Summary
Human immunodeficiency virus (HIV) controllers are patients with different clinical characteristics, including immunologic progression as defined by CD4+ T-cells drop. We defined 2 genetic markers that can be used to segregate HIV controllers with CD4+T-cell counts >500 mm3 over time from those who experience CD4+ T-cell decline.
Keywords: HIV controllers, progression, HLA-B*57, IFNL4.
Abstract
Background.
Human immunodeficiency virus type 1 (HIV-1) controllers maintain HIV-1 viremia at low levels (normally <2000 HIV-RNA copies/mL) without antiretroviral treatment. However, some HIV-1 controllers have evidence of immunologic progression with marked CD4+ T-cell decline. We investigated host genetic factors associated with protection against CD4+ T-cell loss in HIV-1 controllers.
Methods.
We analyzed the association of interferon-lambda 4 (IFNL4)–related polymorphisms and human leukocyte antigen (HLA)-B haplotypes within long-term nonprogressor HIV-1 controllers (LTNP-Cs; defined by maintaining CD4+ T-cells counts >500 cells/mm3 for more than 7 years after HIV-1 diagnosis) vs non-LTNP-Cs who developed CD4+ T-cell counts <500 cells/mm3. Both a Spanish study cohort (n = 140) and an international validation cohort (n = 914) were examined. Additionally, in a subgroup of individuals, HIV-1–specific T-cell responses and soluble cytokines were analyzed.
Results.
HLA-B*57 was independently associated with the LTNP-C phenotype (odds ratio [OR], 3.056 [1.029–9.069]; P = .044 and OR, 1.924 [1.252–2.957]; P = .003) while IFNL4 genotypes represented independent factors for becoming non-LTNP-C (TT/TT, ss469415590; OR, 0.401 [0.171–0.942]; P = .036 or A/A, rs12980275; OR, 0.637 [0.434–0.934]; P = .021) in the Spanish and validation cohorts, respectively, after adjusting for sex, age at HIV-1 diagnosis, IFNL4-related polymorphisms, and different HLA-B haplotypes. LTNP-Cs showed lower plasma induced protein 10 (P = .019) and higher IFN-γ (P = .02) levels than the HIV-1 controllers with diminished CD4+ T-cell numbers. Moreover, LTNP-Cs exhibited higher quantities of interleukin (IL)2+CD57- and IFN-γ +CD57- HIV-1–specific CD8+ T cells (P = .002 and .041, respectively) than non-LTNP-Cs.
Conclusions.
We defined genetic markers able to segregate stable HIV-1 controllers from those who experience CD4+ T-cell decline. These findings allow for identification of HIV-1 controllers at risk for immunologic progression and provide avenues for personalized therapeutic interventions and precision medicine for optimizing clinical care of these individuals.
Human immunodeficiency virus type 1 (HIV-1) controllers are a rare group of individuals who maintain HIV-1 viremia at extremely low (<2000 HIV-RNA copies/mL) or even undetectable levels in the absence of antiretroviral treatment [1]. This group constitutes an important field of study, since they have been proposed as a model for a functional cure. Investigating immune defense mechanisms in these persons may reveal relevant information for HIV-1 vaccine development and eradication strategies [2, 3]. However, HIV-1 controllers are a heterogeneous group composed of subsets with different clinical characteristics. Epidemiological studies showed that about 6%–7% of these persons progressed to AIDS [4, 5], 20% experienced at least 1 non-AIDS–defining event [6], and 30% eventually experienced loss of spontaneous viral control [4, 6]. Importantly, immunologic progression as defined by CD4+ T-cell counts <500 cells/mm3 has been documented in some HIV-1 controllers [7, 8], leading to institution of combination antiretroviral therapy (cART) [4], although treatment-related CD4+ T-cell increases were frequently less pronounced than in noncontrollers [9]. Identifying biological markers of HIV-1 controllers who are at risk for HIV-1 disease progression as defined by declining CD4+ T-cell counts is an important objective, both for improving clinical care of these persons and for extending the conceptual understanding of immune mechanisms involved in HIV spontaneous control.
Single-nucleotide polymorphisms (SNPs), located in the major histocompatibility complex, are the major genetic host factor associated with HIV-1 immune control [10]. Human leukocyte antigen (HLA)-I alleles, in particular HLA-B*57, have been associated with slower progression to AIDS and hence to lower CD4+ T-cell drop in natural history cohorts [11]. Moreover, we have reported an association between the CC variant of a SNP within IL-28B (rs12979860) and HIV-1 spontaneous control in white patients [12], suggesting that polymorphisms at the interferon-lambda (IFNL) gene locus may influence or modulate the dynamics of HIV-1 disease progression in controllers. Prokunina-Olsson et al described the ss469415590 polymorphism within the IFNL region, which is in linkage disequilibrium with rs12979860 and rs12980275 [13, 14]. This novel polymorphism seems to be the functional one of the family, leading to IFNL4 production and modulation of expression of IFN-stimulated genes [13, 15].
Based on these observations, we hypothesized that these host genetic factors could segregate different HIV-1 controller phenotypes and may be correlated with different patterns of HIV-1 disease progression within HIV-1 controllers. Consequently, we tested associations between immunologic progression in HIV-1 controllers (defined by longitudinal evolution of CD4+ T-cell counts) and the presence of HLA-B*57 alleles and IFNL4-related polymorphisms, first in a study cohort and then in a validation cohort of white patients.
METHODS
Study Cohort
HIV-1 controllers were defined exclusively based on virological criteria as patients with plasma HIV viral load (VL) <2000 HIV-RNA copies/mL plasma for at least 1 year in the absence of cART. Long-term nonprogressors (LTNP) were defined exclusively based on immunological criteria as patients with CD4+ T-cell counts >500 cells/mm3 for more than 7 years after HIV diagnosis. HIV-1 controllers who met LTNP criteria were considered as LTNP controllers (LTNP-Cs) and those who did not fulfill this criteria and progressed to CD4+ T-cell counts <500 cells/mm3 in less than 7 years were considered as non-LTNP-Cs. A total of 100 LTNP-Cs and 40 non-LTNP-Cs recruited within the multicenter Spanish AIDS Research Network HIV-Controllers Cohort (ECRIS) were included according to sample availability. HIV-2–infected patients were excluded. Viral load and CD4+ and CD8+ T-cell counts were routinely collected and data recorded in the ECRIS cohort by each collaborating center (Supplementary Materials). Samples were kindly provided by the HIV-1 Biobank integrated within the Spanish AIDS Research Network. Each participant signed an informed consent form. The institutional review boards of the participating hospitals and centers approved the study.
Genotyping
HLA-B alleles and IFNL4 ss469415590 polymorphisms were genotyped, as previously described [12, 16].
Validation Cohort
As a validation cohort, 914 white HIV-1 controllers included in a prior genome-wide association study (GWAS) from the International HIV Controllers Study [10] were analyzed. These HIV controllers were split into LTNP-C and non-LTNP-C in accordance with the above-mentioned criteria. Data on HLA-B alleles and IFNL4-associated polymorphisms were extracted from the prior GWAS study. The IFNL4-associated polymorphism available in the validation cohort was rs12980275, which is in strong linkage disequilibrium with ss469415590 (r2 = 0.901).
Analysis of HIV-1–Specific T-Cell Responses and Soluble Cytokines
Frozen peripheral blood mononuclear cells (PBMCs) were in vitro stimulated with a pool of HIV-1 Gag peptides, stained, and analyzed using flow cytometry as previously described [17].
Soluble plasma concentrations of IFN-gamma (IFN-γ) and IFN-γ–induced protein 10 (IP-10) were assayed in duplicate with a bead-based immunoassay using Luminex technology (HCYTMAG-60K-PX29, Merck Millipore, Darmstadt, Germany).
Statistical Analyses
For the bivariate and multivariate analyses, a logistic regression model was applied. Log-rank Mantel Cox test was used for univariate survival analysis for time to CD4+ T-cell drop <500 cells/mm3, followed by stepwise Cox regression multivariate analyses. All variables with P values < .1 were included in the multivariate analysis, and P < .05 was considered significant. To compare HIV-specific T-cell responses and soluble cytokine levels, Mann-Whitney U tests were used. Statistical analyses were performed using the Statistical Package for Social Sciences software (SPSS 20.0; SPSS Inc., Chicago, Illinois).
RESULTS
LTNP-C Association With HLA*B57 Allele and ss469415590 TT/TT Genotype
The ss469415590 polymorphisms (TT vs ∆G) at the IFN-λ gene locus has previously been found to be strongly associated with natural clearance and clinical responses to IFN-α treatment in patients with hepatitis C virus (HCV) infection [13, 18]. To investigate the impact of these polymorphisms on the dynamics of HIV-1 disease progression in HIV-1 controllers, we analyzed this polymorphism in a cohort of HIV-1 controllers recruited through a multicenter network in Spain. The characteristics of the study cohort are summarized in Table 1. Notably, LTNP-Cs were enriched with men compared with non-LTNP-Cs (69% vs 50%, P = .037) despite an overrepresentation of women in the entire ECRIS cohort [4, 6] and other cohorts [1]. There were no differences in VL between the 2 study groups, expressed as the mean of the logarithm of VL determinations (log mean VL, LTNP-Cs = 2.14 [1.79–2.61] vs non-LTNP-Cs = 2.06 [1.70–2.64]; P = .583). Interestingly, the proportion of persons carrying the homozygous IFNL4 (ss469415590) TT/TT genotype, previously associated with improved clinical responses to exogenous IFN-α treatment in the context of HCV infection, was significantly lower among LTNP-Cs than among non-LTNP-Cs (44% vs 65%; P = .029). In contrast, homozygous carriers of the ∆G/∆G genotype, associated with reduced natural clearance of HCV infection, were significantly more frequent in LTNP-Cs, while being completely absent in non-LNTP-Cs (12.8% vs 0%; P = .019). Notably, the protective HLA class I allele HLA-B*57 was also more frequent among LTNP-Cs than non-LTNP-Cs (29% vs 13%; P = .054). There were no differences in the proportions of HLA-B*27, -B*35, and -B*08 alleles between groups (Table 1).
Table 1.
Characteristics and Bi-and Multivariate Analyses of the Study Cohort
| Characteristic | LTNP-C (n = 100) | Non-LTNP-C (n = 40) | P; OR (95% CI) | P; OR (95% CI) |
|---|---|---|---|---|
| Unadjusted | Adjusted | |||
| Sex (male) | 69 (69) | 20 (50) | .037; 2.226 (1.051–4.716) | .040; 2.467 (1.040–5.852) |
| Age at HIV diagnosis (years) | 26.2 [22.1–31.8] | 29.9 [25.0–34.1] | .038; .950 (.904–.997) | .102; .957 (.908–1.009) |
| Interferon-lambda 4 TT/TT genotypea | 42 (44) | 26 (65) | .029; .427 (.198–.918) | .036; .401 (.171–.942) |
| HLA-B*27b,c | 18 (18) | 7 (18) | .982; .989 (.375–2.607) | |
| HLA-B*57b,c | 29 (29) | 5 (13) | .054; 2.770 (.981–7.824) | .044; 3.056 (1.029–9.069) |
| HLA-B*35b,c | 10 (10) | 7 (18) | .194; .498 (.174–1.426) | |
| HLA-B*08b,c | 5 (5) | 2 (5) | .964; .962 (.178–5.188) |
Qualitative data are number (%) of individuals and quantitative data are median (interquartile range). Significant values are shown in bold.
Abbreviations: CI, confidence interval; HIV, human immunodeficiency virus; HLA, human leukocyte antigen; LTNP-C, long-term nonprogressor HIV-1 controller; OR, odds ratio.
aData were available for 135 patients (95 LTNP-Cs and 40 non-LTNP-Cs).
bData were available for 133 patients (96 LTNP-Cs and 37 non-LTNP-Cs).
cDominant model was applied. There were no homozygous patients for HLA-B*57.
Next, we determined whether the above-mentioned genetic factors were independently associated with the LTNP-C phenotype. Taking LTNP-C as the reference variable, we performed a bi- and multivariate logistic regression analysis, shown in Table 1. After adjusting for sex, age at HIV-1 diagnosis, IFNL4 (ss469415590) TT/TT genotype, and carrier status for HLA-B*27, -B*57, -B*35, and -B*08 alleles, the protective factors independently associated with LTNP-C phenotype were male sex and HLA-B*57 allele carrier. On the other hand, the IFNL4 (ss469415590) TT/TT genotype was an independent factor associated with the non-LTNP-C phenotype (Table 1).
In addition, we also analyzed HLA-B*57 and IFNL4 (ss469415590) genotypes as a composite variable, categorizing individuals in HLA-B*57+/IFNL4 nonTT (ie, TT/G or G/G), HLA-B*57+/IFNL4 TT/TT, HLA-B*57-/IFNL4 nonTT, and HLA-B*57-/IFNL4 TT/TT (Supplementary Table 1). The composite variable was associated with LTNP-C (P = .035). The combined genotype HLA-B*57-/IFNL4 TT/TT was independently associated with developing non-LTNP-C phenotype after adjusting for sex, age, and log mean VL (Supplementary Table 1).
Replication of the Associations Between LTNP-C, HLA-B*57 Allele, and IFNL4-Related Genotype in a Validation Cohort
In order to validate our results, we analyzed available data for 914 white HIV-1 controllers included in a GWAS from the International HIV Controllers study [10]. The characteristics of these patients are summarized in Table 2. We observed 52.8% patients with an LTNP-C phenotype within this cohort. Of the 914 patients, 465 had available data for the rs12980275 genotype, which is in strong linkage disequilibrium with ss469415590 (r2 = 0.901). The proportion of patients with the A/A genotype was 48.2%; 42.2% of individuals had an A/G genotype and 9.7% were carriers of the G/G genotype (Table 2).
Table 2.
Characteristics of the Validation Cohort
| Characteristic | Whole Cohort (N = 914) | Selected Cohort (N = 465) |
|---|---|---|
| Sex (male) | 678 (74.3) | 392 (81.2) |
| Age at HIV diagnosis (years) | 33 (27–40) | 33 (27–40) |
| LTNP-Ca | 468 (52.8) | 241 (51.8) |
| Interferon-lambda 4 A/A genotypeb | NA | 224 (48.2) |
| HLA-B*27 | 127 (14.2) | 94 (20) |
| HLA-B*57 | 268 (29.9) | 134 (28.6) |
| HLA-B*35 | 85 (9.5) | 46 (9.8) |
| HLA-B*08 | 74 (8.3) | 53 (11.3) |
Qualitative data are no. (%) of individuals. and quantitative data are median (interquartile range).
Abbreviations: HIV, human immunodeficiency virus; HLA, human leukocyte antigen; LTNP-C, long-term nonprogressor HIV-1 controller; NA, not available in the whole cohort.
aLTNP-C classification was possible for 887 patients.
bData were available for 465 individuals, for whom characteristics are listed in the right column.
In this cohort, there was a strong association between younger age at HIV-1 diagnosis and development of an LTNP-C phenotype, suggesting that improved regenerative immunological capacities [19] may facilitate preservation of adequate CD4+ T-cell counts in HIV-1 controllers. In contrast to the study cohort, there were no gender differences between LTNP-Cs and non-LTNP-Cs (Table 3). Similar to the observations in the study cohort, we observed an overrepresentation of HLA-B*57 alleles among LTNP-Cs in the validation cohort (35% vs 22%; P = .002). There were no differences in the proportions of HLA-B*27, -B*35, and -B*08 alleles between groups. The rs12980275 A/A genotype was found in elevated frequencies among non-LTNP-Cs in the validation cohort (43% vs 54%; P = .012; Table 3), analogous to what was found with the corresponding SNP in the study cohort. After adjusting for age at HIV-1 diagnosis, rs12980275 A/A genotype, HLA-B*27, -B*57, -B*35, and -B*08 alleles, we replicated the genetic associations found in the study cohort. The protective factors independently associated with LTNP-C phenotype were age at HIV-1 diagnosis and frequency of HLA-B*57 alleles. The IFNL4 (rs12980275) A/A genotype was independently associated with the non-LNTP-C phenotype (Table 3).
Table 3.
Bi- and Multivariate Analysis of Validation Cohort
| Factor | LTNP-C (n = 242) | Non-LTNP-C (n = 222) | P; OR (95% CI) | P; OR (95% CI) |
|---|---|---|---|---|
| Unadjusted | Adjusted | |||
| Sex (male) | 197 (81.4) | 183 (82.1) | .774; .933 (.581–1.498) | |
| Age at HIV diagnosis (years) | 32 (26–38) | 35 (29–41) | .001; .965 (.945–.985) | .001; .962 (.942–.983) |
| Interferon-lambda 4 A/A genotype | 103 (42.6) | 121 (54.3) | .012; .625 (.433–.901) | .021; .637 (.434–.934) |
| HLA-B*27a | 53 (22.3) | 38 (17.9) | .253; 1.312 (.824–2.089) | |
| HLA-B*57a | 84 (35.3) | 47 (22.2) | .002; 1.915 (1.259–2.912) | .03; 1.924 (1.252–2.957) |
| HLA-B*35a | 23 (9.7) | 22 (10.4) | .801; .924 (.499–1.711) | |
| HLA-B*08a | 23 (9.7) | 26 (12.3) | .378; .765 (.422–1.387) |
Qualitative data are number (%) of individuals and quantitative data are median (interquartile range). Significant values are shown in bold.
Abbreviations: CI, confidence interval; HIV, human immunodeficiency virus; HLA, human leukocyte antigen; LTNP-C, long-term nonprogressor HIV-1 controller; OR, odds ratio.
aDominant model was applied. There were only 3 homozygous patients for HLA-B*57.
Although we found differences in VL between the 2 groups in the validation cohort (log mean VL, LTNP-Cs = 2.15 [1.70–2.64] vs non-LTNP-Cs = 2.51 [1.88–2.91]; P = .001), the above-mentioned associations were maintained even after adjusting by log mean VL. In this case the protective factors independently associated with LTNP-C phenotype were the following: lower log mean VL (odds ratio [OR], 0.628; 95% confidence interval [CI], 0.447–0.883; P = .007), younger age at HIV-1 diagnosis (OR, 0.962; 95% CI, 0.941–0.983; P < .0001), and frequency of HLA-B*57 alleles (OR, .869; 95% CI, 1.212–2.883; P = .005). Once more the frequency of IFNL4 (rs12980275) A/A genotype was independently associated with the non-LTNP-C phenotype (OR, 0.644; 95% CI, 0.438–0.948; P = .026).
Again, we analyzed HLA-B*57 and IFNL4 (ss469415590) genotypes as a composite variable, categorizing individuals in HLA-B*57+/IFNL4 (rs12980275) A/A- (ie, A/G or G/G), HLA-B*57+/IFNL4 (rs12980275) A/A+, HLA-B*57-/IFNL4 (rs12980275) A/A-, and HLA-B*57-/IFNL4 (rs12980275) A/A+. The composite variable was associated with LTNP-C (P = .001). The combined genotype HLA-B*57-/IFNL4 A/A+ was independently associated with developing non-LTNP-C phenotype after adjusting for age and log mean VL (Supplementary Table 2).
HLA-B*57 Allele and IFNL4-Related Genotypes Are Independently Associated With Faster CD4+ T-Cell Loss Below 500 cell/mm3
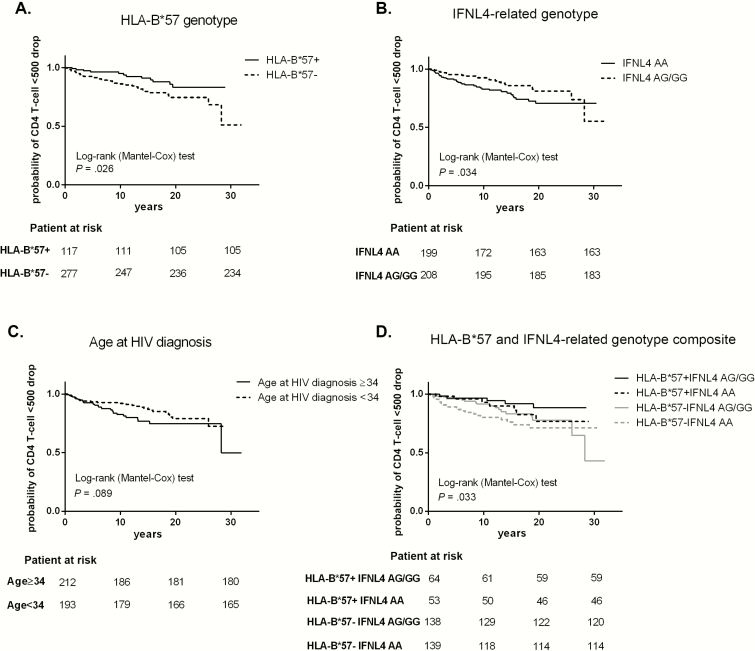
Next, we analyzed variables associated with the time to CD4+ T-cell loss below 500 cells/mm3. This analysis was only performed in the validation cohort; there was not enough statistical power to perform the analysis in the study cohort. HLA-B*57 was a protective factor independently associated with the time to CD4+ T-cells below 500 cell/mm3 (Figure 1A), while the frequency of IFNL4 (rs12980275) A/A genotypes (Figure 1B) and age at HIV-1 diagnosis (Figure 1C) represented risk factors for accelerated CD4+ T-cell decline (Table 4). After adjusting by log mean VL, HLA-B*57 remained a protective factor (OR, 0.493; 95% CI, 0.259–0.941; P = .032) and higher log mean VL was a risk factor (OR, 3.057; 95% CI, 1.694–5.518; P = <.0001). In this analysis the frequency of IFNL4 (rs12980275) A/A genotype and age at diagnosis showed a trend toward being a risk factor for accelerated CD4+ T-cell decline (P = .093 and P = .059, respectively).
Figure 1.
Kaplan-Meier curves for factors associated with CD4+ T-cell drop <500 cell/mm3. Variables with P < .1 in the Cox proportional, hazards bivariate regression are represented. A, human leukocyte antigen (HLA)-B*57, B, interferon-lambda 4 (IFNL4)–related genotype, C, age at diagnosis, and D, HLA-B*57 and IFNL4-related genotype composite variable expressed as the 4 possible combinations. Log-rank test bivariate analysis was performed to assess statistical differences among the curves. Abbreviations: HIV, human immunodeficiency virus.
Table 4.
Time to CD4+ <500 cell/mm3 Drop Analysis of Validation Cohort
| Factor | P; HR (95% CI) | P; HR (95% CI) |
|---|---|---|
| Unadjusted | Adjusted | |
| Sex (male) | .066; .585 (.330–1.036) | |
| Age at human immunodeficiency virus diagnosis (years) | .091; 1.562 (.931–2.621) | .043; 1.770 (1.019–3.076) |
| Interferon-lambda 4 A/A genotype | .038; 1.717 (1.030–2.865) | .037; 1.790 (1.035–3.096) |
| HLA-B*27 | .972; 1.041 (.540–1.894) | |
| HLA-B*57 | .029; .490 (.257–.935) | .045; .510 (.271–.985) |
| HLA-B*35 | .425; .661 (.258–1.835) | |
| HLA-B*08 | .157; 1.725 (.811–3.662) |
Significant values are shown in bold.
Abbreviations: HR, hazard ratio; CI, confidence interval; HLA, human leukocyte antigen.
We also analyzed HLA-B*57 and IFNL4 (rs12980275) genotypes as a composite variable, as mentioned above. The composite variable was associated with the time to event (Figure 1D). The combined genotype HLA-B*57-/IFNL4 (rs12980275) A/A+ was a risk factor independently associated with the time to CD4+T-cell loss below 500 cell/mm3 after adjusting for gender, age, and log mean VL (Supplementary Table 3).
HIV-Specific T-Cell Response and Plasma Soluble Cytokines
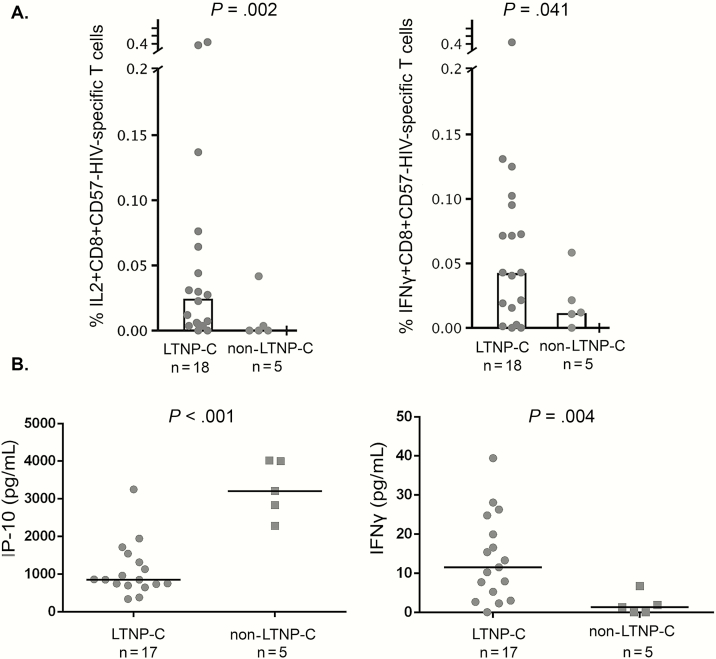
We analyzed HIV-1–specific T-cell responses and plasma soluble cytokines in a subgroup of LTNP-Cs (n = 18) and non-LTNP-Cs (n = 5) with available plasma and PBMC samples and undetectable plasma VL. LTNP-C exhibited a higher percentage of IL2+CD8+CD57- and IFNγ+CD8+CD57- HIV-1–specific T cells (P = .002 and 0.041, respectively) than non-LTNP-Cs (Figure 2A). Moreover, LTNP-C showed higher IFN-γ and lower IP-10 plasma levels (P = .004 and P < .001, respectively) than non-LTNP-Cs (Figure 2B).
Figure 2.
Human immunodeficiency virus (HIV)–specific T-cell response and plasma soluble cytokine levels in long-term nonprogressor HIV-1 controller (LTNP-C) and non-LTNP-C subgroups. A, IL2+CD8+CD57- (left panel) and interferon-gamma (IFNγ+)CD8+CD57-HIV-specific T-cell response (right panel) were assayed in 18 LTNP-Cs and 5 non-LTNP-Cs with peripheral blood mononuclear cell samples available. B, induced protein 10 (left panel) and IFN-γ (right panel) levels were assayed in 17 LTNP-Cs and 5 non-LTNP-Cs with plasma samples available. Abbreviations: HIV, human immunodeficiency virus; IFNγ, interferon-gamma; IL2, interleukin-2; IP-10, induced protein 10; LTNP-C, long-term nonprogressor HIV-1 controller.
DISCUSSION
Here, we show that the dynamics of CD4+ T-cell decline among HIV-1 controllers is associated with HLA-B*57 and IFNL4-related genotypes. HIV-1 controllers with stable CD4+ T-cell counts, termed here LTNP-Cs, were more likely to carry the HLA-B*57 allele and less likely to express the IFNL4-related genotypes TT/TT (ss469415590) or A/A (rs12980275). The consistency of these results was further confirmed in an independent validation cohort with 3.5-fold larger patient numbers. Notably, these associations were true not only when taking the definition of LTNP-C as an endpoint but also in the subgroup longitudinally studied, when considering the time-to-event analysis, strengthening the findings of this study.
HLA-B*57 has been associated with delayed progression to AIDS and CD4+ T-cell losses in several prior natural history cohorts [11, 20]. This observation is consistent with our finding that LTNP-Cs were enriched with HLA-B*57 alleles. Interestingly, despite the known influence of HLA-B*57 on HIV viremia [10, 21], the associations between HLA-B*57 and reduced CD4+ T-cell number remained independent even after adjusting for VL in our model. The precise mechanism by which HLA-B*57 protects against CD4+ T-cell losses in HIV-1 patients is unknown but likely includes the restriction of HIV-1–specific CD8+ T-cell responses [22] with improved antiviral functions (eg, IL2 and IFN-γ production among HLA-B*57–restricted HIV-specific T cells [23]) as well as altered immunoregulatory interactions with HLA class I receptors expressed on innate immune cells [24].
Previously we communicated that the CC genotype of rs12979860 polymorphisms was associated with spontaneous control of HIV-1 in white patients recruited in Spain [12]. This SNP is in strong linkage disequilibrium with rs12980275 [14] and ss469415590 [13]. The ∆G allele of this polymorphism encodes for the IFNL4 protein [13] and seems to be associated with lower expression of IL28 mRNA and IP-10 than wild-type allele carriers [15]. Interestingly, IP-10, a proinflammatory chemokine related to immune activation, has been reported to negatively correlate with CD4+ T-cell counts in HIV-1 controllers [25]. This observation is consistent with our findings, as LTNP-Cs were more likely to carry the ∆G mutant allele and have lower levels of soluble IP-10. These findings are also in accordance with the time-to-event analysis presented in this work using HLA-B*57 and IFNL4-related genotype as a composite variable. Together, these data raise the provocative hypothesis that patients with the IFNL4 TT/TT genotype may have higher antiviral capacity mediated by type I IFN responses [26] and IFN-stimulated gene upregulation [15], possibly through an enhanced plasmacytoid dendritic cell (pDC) function [27]. This robust IFN response may have a role in supporting the ability to naturally control HIV-1 replication for a limited period of time or to clear nonintegrating viruses such as HCV. However, the response may be achieved at the expense of increased T-cell activation [25] and IFN-mediated thymic dysfunction [28] that eventually may contribute to CD4+ T-cell drop and a loss of the HIV-1 controller status. This fact could explain the overrepresentation of women in the non-LTNP-C group, as it is known that women exhibit higher TLR-mediated pDC responses and the subsequently higher CD8+ T-cell activation [29].
Defining heterogeneous phenotypes among HIV-1 controllers could help to clarify some controversial results reported in the literature. For example, the above-mentioned association between the CC genotype of rs12979860 and HIV-1 control in a white cohort [12] was not found by other authors [30]. In addition to ethnic differences in the cohorts, these contradictory data could be due to the heterogeneity of controller cohorts being analyzed. We noticed that in our previous work, the cohort was enriched with non-LTNP-Cs (about 75%). In contrast, other studies, in particular the cohort described by Salgado et al [30], consisted predominantly (>90%) of LTNP-Cs. Together, these data argue that investigating genetic polymorphisms associated with an HIV-1–controller status will require more robust definitions of clinical phenotypes that do not rely on VL as the only criteria.
The segregation between LTNP-Cs and non-LTNP-Cs may have important implications. First, controllers who are at risk of disease progression [4–8] could possibly be identified at earlier stages and may benefit from more rapid cART initiation or other immunotherapeutic interventions. In HIV-1 controllers genetically predisposed to maintain an LTNP-C phenotype, the risk-to-benefit ratio for institution of cART may be less definitive. Second, HIV-1 controllers have been proposed as a model for functional cure and effective viral eradication strategies [2, 3]. The subgroup of HIV-1 controllers with no detectable viremia, high CD4+ T-cell counts, and no signs of disease progression may represent the most informative patient populations in this regard. So, it is pivotal to accomplish a more precise definition of controller phenotypes in order to avoid spurious results and to properly identify immune correlates of persistent spontaneous viral control in the search for the right model of a functional cure or “spontaneous eradicators.”
Our work has some limitations. First, both cohorts were composed of seroprevalent patients, for which precise times of HIV-1 seroconversion or viral transmission could not be firmly established. Consequently, follow-up times were calculated based on the time since HIV-1 diagnosis; hence, initial VL and CD4+ T-cell evolution prior to entry into the cohort were missed. However, the confirmation of the results in a large validation cohort supports the appropriateness of the LTNP-C classification criteria. Unfortunately, we had few samples available to perform functional analyses. Although results were consistent with those reported in the literature, further analyses should be performed with a larger number of patients. Last, we only analyzed HLA-B and IFNL4-related genotypes. It could be desirable to perform more comprehensive genetic analysis to segregate HIV-1 controllers in different phenotypes and/or genotypes in order to propose an algorithm with improved discriminatory capacity.
In conclusion, we segregated 2 HIV-1 controller phenotypes based on 2 genetic markers for which established laboratory tests are available and which are routinely used in HIV-1 clinical practice. The better characterization of distinct HIV-1 controller phenotypes may allow improved clinical care of this specific patient population and enhance our understanding of how long-term spontaneous control of HIV-1 infection can be naturally achieved.
Supplementary Material
Notes
Acknowledgments. This study would not have been possible without the collaboration of the patients, medical and nursery staff, and data managers who took part in the project.
Financial support. This work was supported by the Instituto de Salud Carlos III Red Temática de Investigación Cooperativa en Sindrome de inmunodeficiencia humana (SIDA) (RD12/0017, RD16/0025/0020 and CPII014/00025 to E. R.-M.); the Spanish Ministry of Education (FPU13/02451 to B. D. M.); Ministerio de Economia y Competitividad (MINECO)/Fondos Europeos para el Desarrollo Regional (FEDER; SAF2013-48754-C2-1-R to M. D. V.); Fondo de Investigacion Sanitaria, Instituto de Salud Carlos III FEDER, Programa de Suport als Grups de Recerca AGAUR (Agencia de Gestio d'Ajuts Universitaris i de Recerca), Gilead Fellowship Program (PI13/00796, PI16/00503, 2014SGR250, GLD14/293 to E. R.-G. and F.V.); and a National Institutes of Health grant (AI098487 and AI106468 to Y. X. and M. L., and AI116228, HL134539, and HL126554 to Y.X.).
Potential conflicts of interest. All authors: No potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Madec Y, Boufassa F, Rouzioux C, Delfraissy JF, Meyer L; SEROCO Study Group Undetectable viremia without antiretroviral therapy in patients with HIV seroconversion: an uncommon phenomenon? Clin Infect Dis 2005; 40:1350–4. [DOI] [PubMed] [Google Scholar]
- 2. Shasha D, Walker BD. Lessons to be learned from natural control of HIV—future directions, therapeutic, and preventive implications. Front Immunol 2013; 4:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cockerham LR, Hatano H. Elite control of HIV: is this the right model for a functional cure? Trends Microbiol 2015; 23:71–5. [DOI] [PubMed] [Google Scholar]
- 4. Leon A, Perez I, Ruiz-Mateos E, et al. ; EC and Immune Pathogenesis Working Group of the Spanish AIDS Research Network Rate and predictors of progression in elite and viremic HIV-1 controllers. AIDS 2016; 30:1209–20. [DOI] [PubMed] [Google Scholar]
- 5. Ruiz-Mateos E, Ferrando-Martinez S, Machmach K, et al. High levels of CD57+CD28- T-cells, low T-cell proliferation and preferential expansion of terminally differentiated CD4+ T-cells in HIV-elite controllers. Curr HIV Res 2010; 8:471–81. [DOI] [PubMed] [Google Scholar]
- 6. Dominguez-Molina B, Leon A, Rodriguez C, et al. ; Spanish AIDS Research Network HIV Controllers Cohort Analysis of non-AIDS-defining events in HIV controllers. Clin Infect Dis 2016; 62:1304–9. [DOI] [PubMed] [Google Scholar]
- 7. Okulicz JF, Marconi VC, Landrum ML, et al. ; Infectious Disease Clinical Research Program HIV Working Group Clinical outcomes of elite controllers, viremic controllers, and long-term nonprogressors in the US Department of Defense HIV natural history study. J Infect Dis 2009; 200:1714–23. [DOI] [PubMed] [Google Scholar]
- 8. Boufassa F, Saez-Cirion A, Lechenadec J, et al. ; ANRS EP36 HIV Controllers Study Group CD4 dynamics over a 15-year period among HIV controllers enrolled in the ANRS French observatory. PLoS One 2011; 6:e18726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boufassa F, Lechenadec J, Meyer L, et al. ; ANRS CO18 HIV Controllers Cohort; Cascade Collaboration in Eurocoord; SCOPE Cohort; International HIV Controllers Study Blunted response to combination antiretroviral therapy in HIV elite controllers: an international HIV controller collaboration. PLoS One 2014; 9:e85516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pereyra F, Jia X, Mclaren PJ, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 2010; 330:1551–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med 2003; 54:535–51. [DOI] [PubMed] [Google Scholar]
- 12. Machmach K, Abad-Molina C, Romero-Sánchez MC, et al. ; HIV Controllers Consortium of the AIDS Spanish Network IL28B single-nucleotide polymorphism rs12979860 is associated with spontaneous HIV control in white subjects. J Infect Dis 2013; 207:651–5. [DOI] [PubMed] [Google Scholar]
- 13. Prokunina-Olsson L, Muchmore B, Tang W, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet 2013; 45:164–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet 2009; 41:1105–9. [DOI] [PubMed] [Google Scholar]
- 15. Bibert S, Roger T, Calandra T, et al. ; Swiss Hepatitis C Cohort Study IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med 2013; 210:1109–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Machmach K, Abad-Molina C, Romero-Sánchez MC, et al. IFNL4 ss469415590 polymorphism is associated with unfavourable clinical and immunological status in HIV-infected individuals. Clin Microbiol Infect 2015; 21:289.e1–4. [DOI] [PubMed] [Google Scholar]
- 17. Ferrando-Martínez S, Casazza JP, Leal M, et al. Differential Gag-specific polyfunctional T cell maturation patterns in HIV-1 elite controllers. J Virol 2012; 86:3667–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aka PV, Kuniholm MH, Pfeiffer RM, et al. Association of the IFNL4-ΔG allele with impaired spontaneous clearance of hepatitis C virus. J Infect Dis 2014; 209:350–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang Y, Al-Mozaini M, Buzon MJ, et al. CD4 T-cell regeneration in HIV-1 elite controllers. AIDS 2012; 26:701–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Borghans JA, Mølgaard A, de Boer RJ, Keşmir C. HLA alleles associated with slow progression to AIDS truly prefer to present HIV-1 p24. PLoS One 2007; 2:e920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McLaren PJ, Coulonges C, Bartha I, et al. Polymorphisms of large effect explain the majority of the host genetic contribution to variation of HIV-1 virus load. Proc Natl Acad Sci U S A 2015; 24:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sáez-Cirión A, Sinet M, Shin SY, et al. ; ANRS EP36 HIV Controllers Study Group Heterogeneity in HIV suppression by CD8 T cells from HIV controllers: association with gag-specific CD8 T cell responses. J Immunol 2009; 182:7828–37. [DOI] [PubMed] [Google Scholar]
- 23. Lécuroux C, Sáez-Cirión A, Girault I, et al. Both HLA-B*57 and plasma HIV RNA levels contribute to the HIV-specific CD8+ T cell response in HIV controllers. J Virol 2014; 88:176–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bashirova AA, Martin-Gayo E, Jones DC, et al. LILRB2 interaction with HLA class I correlates with control of HIV-1 infection. PLoS Genet 2014; 10:e1004196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Noel N, Boufassa F, Lécuroux C, et al. ; ANRS C021 CODEX Study Group Elevated IP10 levels are associated with immune activation and low CD4⁺ T-cell counts in HIV controller patients. AIDS 2014; 28:467–76. [DOI] [PubMed] [Google Scholar]
- 26. Hou W, Wang X, Ye L, et al. Lambda interferon inhibits human immunodeficiency virus type 1 infection of macrophages. J Virol 2009; 83:3834–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Machmach K, Leal M, Gras C, et al. Plasmacytoid dendritic cells reduce HIV production in elite controllers. J Virol 2012; 86:4245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stoddart CA, Keir ME, McCune JM. IFN-alpha-induced upregulation of CCR5 leads to expanded HIV tropism in vivo. PLoS Pathog 2010; 6:e1000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meier A, Chang JJ, Chan ES, et al. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat Med 2009; 15:955–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Salgado M, Kirk GD, Cox A, et al. Protective interleukin-28B genotype affects hepatitis C virus clearance, but does not contribute to HIV-1 control in a cohort of African-American elite controllers/suppressors. AIDS 2011; 25:385–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.