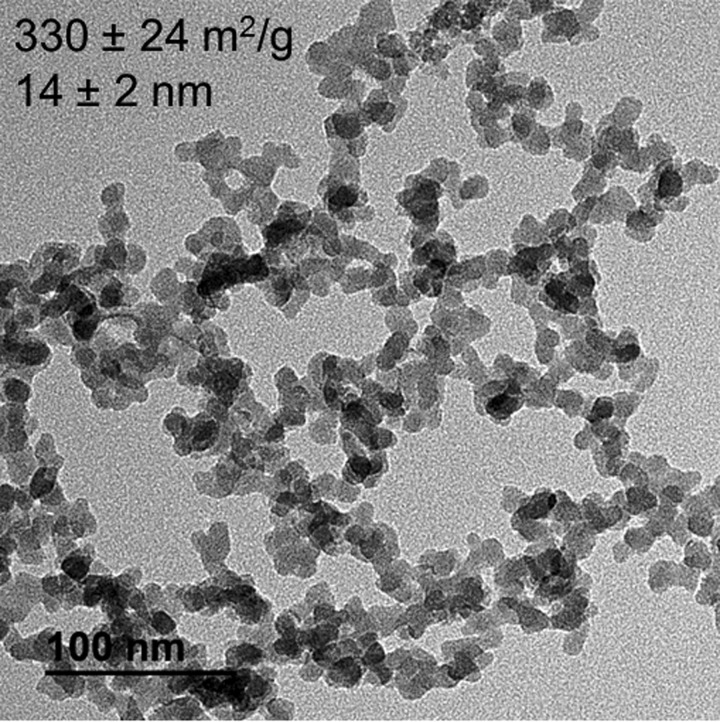
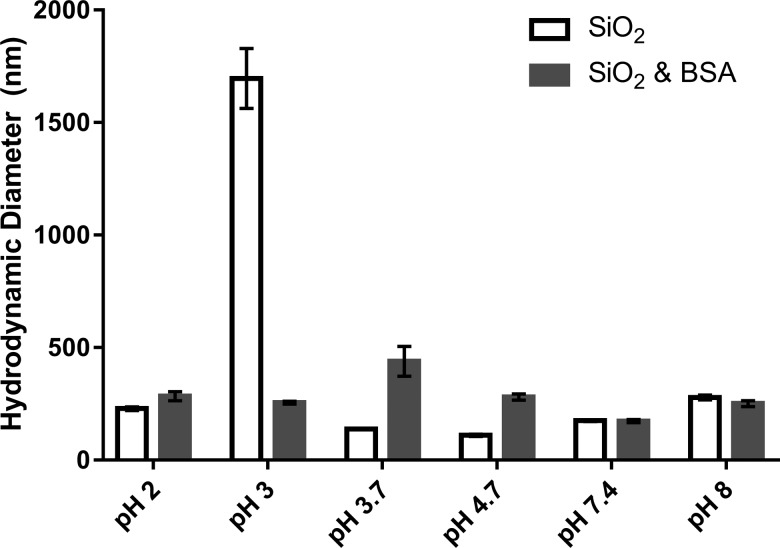
Nanoparticles were characterized according to their size, shape, and crystallinity via TEM (Fig. 2). Additionally, TEM confirmed that the nanoparticles were present as aggregates of approximately 250 nm. Analysis using ImageJ over 50 particles indicated that the average diameter of primary particles was 14 ± 2 nm. The specific surface area using BET analysis of three samples each weighing approximately 170 mg was determined to be 330 ± 24 m2/g. The hydrodynamic diameter provided information on aggregate sizes in aqueous solution. At most pH values, the hydrodynamic diameter was between 200 and 300 nm regardless of the presence or the absence of BSA (Fig. 3). However, at or near the isoelectric point, a large degree of aggregation occurred between particles due to decreased electrostatic interactions, or less repulsion, and subsequent charge stabilization. This occurred at pH 3.0 for pristine SiO2 nanoparticles and at pH 3.7 for BSA coated SiO2 nanoparticles.
Fig. 2.
TEM image of Cab-O-Sil HS-5; the scale bar is equal to 100 nm; the average particle size is 14 ± 2 nm based on fifty distinct particles, and the BET specific surface area is equal to 330 ± 24 m2/g.
Fig. 3.
Hydrodynamic diameter of bare SiO2 nanoparticles (white) and BSA-coated SiO2 nanoparticles (black) as a function of pH. The values reported are the mean of three trials, and error bars represent one standard deviation from the mean.
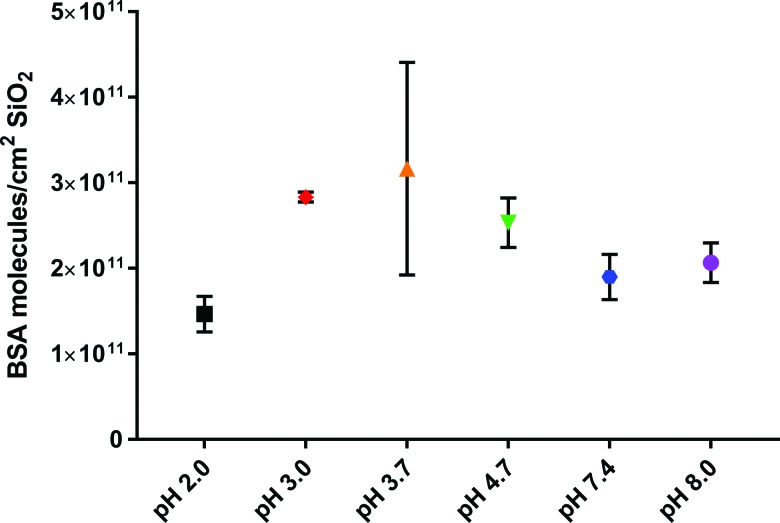
The nanoparticle zeta potential was measured over a range of pH values, and it was confirmed that pH 3.0 and 3.7 are closest to the isoelectric points of SiO2 and BSA-SiO2, respectively (Table I). It has been reported that at the isoelectric point of the nanoparticle, surface adsorption was enhanced.53,54 For bare silica, the isoelectric point occurred at pH 3.0. Other groups found that the protein isoelectric point was optimal for adsorption, which occurs at pH 4.7.37,55 Therefore, it was expected that either pH 3.0 or 4.7 would result in the greatest degree of adsorption. Here, we find that the greatest amount of protein adsorption, measured quantitatively from TGA (Fig. 4 and Table S1),62 occurs in the pH range of 3–4.7 with the value of the surface coverage being the greatest at pH 3.7, the isoelectric point of the nanoparticle–protein complex. However, that value also has the greatest error associated with it. This may be due to the fact that aggregates are the largest at this value and aggregation can play a role in the available surface area and can impact these measurements.
Table I.
Zeta (ζ) potential of native SiO2 nanoparticles and BSA-coated SiO2 nanoparticles (1 mg/ml SiO2, BSA) Samples were run in triplicate and the mean and standard deviation are reported.
| pH | ζ-potential (mV) SiO2 | ζ-potential (mV) BSA-SiO2 |
|---|---|---|
| 2.0 | 2.4 ± 0.5 | 23.9 ± 2.8 |
| 3.0 | 0.9 ± 0.1 | 27.1 ± 0.7 |
| 3.7 | −4.7 ± 0.2 | 0.4 ± 0.1 |
| 4.7 | −7.3 ± 1.2 | −24.3 ± 1.2 |
| 7.4 | −22.6 ± 0.5 | −22.3 ± 2.7 |
| 8.0 | −18.6 ± 2.2 | −30.5 ± 1.9 |
Fig. 4.
Surface coverage of BSA on SiO2 nanoparticles determined from thermogravimetric analysis. The reported values are mean ± standard deviation.
The zeta potential of the SiO2 particles changes in the presence of BSA at all pH, except pH 7.4, the physiological pH (Table I). In general, above the isoelectric point, the zeta potential will be negative, whereas below the isoelectric point, the zeta potential will be positive. The zeta potential was measured as a function of pH and provided as a plot previously.56 The measured zeta potentials are not linear.37 This suggests that there are compounding effects. One such effect is the protonation and deprotonation of functional groups in the amino acid residues within BSA as a function of pH. Furthermore, the protonation and deprotonation of amino acids changes as a function of pH, which may influence how BSA binds to the nanoparticle surface as well as the surface charge.
B. Protein adsorption as a function of time and pH
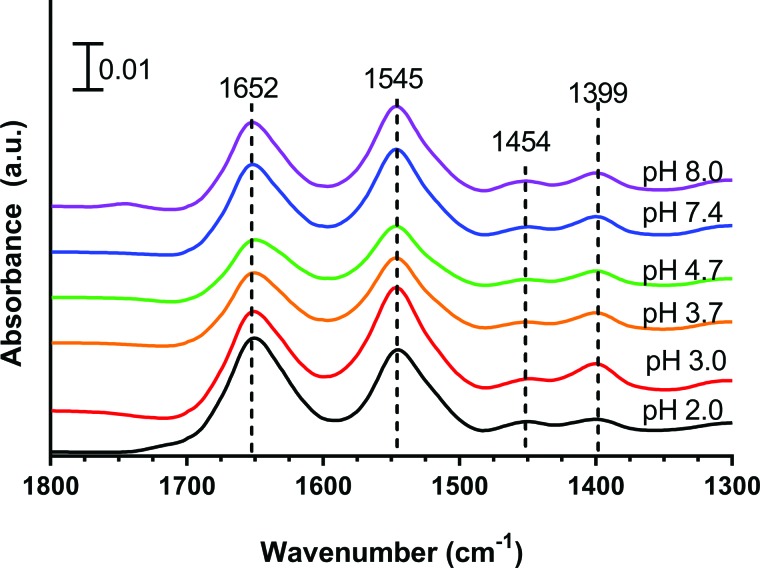
ATR-FTIR spectra of solution phase BSA at 10 mg/ml were collected over a range of pH values to determine peak locations for BSA (Fig. 5). The characteristic IR absorption bands of proteins are commonly referred to as the amide I, II, and III regions. The amide I region extends from 1600 to 1700 cm−1. The amide II and III regions arise from the combined motions of the C-N stretching and the N-H bending and range from 1500 to 1600 cm−1 and 1200 to 1350 cm−1, respectively. Peaks above 1700 cm−1 are due to the stretching motion of the C=O for protonated carboxylic acid groups of the amino acid residues. The band at around 1454 cm−1 is due to the CH2 scissoring motion of these methylene groups.
Fig. 5.
ATR-FTIR spectra of 10 mg/ml aqueous BSA as a function of pH. The characteristic IR absorption bands of proteins are associated with the amide I, II, and III regions. The amide I, II, and III regions extend from 1600–1700, 1500–1600, and 1200–1350 cm−1, respectively. The 1454 cm−1 band is typically associated with δ(CH2) vibrations. The band above 1700 cm−1 is due to the stretching motion of the C=O, for protonated carboxylic acid groups. Peak frequencies of the amide I band vary by approximately 2 cm−1, and the peak position of the amide II band varies within 1 cm−1 as a function of pH.
As proteins adsorb onto nanoparticle surfaces, they may undergo conformational changes.13,46 In some cases, this can be seen by frequency shifts of the absorbance bands to higher or lower wavenumbers. In particular, peak frequency shifts to lower wavenumbers in the amide I band are indicative of increasing hydrogen bonds.49 BSA in the solution phase exhibited very small peak shifts of ±3 cm−1 in the amide I and amide II regions as a function of pH (Fig. 5).
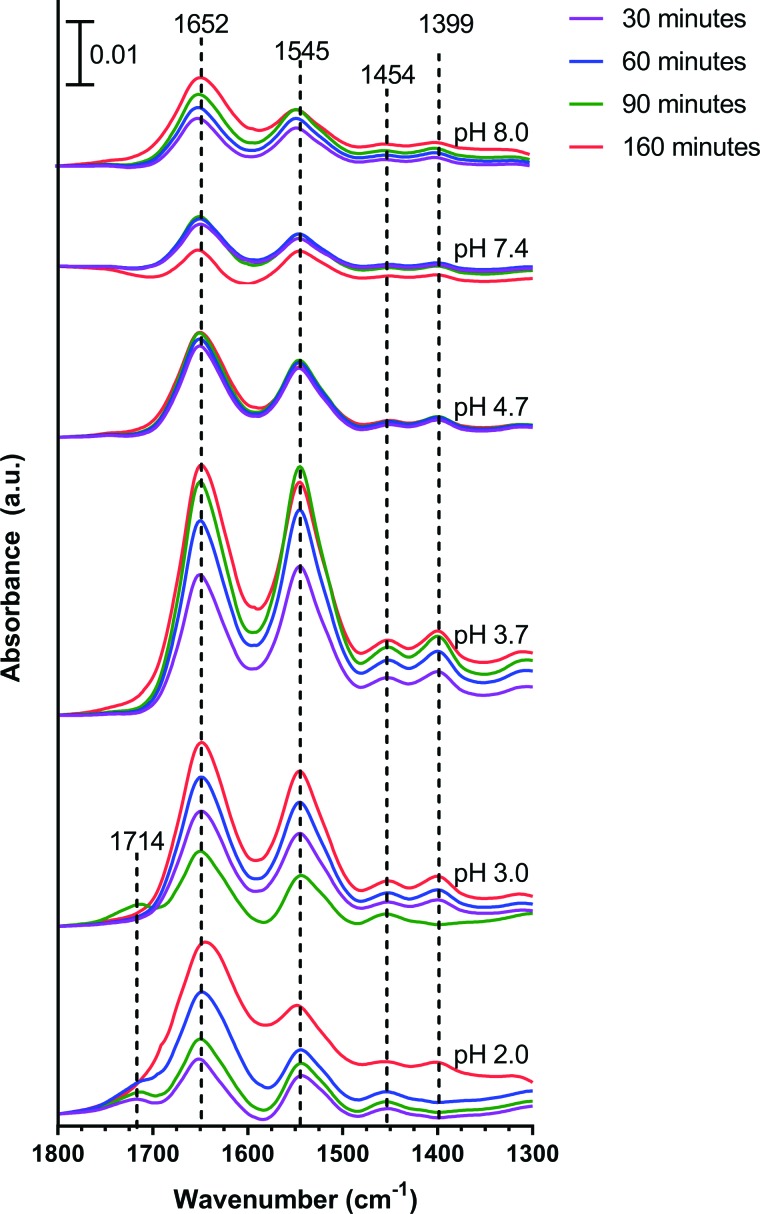
In ATR-FTIR experiments to investigate surface adsorption, 1 mg/ml BSA was used. At this concentration, BSA has a very weak signal on the bare crystal, but the signal is enhanced by adsorption to the nanoparticle thin film. Upon adsorption to the nanoparticle surface, frequency shifts in the peak location were also minimal (Fig. 6), indicating small changes in the protein structure. At pH 2.0, adsorbed BSA has an additional peak at 1714 cm−1 due to the protonated carboxyl group, and the peak at 1400 cm−1 disappears. The remaining peaks shift only slightly over time compared to the solution phase BSA at the same pH.
Fig. 6.
ATR-FTIR spectra of 1 mg/ml BSA adsorbed onto the SiO2 nanoparticle surface. Representative adsorption spectra at 30 min (purple), 60 min (blue), and 90 min (green) and desorption spectra after 60 min of desorption (red) are provided. The characteristic IR absorption bands for adsorbed proteins are associated with the amide I, II, and III regions. The amide I, II, and III regions extend from 1600–1700, 1500–1600 and 1200–1350 cm−1, respectively. The band above 1700 cm−1 is due to the stretching motion of the C=O for protonated carboxylic acid groups. It can be seen in the spectra that both the peak position and the absorbance intensity vary as a function of pH; the peak position of the amide I band varies from 1650 to 1653 cm−1, and the amide II band varies from 1543 to 1546 cm−1.
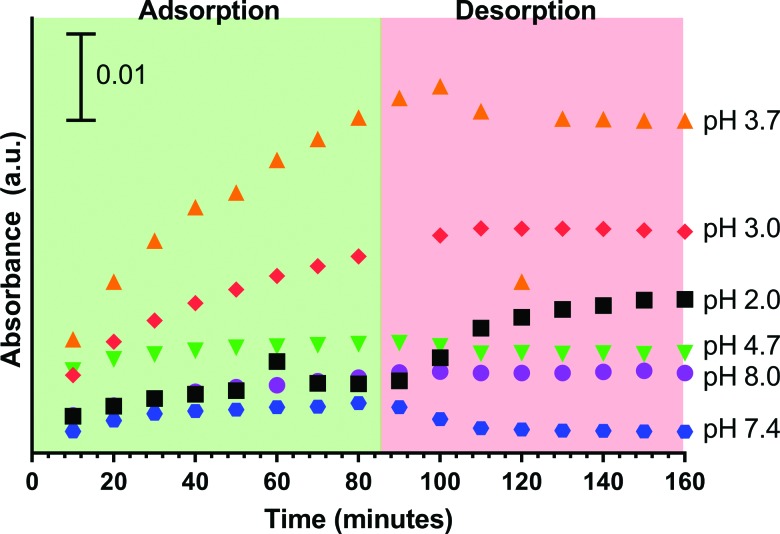
The peak intensity of the amide II band was recorded to provide insights into the adsorption of BSA onto the SiO2 surface (Fig. 7). Each measurement was taken at 1545 cm−1 for all the pH values and all the times. This standard was used because the native amide II peak appeared at 1545 cm−1, and data were reported at the chosen wavenumber, not interpolated between wavenumbers. From this plot, it is clear that pH 3.7 yields the greatest intensity and amount of BSA adsorbed, which is consistent with the TGA results (Table I). These results agree with previous studies that show the greatest protein adsorption at either the isoelectric point of the protein or the surface, as already determined from TGA measurements.37,55
Fig. 7.
Peak height of the amide II band as a function of time and pH for adsorbed BSA with values taken at approximately 1545 cm−1 for all pH.
All desorption measurements were done under the same conditions—circumneutral (pH 6.5). At most pH values, there was a small decrease in absorbance intensity. However, at pH 2, there was an increase in absorbance intensity during desorption. The amide bands are strongly dependent on the hydrogen bonds between the protein and the substrate. Stronger hydrogen bonds alter the peptide chain, for example, denature proteins, which would also result in a shift in the absorbance.48 Thus, it can be inferred that at pH 2.0, there are conformational changes occurring within the protein as a result of the higher solution pH.
C. Protein secondary structure as a function of pH
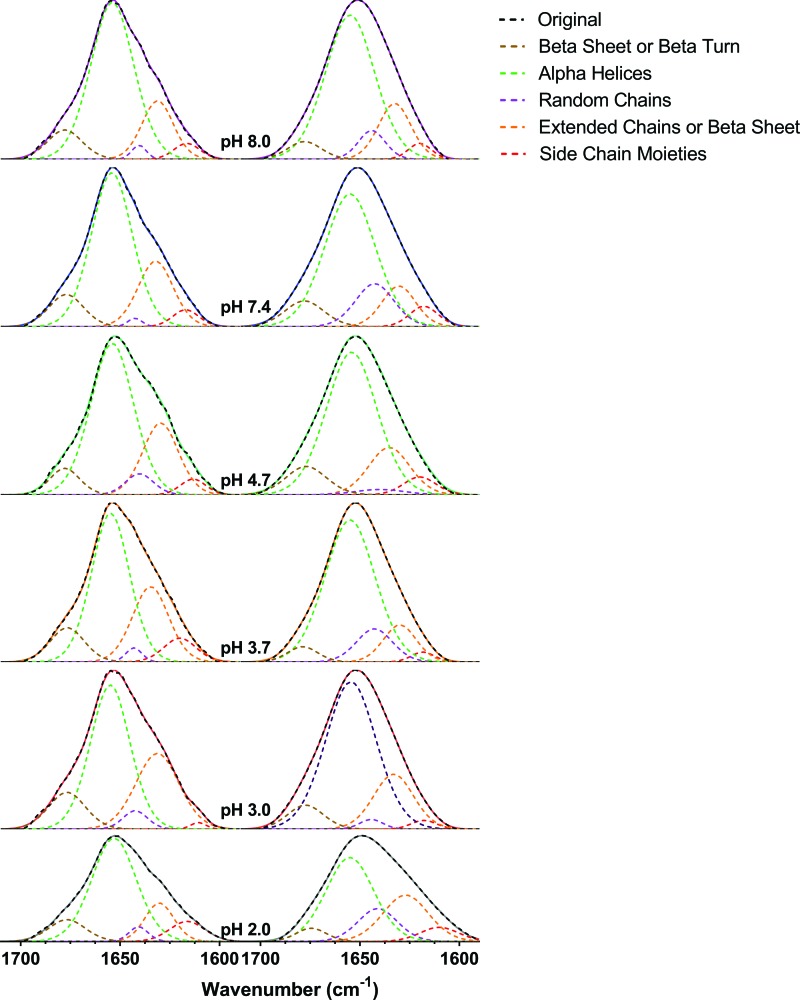
The amide I bands of the ATR spectra in Fig. 5 and the final adsorption spectrum at each pH from Fig. 6 were curve fitted with five components (Table II, Fig. 8),13,44,46 representing the secondary structure of BSA with quantitative values presented in Table III. Once fitted, these components provided information about the changing secondary structure of BSA. The protein is predominantly composed of α-helices which are also the most unstable of the secondary structure.13 If BSA unfolds on the nanoparticle surface, the alpha helix content is expected to change most of all the secondary structural elements.
Table II.
Vibrational frequency range for secondary structural elements in the amide I region (Refs. 7, 18, and 34).
| Secondary structure element | Vibrational frequency range (cm−1) |
|---|---|
| β-sheets/turns | 1685–1663 |
| α-helices | 1655–1650 |
| Random chains | 1648–1644 |
| Extended chains/β-sheets | 1639–1621 |
| Side chain moieties | 1616–1600 |
Fig. 8.
Curve fitting components of the amide I band for free BSA (a) and adsorbed BSA (b) as a function of pH. All the peaks were normalized to a linear baseline at zero and a maximum height of one absolute absorbance unit. Five different components used are shown in the figure inset. The term “Original” represents the total fit, the sum of all of the components, that is seen to match each of the original spectra.
Table III.
Percentage of the different secondary structural elements of solution phase BSA as a function of pH compared to BSA adsorbed onto the SiO2 nanoparticle surface. Also, shown is the difference between the structure of adsorbed and solution phase BSA. In some cases, due to rounding errors, not all values add up to exactly 100%.
| pH | Secondary structure | Solution BSA | Adsorbed BSA | (Δ from solution) |
|---|---|---|---|---|
| 8.0 | β-sheets/turns | 10.1 | 5.6 | ↑4.5 |
| α-helices | 66.0 | 66.5 | ↓0.5 | |
| Random chains | 2.0 | 7.3 | ↓5.3 | |
| Extended chains/β-sheets | 18.0 | 17.1 | ↑0.9 | |
| Side chain moieties | 4.1 | 3.5 | ↑0.6 | |
| 7.4 | β-sheets/turns | 10.9 | 9.0 | ↑1.9 |
| α-helices | 61.2 | 59.2 | ↑2.0 | |
| Random chains | 1.2 | 14.6 | ↓13.4 | |
| Extended chains/β-sheets | 22.4 | 12.1 | ↑10.3 | |
| Side chain moieties | 4.3 | 5.2 | ↓0.9 | |
| 4.7 | β-sheets/turns | 7.4 | 10.4 | ↓3.0 |
| α-helices | 59.9 | 64.5 | ↓4.6 | |
| Random chains | 5.5 | 2.4 | ↑3.1 | |
| Extended chains/β-sheets | 23.6 | 17.4 | ↑6.2 | |
| Side chain moieties | 3.7 | 5.3 | ↓1.6 | |
| 3.7 | β-sheets/turns | 11.2 | 4.6 | ↑6.6 |
| α-helices | 53.4 | 69.5 | ↓16.1 | |
| Random chains | 2.2 | 11.7 | ↑9.5 | |
| Extended chains/β-sheets | 25.9 | 11.9 | ↑14 | |
| Side chain moieties | 7.4 | 2.3 | ↑5.1 | |
| 3.0 | β-sheets/turns | 12.6 | 7.9 | ↑4.7 |
| α-helices | 52.3 | 68.0 | ↓15.7 | |
| Random chains | 4.2 | 2.1 | ↑2.1 | |
| Extended chains/β-sheets | 30.1 | 20.0 | ↑10.1 | |
| Side chain moieties | 0.9 | 2.1 | ↓1.2 | |
| 2.0 | β-sheets/turns | 10.7 | 4.8 | ↑5.9 |
| α-helices | 61.2 | 47.9 | ↑13.3 | |
| Random chains | 3.8 | 15.5 | ↓11.7 | |
| Extended chains/β-sheets | 14.8 | 25.2 | ↓10.4 | |
| Side chain moieties | 9.4 | 6.6 | ↑2.8 |
In the solution phase, minimal changes in the structure were observed as a function of pH. This is consistent with the peak locations in the overall ATR spectra. However, when the curve fitting was compared from the solution phase, 10 mg/ml BSA, to the adsorbed phase, 1 mg/ml BSA after 90 min, the α-helix content (brown) does not necessarily decrease upon protein adsorption to the nanoparticle surface. Additionally, the extended chain/β-sheet content (blue) decreases for most pH values, while the random chain content (purple) generally increases upon adsorption. These are the expected responses according to the literature. However, as they are not universally observed, there may be some pH-dependence in this transition. At pH 2.0, the extended chain/β-sheet content increases and the β-sheet/β-turn (green) decreases; at other pH values, these trends are reversed.
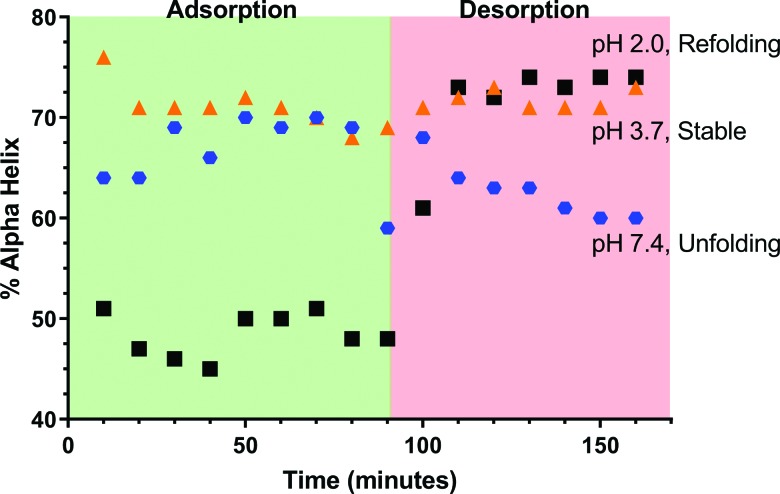
A comparison in the α-helix content between the adsorption and desorption phases was also conducted (Fig. 9). The complete range of pH values can be found in Fig. S1.62 Effects toward the alpha helix content can be noted due to pH (early time points) and nanoparticle adsorption (later time points). During the desorption phase, there was no additional BSA added to the system and all the samples were in pure water with no pH adjustment, which had measured pH values from pH 5.8 to 6.5. Thus, the effects noted during the desorption phase must be a result of protein structural changes as no additional BSA was added to the system.
Fig. 9.
Alpha helix content of adsorbed BSA as a function of time for three select pH values during adsorption at a BSA concentration of 1 mg/ml. During the desorption phase, the solution was then changed to circumneutral pH and contained no BSA. Following this change, each pH demonstrates a unique phenomenon of refolding (pH 2.0), unfolding (pH 7.4), or no changes in folding (pH 3.7) when no additional BSA is being added to the ATR-FTIR flow system.
Jachimska and Pajor reported the native α-helix content of BSA as a function of BSA conformation and found that the extended form exhibited the lowest percentage of α-helix content (35%) and the normal form exhibited the highest (55%).57 Bouhekka and Bürgi noted that BSA adsorption on TiO2 resulted in a decreased α-helix content to 23%.24 Lehman et al. reported a total α-helix content of 10% for BSA on nonporous SiO2 and 12% on mesoporous SiO2, compared to 57% for BSA in solution.40 Catalano et al. reported an alpha helix content of approximately 36% for BSA adsorbed onto pyrolytic SiO2 nanoparticles.58 All previously reported results have a much lower content than what was measured in these studies, which can be due to the different nanoparticles used. Neither Lehman et al. nor Catalano et al. used water as the dispersant,40,58 and Catalano et al. reported that the media influence protein–nanoparticle interactions.58 The porosity of the nanoparticles may influence the adsorption of BSA to nanoparticle surfaces.59 A comparison study between TiO2 and SiO2 nanoparticles found that the surface hydroxyl groups available for nanoparticles differ,56 which change depending on the generation method for SiO2 nanoparticles.60
In addition to the differences in the alpha helix content compared to those in the literature, three phenomena were noted between the adsorption and desorption phases: refolding, unfolding, and structural stability. Unfolding is the linearization of the protein molecule on the surface, while refolding generates a more compact protein structure on the surface. This naturally occurs as a function of pH with the different conformations of BSA15,16 and may also occur as a result of adsorption.13,27 This suggests that although the isoelectric point may mediate protein adsorption as a function of pH,34–37,48,50 there are also other effects driving nanoparticle–protein interactions. At pH 2.0, the greatest change in the alpha-helix content is observed. In the solution phase, the alpha helix content is 60%, in the adsorbed phase, it is about 50%, and in the desorbed phase, it is approximately 75%. The increase in the alpha helix content in the adsorbed phase is expected to result from a change in intermolecular hydrogen bonds, which is consistent with the amide I peak shifts.
The purpose of the desorption studies is to determine if protein adsorption is reversible on the nanoparticle surface.40,61 The reversibility is generally determined by comparing the absolute absorbance intensity over time, such as in Fig. 6. However, this may be insufficient if the changing protein conformation alters the spectra, as with pH 2. Thus, comparing the secondary structure of BSA provides additional information on the ability of BSA to return to its native conformation and presumably its native function.
IV. CONCLUSION
The nanoparticle–protein corona is influenced by pH and can impact the aggregate size, surface charge, adsorbed protein quantity, and protein secondary structure. We found that:
-
(1)
The quantity of BSA adsorbed as a function of pH changes and is the greatest at the isoelectric point of the BSA-SiO2 nanoparticle complex.
-
(2)
The secondary structure of adsorbed BSA differs from that of the solution phase BSA.
-
(3)
The protein structure changes upon adsorption relative to the solution phase with the greatest changes seen at the lowest pH, pH 2.
-
(4)
BSA protein adsorbed at pH 2 can change its structure when the solution pH increases back to circumneutral pH. Thus, in some cases, these pH-dependent structural changes are reversible.
Further studies of the nanoparticle–protein corona—structure and surface coverage—and how it changes with pH are important to understand as nanoparticles translocate to different organs or present in different soils or water in the environment with different pH values as this will impact nanoparticle behavior.
ACKNOWLEDGMENTS
Transmission electron microscopy was completed using the Central Microscopy Facility at the University of Iowa. Thermogravimetric analysis was done using the instrumentation in C. Allan Guymon laboratory at the University of Iowa in the Department of Chemical and Biochemical Engineering; assistance and training were provided by Jacob McLaughlin. This work was supported by the NSF CBET Award (NSF-1424502 and 1640936). Nina D. Diklich was supported by NSF REU Grant No. 1359063. Additional support for Brittany E. Givens was provided by the Alfred P. Sloan Foundation through the University of Iowa Center for Exemplary Mentoring. The authors also acknowledge helpful discussions with Zhenzhu Xu, Sarah C. Larsen, Imali A. Mudunkotuwa, and Sean E. Lehman.
References
- 1. Lin W., Huang Y. W., Zhou X. D., and Ma Y., Toxicol. Appl. Pharmacol. 217, 252 (2006). 10.1016/j.taap.2006.10.004 [DOI] [PubMed] [Google Scholar]
- 2. Patwardhan S. V., Emami F. S., Berry R. J., Jones S. E., Naik R. R., Deschaume O., Heinz H., and Perry C. C., J. Am. Chem. Soc. 134, 6244 (2012). 10.1021/ja211307u [DOI] [PubMed] [Google Scholar]
- 3. Uboldi C., Giudetti G., Broggi F., Gilliland D., Ponti J., and Rossi F., Mutat. Res. 745, 11 (2012). 10.1016/j.mrgentox.2011.10.010 [DOI] [PubMed] [Google Scholar]
- 4. Xu M., Li J., Iwai H., Mei Q., Fujita D., Su H., Chen H., and Hanagata N., Sci. Rep. 2, 406 (2012). 10.1038/srep00406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tso C. P., Zhung C. M., Shih Y. H., Tseng Y. M., Wu S. C., and Doong R. A., Water Sci. Technol. 61, 127 (2010). 10.2166/wst.2010.787 [DOI] [PubMed] [Google Scholar]
- 6. Kim I. S., Baek M., and Choi S. J., J. Nanosci. Nanotechnol. 10, 3453 (2010). 10.1166/jnn.2010.2340 [DOI] [PubMed] [Google Scholar]
- 7. Bian S. W., Mudunkotuwa I. A., Rupasinghe T., and Grassian V. H., Langmuir 27, 6059 (2011). 10.1021/la200570n [DOI] [PubMed] [Google Scholar]
- 8. Napierska D., Thomassen L. C., Lison D., Martens J. A., and Hoet P. H., Part. Fibre Toxicol. 7, 39 (2010). 10.1186/1743-8977-7-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Keller A. A., McFerran S., Lazareva A., and Suh S., J. Nanopart. Res. 15, 1692 (2013). 10.1007/s11051-013-1692-4 [DOI] [Google Scholar]
- 10. Lynch I. and Dawson K. A., Nano Today 3, 40 (2008). 10.1016/S1748-0132(08)70014-8 [DOI] [Google Scholar]
- 11. Monopoli M. P., Aberg C., Salvati A., and Dawson K. A., Nat. Nanotechnol. 7, 779 (2012). 10.1038/nnano.2012.207 [DOI] [PubMed] [Google Scholar]
- 12. Saptarshi S. R., Duschl A., and Lopata A. L., J. Nanobiotechnol. 11, 26 (2013). 10.1186/1477-3155-11-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roach P., Farrar D., and Perry C. C., J. Am. Chem. Soc. 127, 8168 (2005). 10.1021/ja042898o [DOI] [PubMed] [Google Scholar]
- 14. Dominguez-Medina S., Blankenburg J., Olson J., Landes C. F., and Link S., ACS Sustainable Chem. Eng. 1, 833 (2013). 10.1021/sc400042h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. T. Peters, Jr. , All About Albumin: Biochemistry, Genetics, and Medical Applications ( Academic, San Diego, CA, 1995). [Google Scholar]
- 16. Wright A. K. and Thompson M. R., Biophys. J. 15, 137 (1975). 10.1016/S0006-3495(75)85797-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carter D. C. and Ho J. X., Adv. Protein Chem. 45, 153 (1994). 10.1016/S0065-3233(08)60640-3 [DOI] [PubMed] [Google Scholar]
- 18. Dockal M., Carter D. C., and Ruker F., J. Biol. Chem. 275, 3042 (2000). 10.1074/jbc.275.5.3042 [DOI] [PubMed] [Google Scholar]
- 19. Geisow M., Nature 270, 476 (1977). 10.1038/270476a0 [DOI] [Google Scholar]
- 20. Khan M. Y. and Salahuddin A., Eur. J. Biochem. 141, 473 (1984). 10.1111/j.1432-1033.1984.tb08216.x [DOI] [PubMed] [Google Scholar]
- 21. Khan M. Y., Biochem. J. 236, 307 (1986). 10.1042/bj2360307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Slayter E. M., J. Mol. Biol. 14, 443 (1965). 10.1016/S0022-2836(65)80194-2 [DOI] [PubMed] [Google Scholar]
- 23. Bujacz A., Acta Crystallogr., Sect. D 68, 1278 (2012). 10.1107/S0907444912027047 [DOI] [PubMed] [Google Scholar]
- 24. Bouhekka A. and Bürgi T., Appl. Surf. Sci. 261, 369 (2012). 10.1016/j.apsusc.2012.08.017 [DOI] [Google Scholar]
- 25. Murayama K., Wu Y. Q., Czarnik-Matusewicz B., and Ozaki Y., J. Phys. Chem. B 105, 4763 (2001). 10.1021/jp004537a [DOI] [Google Scholar]
- 26. Reed R. G., Feldhoff R. C., Clute O. L., and T. Peters, Jr. , Biochemistry 14, 4578 (1975). 10.1021/bi00692a004 [DOI] [PubMed] [Google Scholar]
- 27. Marquez A., Berger T., Feinle A., Husing N., Himly M., Duschl A., and Diwald O., Langmuir 33, 2551 (2017). 10.1021/acs.langmuir.6b03785 [DOI] [PubMed] [Google Scholar]
- 28. Ball A. and Jones R. A. L., Langmuir 11, 3542 (1995). 10.1021/la00009a042 [DOI] [Google Scholar]
- 29. Giacomelli C. E., Bremer M. G. E. G., and Norde W., J. Colloid Interface Sci. 220, 13 (1999). 10.1006/jcis.1999.6479 [DOI] [PubMed] [Google Scholar]
- 30. Glassford S. E., Byrne B., and Kazarian S. G., Biochim. Biophys. Acta 1834, 2849 (2013). 10.1016/j.bbapap.2013.07.015 [DOI] [PubMed] [Google Scholar]
- 31. McClellan S. J. and Franses E. I., Colloids Surf., A 260, 265 (2005). 10.1016/j.colsurfa.2005.03.017 [DOI] [Google Scholar]
- 32. Tofan-Lazar J. and Al-Abadleh H. A., J. Phys. Chem. A 116, 10143 (2012). 10.1021/jp308913j [DOI] [PubMed] [Google Scholar]
- 33. Zeng H., Chittur K. K., and Lacefield W. R., Biomaterials 20, 377 (1999). 10.1016/S0142-9612(98)00184-7 [DOI] [PubMed] [Google Scholar]
- 34. Park Y. J., Kim K. H., Lim D. W., and Lee E. K., Process Biochem. 50, 1379 (2015). 10.1016/j.procbio.2015.05.018 [DOI] [Google Scholar]
- 35. Su T. J., Lu J. R., Thomas R. K., and Cui Z. F., J. Phys. Chem., B 103, 3727 (1990). 10.1021/jp983580j [DOI] [Google Scholar]
- 36. Su T. J., Lu J. R., Thomas R. K., Cui Z. F., and Penfold J., J. Phys. Chem., B 102, 8100 (1998). 10.1021/jp981239t [DOI] [Google Scholar]
- 37. Wiśniewska M., Szewczuk-Karpisz K., and Sternik D., J. Therm. Anal. Calorim. 120, 1355 (2014). 10.1007/s10973-014-4300-7 [DOI] [Google Scholar]
- 38. Aggarwal N., Lawson K., Kershaw M., Horvath R., and Ramsden J., Appl. Phys. Lett. 94, 083110 (2009). 10.1063/1.3078397 [DOI] [Google Scholar]
- 39. Chittur K. K., Biomaterials 19, 357 (1998). 10.1016/S0142-9612(97)00223-8 [DOI] [PubMed] [Google Scholar]
- 40. Lehman S. E., Mudunkotuwa I. A., Grassian V. H., and Larsen S. C., Langmuir 32, 731 (2016). 10.1021/acs.langmuir.5b03997 [DOI] [PubMed] [Google Scholar]
- 41. Mudunkotuwa I. A. and Grassian V. H., Langmuir 30, 8751 (2014). 10.1021/la500722n [DOI] [PubMed] [Google Scholar]
- 42. Mudunkotuwa I. A., Al Minshid A. A., and Grassian V. H., Analyst 139, 870 (2014). 10.1039/C3AN01684F [DOI] [PubMed] [Google Scholar]
- 43. Buijs J., Norde W., and Lichtenbelt J. W. T., Langmuir 12, 1605 (1996). 10.1021/la950665s [DOI] [Google Scholar]
- 44. Byler D. M. and Susi H., Biopolymers 25, 469 (1986). 10.1002/bip.360250307 [DOI] [PubMed] [Google Scholar]
- 45. Huang H., Xie J., and Chen H., Analyst 136, 1747 (2011). 10.1039/c0an00890g [DOI] [PubMed] [Google Scholar]
- 46. Roach P., Farrar D., and Perry C. C., J. Am. Chem. Soc. 128, 3939 (2006). 10.1021/ja056278e [DOI] [PubMed] [Google Scholar]
- 47. Shivu B., Seshadri S., Li J., Oberg K. A., Uversky V. N., and Fink A. L., Biochemistry 52, 5176 (2013). 10.1021/bi400625v [DOI] [PubMed] [Google Scholar]
- 48. Qing H., Yanlin H., Fenlin S., and Zuyi T., Spectrochim. Acta Mol. Biomol. Spectrosc. 52, 1795 (1996). 10.1016/S0584-8539(96)01726-6 [DOI] [Google Scholar]
- 49. Barth A., Biochim. Biophys. Acta 1767, 1073 (2007). 10.1016/j.bbabio.2007.06.004 [DOI] [PubMed] [Google Scholar]
- 50. Wassell D. T. and Embery G., Biomaterials 17, 859 (1996). 10.1016/0142-9612(96)83280-7 [DOI] [PubMed] [Google Scholar]
- 51. Tantipolphan R., Rades T., McQuillan A. J., and Medlicott N. J., Int. J. Pharm. 337, 40 (2007). 10.1016/j.ijpharm.2006.12.021 [DOI] [PubMed] [Google Scholar]
- 52. Lu R., Li W. W., Katzir A., Raichlin Y., Yu H. Q., and Mizaikoff B., Analyst 140, 765 (2015). 10.1039/C4AN01495B [DOI] [PubMed] [Google Scholar]
- 53. Demanèche S., Chapel J.-P., Monrozier L. J., and Quiquampoix H., Colloids Surf., B 70, 226 (2009). 10.1016/j.colsurfb.2008.12.036 [DOI] [PubMed] [Google Scholar]
- 54. Kosmulski M., J. Colloid Interface Sci. 253, 77 (2002). 10.1006/jcis.2002.8490 [DOI] [PubMed] [Google Scholar]
- 55. Fukuzaki S., Urano H., and Nagata K., J. Ferment. Bioeng. 81, 163 (1996). 10.1016/0922-338X(96)87596-9 [DOI] [Google Scholar]
- 56. Givens B. E., Xu Z., Fiegel J., and Grassian V. H., J. Colloid Interface Sci. 493, 334 (2017). 10.1016/j.jcis.2017.01.011 [DOI] [PubMed] [Google Scholar]
- 57. Jachimska B. and Pajor A., Bioelectrochemistry 87, 138 (2012). 10.1016/j.bioelechem.2011.09.004 [DOI] [PubMed] [Google Scholar]
- 58. Catalano F., Alberto G., Ivanchenko P., Dovbeshko G., and Marta G., J. Phys. Chem. C 119, 26493 (2015). 10.1021/acs.jpcc.5b07764 [DOI] [Google Scholar]
- 59. Hwang M.-J., Kim O.-H., Shim W.-G., and Moon H., Microporous Mesoporous Mater. 182, 81 (2013). 10.1016/j.micromeso.2013.08.022 [DOI] [Google Scholar]
- 60. Mueller R., Kammler H. K., Wegner K., and Pratsinis S. E., Langmuir 19, 160 (2003). 10.1021/la025785w [DOI] [Google Scholar]
- 61. Mudunkotuwa I. A. and Grassian V. H., Environ. Sci.: Nano 2, 5 (2015). 10.1039/c4en00215f [DOI] [Google Scholar]
- 62.See supplementary material at http://dx.doi.org/10.1116/1.4982598 E-BJIOBN-12-314702 for complete secondary structure of solution phase BSA and the alpha helix content as a function of time for all pH values.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1116/1.4982598 E-BJIOBN-12-314702 for complete secondary structure of solution phase BSA and the alpha helix content as a function of time for all pH values.