Summary
Whole genome sequencing of Methicillin-Resistant Staphylococcus aureus (MRSA) from military trainees with skin and soft tissue infection revealed patterns of intra- and interclass disease transmission. A phylogenetic cluster stemming from 2 training classes separated by 1 year suggested a long-term reservoir for MRSA
Keywords: methicillin-resistant Staphylococcus aureus (MRSA), skin and soft tissue infection, whole genome sequencing, genomic epidemiology, military.
Abstract
Background.
Military trainees are at increased risk for methicillin-resistant Staphylococcus aureus (MRSA) skin and soft tissue infection (SSTI). Whole genome sequencing (WGS) can refine our understanding of MRSA transmission and microevolution in congregate settings.
Methods.
We conducted a prospective case-control study of SSTI among US Army infantry trainees at Fort Benning, Georgia, from July 2012 to December 2014. We identified clusters of USA300 MRSA SSTI within select training classes and performed WGS on clinical isolates. We then linked genomic, phylogenetic, epidemiologic, and clinical data in order to evaluate intra- and interclass disease transmission. Furthermore, among cases of recurrent MRSA SSTI, we evaluated the intrahost relatedness of infecting strains.
Results.
Nine training classes with ≥5 cases of USA300 MRSA SSTI were selected. Eighty USA300 MRSA clinical isolates from 74 trainees, 6 (8.1%) of whom had recurrent infection, were subjected to WGS. We identified 2719 single nucleotide variants (SNVs). The overall median (range) SNV difference between isolates was 173 (1–339). Intraclass median SNV differences ranged from 23 to 245. Two phylogenetic clusters were suggestive of interclass MRSA transmission. One of these clusters stemmed from 2 classes that were separated by a 13-month period but housed in the same barracks. Among trainees with recurrent MRSA SSTI, the intrahost median SNV difference was 7.5 (1–48).
Conclusions.
Application of WGS revealed intra- and interclass transmission of MRSA among military trainees. An interclass cluster between 2 noncontemporaneous classes suggests a long-term reservoir for MRSA in this setting.
Methicillin-resistant Staphylococcus aureus (MRSA) is a major cause of skin and soft tissue infection (SSTI). Individuals in congregate settings (eg, athletes, incarcerated persons, and military personnel) are at increased risk for MRSA colonization and disease. In these settings, a predominance of risk factors, including crowding, infrequent hand washing/bathing, damaged skin, and environmental contamination, favors S. aureus transmission and leads to SSTI outbreaks [1–3].
Among military personnel, SSTI rates are highest among trainees. High rates of MRSA SSTI have been reported at Fort Benning, Georgia, a large training base for the US Army [4–6]. Annually, approximately 35000 soldiers cycle through Fort Benning for Infantry One station unit training (OSUT), which lasts 14 weeks. During OSUT, trainees are grouped into companies (ie, classes) of approximately 200 soldiers each, where a strict daily/weekly regimen is followed, sleeping/living quarters are subgrouped by platoon (approximately 50 soldiers each), and interactions with others outside of the company are infrequent.
Whole genome sequencing (WGS) as an epidemiologic tool has been increasingly used to describe community-associated MRSA transmission dynamics, as well as intrahost evolution [7–10]. Incorporating WGS in the study of military SSTI clusters may refine our understanding of SSTI epidemiology and lend guidance to future control and prevention measures in these and other congregate settings. Here, we describe the genomic characteristics of MRSA isolates within and between military training classes and within individuals with recurrent infection in the OSUT population at Fort Benning.
METHODS
From July 2012 through December 2014, we conducted a prospective SSTI case-control study among soldiers undergoing OSUT at Fort Benning. The trainee population was all male, aged between 17 and 42 years, ethnically diverse, and in generally good physical condition. Trainees seeking medical care for any condition were referred to a single troop medical clinic where study recruitment occurred.
All patients with SSTI and a sample of trainees without SSTI (ie, controls) were recruited for participation. A case was defined as a trainee who presented with cellulitis, abscess, folliculitis, impetigo, paronychia, infected blister, or pilonidal cyst. An S. aureus SSTI case was defined as an SSTI with a positive culture for S. aureus from the infection site. Recurrent SSTI was defined as a subsequent SSTI at a different anatomic site ≥30 days after the first infection.
Demographic, class (ie, platoon, company, battalion, and similar groupings), and risk factor information was collected at enrollment. Participants had S. aureus culture swabs taken at several anatomic sites (eg, nasal, oropharynx, inguinal, perianal regions). Clinical data were abstracted from electronic medical records (Armed Forces Health Longitudinal Technology Application).
Cultures were processed by the Martin Army Community Hospital clinical microbiology laboratory using standard protocols. All S. aureus isolates underwent identification and susceptibility testing using Microscan Walk-Away-96 (Dade Behring Inc.; Deerfield, Illinois), according to Clinical Laboratory Standards Institute methods. Mupirocin (mupA) and chlorhexidine (qacA/B) resistance markers were assessed using polymerase chain reaction (PCR) [11]. Plasmid-encoded mupA was later confirmed by sequence analysis. Staphylococcus aureus isolates were typed by pulsed-field gel electrophoresis (PFGE). Control strains of known pulsed-field types (PFT) were obtained from the Biodefense and Emerging Infections Research Resources Repository [12]. PFGE findings were resolved and analyzed using BioNumerics (Applied Math; Austin, Texas).
For the genomic analysis of MRSA clusters, we selected classes with ≥5 cases of USA300 MRSA SSTI. Among 9 classes, we identified 74 patients, 6 (8.1%) of whom had recurrent SSTI during training.
DNA extraction was performed using the Wizard Kit (Promega; Madison, Wisconsin) and libraries produced using the Next-era XT DNA Library Preparation Kit (Illumina, Inc.; San Diego, California) according to the manufacturer’s instructions. Libraries were multiplexed and sequenced using an Illumina MiSeq 600-cycle kit and 2 × 300 basepair read lengths. Sequence read quality was analyzed with FastQC [13], and low-quality bases were trimmed with Sickle [14]. In order to determine the closest reference for single nucleotide variant (SNV) analysis, sequence reads were assembled using SPAdes [15], and the longest contig from each assembly was aligned against the National Center for Biotechnology Information (NCBI) nucleotide database using BLAST [16]. Staphylococcus aureus USA300 strain TCH1516 (NCBI accession number CP000730) was selected as the reference.
SNV data were analyzed using the Bacterial and Archaeal Genome Analyser [17], a wrapper for proven third-party bioinformatics tools. Sequence reads were mapped to the reference using bwa [18], and variant calls and filtering were performed with GATK [19]. Genomics regions that contained insertions/deletions (indels), potential chromosomal rearrangements, and sequence repeats known to increase the likelihood of false-positive variant calls were excluded from the SNV set. A multiple sequence alignment (MSA) was created from nucleotide substitutions, small deletions called by GATK, and putative large deletions detected in the bwa sequence alignments where no reads mapped. A maximum likelihood tree was constructed from the nucleic acid MSA using PhyML [20] tree search with the GTR substitution model. Sequence data were submitted to NCBI under BioProject PRJNA356758.
Statistical analyses were performed in SAS (SAS, version 9.3; SAS Institute; Cary, North Carolina). The Uniformed Services University Infectious Disease Institutional Review Board (IDCRP-074) approved this study.
RESULTS
Study Population
We approached 2220 SSTI patients; of these, 212 (9.5%) declined to participate. Of 2008 enrollees, 1140 (56.8%) had purulent SSTI, from which 602 (52.8%) were positive for S. aureus. MRSA comprised 49.6% (n = 299) of isolates. The overwhelming majority (91%; n = 271) of these were PFT USA300.
Temporal-Spatial Distribution of Cases
A schematic of the military training structure is shown in Supplementary Figure 1. The 271 cases of USA300 MRSA SSTI were distributed across 121 classes. Nine (7.4%) classes had ≥5 cases (Table 1). Among these classes, the first and last SSTI cases spanned a 28-month period; the training periods of 6 classes (A, C, E, G, H, and I) spanned a 12-month period (Supplementary Figure 2). Four classes (D, E, F, and G) were from the same battalion and housed in the same barracks. Classes A and I were from the same battalion but housed in separate barracks.
Table 1.
Characteristics of 9 Army Infantry Training Classes Included in Genomic Analysis of USA300 Methicillin-Resistant Staphylococcus aureus from Clusters of Skin and Soft Tissue Infection
| Class | Training Period, Start– Finish (month/year) | Total Number of Traineesa | Number (%) of SSTI Casesb | Number (%) of Purulent SSTI Casesc | USA300 Methicillin-Resistant Staphylococcus aureus SSTI | ||
|---|---|---|---|---|---|---|---|
| Number (%) of Casesd | Median (range) Number of Days from Training Start to Presentation | Number of Days Between First and Last Case in Class | |||||
| A | 2/13–5/13 | 222 | 28 (13) | 16 (57) | 13 (81) | 36.5 (13–75) | 62 |
| B | 7/14–10/14 | 230 | 31 (13) | 21 (68) | 11 (52) | 60 (20–90) | 70 |
| C | 11/12–2/13 | 227 | 19 (8) | 14 (74) | 11 (79) | 42.5 (28–93) | 65 |
| D | 4/14–7/14 | 218 | 21 (10) | 14 (67) | 9 (64) | 76.5 (17–87) | 70 |
| E | 11/12–3/13 | 208 | 18 (9) | 11 (61) | 8 (72) | 19.5 (10–88) | 78 |
| F | 5/12–8/12 | 231 | 11 (5) | 8 (73) | 8 (100) | 64 (10–91) | 81 |
| G | 6/13–9/13 | 237 | 22 (9) | 14 (64) | 7 (50) | 66 (45–75) | 30 |
| H | 6/13–8/13 | 215 | 28 (13) | 15 (54) | 7 (47) | 41 (10–54) | 44 |
| I | 3/13–6/13 | 229 | 8 (3) | 6 (75) | 6 (100) | 77.5 (14–91) | 77 |
Abbreviation: SSTI, skin and soft tissue infection.
aNumber at start of training period.
bProportion of training class who developed a SSTI.
cProportion with purulent SSTI among those with SSTI.
dProportion with USA300 methicillin-resistant Staphylococcus aureus (MRSA) SSTI among those with purulent SSTI; 6 individuals with recurrent USA300 MRSA SSTI were in classes F (n = 2), A (n = 1), C (n = 1), D (n = 1), and I (n = 1).
Case Characteristics
We identified 74 trainees with USA300 MRSA SSTI, 6 (8.1%) of whom had recurrent USA300 MRSA SSTI. The median (range) number of cases per class was 8 (6–13) (Table 1). Cases presented for care after a median of 54 (10–93) days of training. Within classes, a median of 70 (30–81) days spanned the first and last cases of USA300 MRSA SSTI. Case chronology by class is shown in Supplementary Table 2.
The demographic and clinical characteristics of cases are presented in Table 2. The median age was 20 (17–29) years. Cases were male and predominantly white (70.3%). The most frequent diagnoses were abscess (63.5%) and purulent cellulitis (54.1%). Most (58.1%) infections occurred on the lower extremities. With respect to SSTI risk factors, 14.5% reported antibiotic use in the past 6 months, 7.2% reported a known/suspected SSTI in the past year, and 2.9% reported having a preceding medically attended SSTI while at Fort Benning.
Table 2.
Demographic, Epidemiologic, and Clinical Characteristics of Infantry Trainees With USA300 Methicillin-Resistant Staphylococcus aureus Skin and Soft Tissue Infection
| Characteristic | Number (%), n = 74 |
|---|---|
| Median (range) age, y | 20 (17–29) |
| Race/Ethnicity | |
| White, Non-Hispanic | 52 (70.3) |
| Hispanic | 16 (21.6) |
| Black, Non-Hispanic | 4 (5.4) |
| Others, Non-Hispanic | 2 (2.7) |
| Median (range) number of days from training start to presentation for first SSTI | 54 (10–103) |
| Clinical Diagnosis | |
| Abscess | 47 (63.5) |
| Purulent cellulitis | 40 (54.1) |
| Infected blister | 6 (8.1) |
| Folliculitis | 5 (6.8) |
| Site of Infection | |
| Lower extremity | 43 (58.1) |
| Upper extremity | 23 (31.1) |
| Head | 7 (9.5) |
| Thorax | 4 (5.4) |
| Groin/Inguinal/Perianal | 4 (5.4) |
| Self-Reported Risk Factors | |
| Known or suspected SSTI in past year | 5 (7.2) |
| Prior medically attended SSTI at Fort Benning | 2 (2.9) |
| Antibiotic use in prior 6 months | 10 (14.5) |
| Contact with a person with SSTI at Fort Benning | 2 (2.9) |
Abbreviation: SSTI, skin and soft tissue infection.
Antibiotic Resistance
The antibiotic resistance profile of isolates by class is presented in Table 3. Six isolates from 2 classes were PCR positive for mupA, the gene associated with high-level mupirocin resistance [21]. Four were from class A and 2 were from class I, accounting for 30.7% and 33.3% of isolates in each class, respectively. In class A, these isolates were from the fourth (patient 1557.C01) and eighth through tenth (patients 1617.C01, 1624.C01, and 1641.C01) case of 13 total cases of USA300 MRSA SSTI in the class. These 4 trainees, all with abscess, presented for care on days 19, 45, 46, and 52 of training, respectively. With regards to mupirocin use, 6 (2.7%) of 222 trainees in class A had been prescribed mupirocin, including 3 cases with USA300 MRSA SSTI (patients 1552.C01, 1581.C01, and 1617.C01), one of whom (patient 1617.C01) had a mupA-positive isolate.
Table 3.
Prevalence of Antibiotic Resistance Among USA300 Methicillin-Resistant Staphylococcus aureus Clinical Isolates by Infantry Training Class
| Resistant to: | Class | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | |
| n = 13 | n = 11 | n = 11 | n = 9 | n = 8 | n = 8 | n = 7 | n = 7 | n = 6 | |
| Trimethoprim–sulfamethoxazole | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Linezolid | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Erythromycin | 1 (7.7) | 11 (100) | 11 (100) | 8 (88.8) | 1 (12.5) | 6 (75) | 4 (57) | 7 (100) | 4 (66.7) |
| Levofloxacin/Ciprofloxacin | 13 (100) | 1 (9) | 0 (0) | 8 (88.8) | 6 (75) | 2 (25) | 4 (57) | 5 (71.4) | 3 (50) |
| Tetracycline | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 6 (75) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Clindamycin | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (14.3) | 0 (0) | 1 (16.7) |
| Chlorhexidine | 0 (0) | 1 (9) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Mupirocin | 4 (30.7) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (33.3) |
A total of 79 (99%) of the isolates were ST-8, and 1 (1%; 2698.C01) did not match to any isolates in the multilocus sequence typing database. All were Panton-Valentine leukocidin positive by polymerase chain reaction.
The 2 mupA-positive isolates from class I (patients 1768.C01 and 1772.C01) were from the fifth and sixth case, respectively, of 6 total cases of USA300 MRSA SSTI in the class. These trainees presented for care on days 90 (abscess) and 91 (abscess/cellulitis) of training, respectively. Two (0.8%) of 229 trainees in class I had been prescribed mupirocin, 1 of whom (patient 1592.C01) had USA300 MRSA SSTI due to a mupA-negative isolate. Of note, classes A and I were from the same battalion but were housed in separate barracks. The training start dates of these 2 classes differed by 21 days.
One isolate, from patient 2880.C01 in class B, was PCR positive for qacA/B, the gene associated with decreased susceptibility to chlorhexidine. This patient presented for treatment of abscess/cellulitis on day 84 of training. He was the eighth of 11 cases of USA300 MRSA SSTI in the class, and his was the only isolate that was PCR positive for the qacA/B gene.
Genomic Characteristics
The average (min-max) read depth was 264X (86X-888X). Sequence data revealed the following sequence types (STs): 79 (99%) were ST-8 and 1 (1%; 2698.C01) did not match to any isolates in the multilocus sequence typing database. A total of 2719 SNVs were identified. The overall median (range) SNV difference between isolates was 173 (1–339).
Intra- and Interclass Relatedness of MRSA
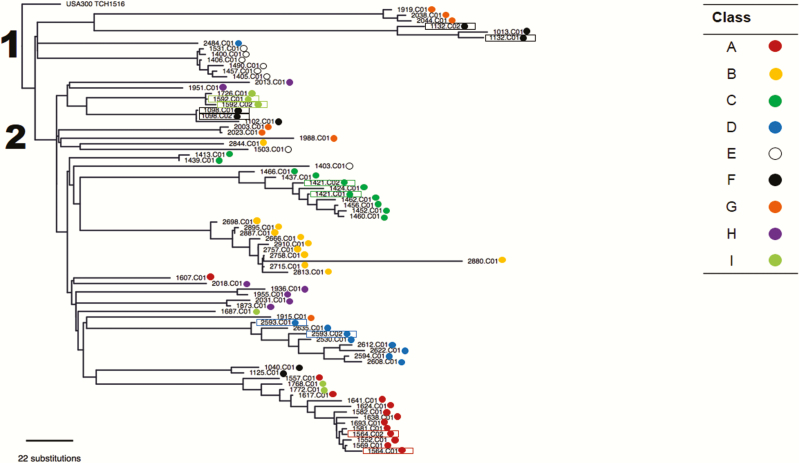
The phylogenetic tree revealed 2 major clades (Figure 1). Intraclass median SNV differences are presented in Table 4. Isolates within each of 5 classes (A–E) were closely related (ie, intraclass median <40), whereas isolates from the other classes (F–I) were more diverse (ie, intraclass median >140), suggesting that a mixed lineage of MRSA, presumably caused by multiple introductions, was responsible for MRSA SSTI within these classes.
Figure 1.
Dendrogram of USA300 methicillin-resistant Staphylococcus aureus (MRSA) isolates from clusters of skin and soft tissue infection in 9 military training classes at Fort Benning, Georgia. The numbers denote the 2 major clades of MRSA identified in the sample, all but 1 of which were ST8.
Table 4.
Single Nucleotide Variants Between USA300 Methicillin-Resistant Staphylococcus aureus Clinical Isolates to Describe Strain Relatedness Within 9 Infantry Training Classes
| Class | Median (range) |
|---|---|
| A | 32 (8–152) |
| B | 30 (7–239) |
| C | 35 (5–163) |
| D | 36 (17–196) |
| E | 23 (5–196) |
| F | 245 (5–277) |
| G | 142 (13–176) |
| H | 214.5 (1–264) |
| I | 148 (5–167) |
Data represent the number of single nucleotide variants between strains within a given training class.
The relatedness of mupA-positive isolates (ie, 1557.C01, 1617.C01, 1624.C01, and 1641.C01 from class A and 1768.C01 and 1772.C01 from class I) was noted. These 6 isolates were collected within a 3-month period and had a maximum pairwise SNV difference of 45. A common lineage was also demonstrated in a cluster of cases from classes F and G. The maximum pairwise SNV difference among the 6 isolates (ie, patients 1132.C01, 1132.C02 and 1013.C01 from class F and 1919.C01, 2038.C01, and 2044.C01 from class G) in this cluster was 295. Of note, these classes belonged to the same battalion and were housed in the same barracks but were separated in time by 13 months.
Recurrent USA300 MRSA SSTI
Among trainees with recurrent infection, the median interval between episodes was 63.5 (49–73) days. Details on clinical characteristics and antibiotic treatment are presented in Table 5. A median of 7.5 (1–48) SNV differences was observed for each pair. There was no correlation between pairwise SNV differences and number of days between episodes. Genomic characteristics of pairwise SNV differences are listed in Supplementary Table 1. Paired isolates from patient 1098 differed by 1 nucleotide in the gene encoding the IS6 family transposase. Similar variants of this gene were found in paired isolates from patients 1421 (3 SNVs) and 1592 (5 SNVs).
Table 5.
Clinical and Bacterial Genomic Characteristics of Recurrent USA300 Methicillin-Resistant Staphylococcus aureus Skin and Soft Tissue Infection Among Infantry Trainees
| Patient ID | Class | First Infection | Second Infection | Number of Days Between Episodes | Number of Single Nucleotide Variants Between Isolates | ||||
|---|---|---|---|---|---|---|---|---|---|
| Diagnosis | Body Location | Treatment | Diagnosis | Body Location | Treatment | ||||
| 1098 | F | Cellulitis | Knee | TMP/SMX | Cellulitis | Forearm | I&D | 67 | 1 |
| TMP/SMX | |||||||||
| Amoxicillin | |||||||||
| Clindamycin | |||||||||
| 1132 | F | Cellulitis | Forearm | TMP/SMX | Abscess | Forearm | TMP/SMX | 73 | 48 |
| Mupirocin | Bacitracin | ||||||||
| 1421 | C | Cellulitis | Neck | Doxycycline | Abscess | Buttock | I&D | 60 | 4 |
| TMP/SMX | |||||||||
| Clindamycin | |||||||||
| 1564 | A | Abscess, Folliculitis | Abdomen, Leg | I&D | Abscess | Forearm | TMP/SMX | 49 | 9 |
| TMP/SMX | |||||||||
| Mupirocin | |||||||||
| 2593 | D | Abscess, Cellulitis | Thigh | I&D | Abscess, Cellulitis | Thigh | I&D | 59 | 25 |
| TMP/SMX | TMP/SMX | ||||||||
| Mupirocin | Cephalexin | ||||||||
Cases with a primary diagnosis of cellulitis also had an abscess.
Abbreviations: I&D, incision and drainage; TMP-SMX, trimethoprim–sulfamethoxazole.
DISCUSSION
Military trainees are at increased risk for acute respiratory infection, gastrointestinal infection, and SSTI [3, 22, 23]. In our study of MRSA SSTI clusters among US Army infantrymen, we used whole genome sequencing to describe the relatedness of clinical isolates and thereby elucidate patterns of disease transmission in these close-quartered cohorts. As anticipated, the majority of isolates within classes were highly related (ie, median SNV differences <40), lending evidence for person-to-person transmission. By contrast, some intraclass clusters were from a mixed lineage, suggesting that multiple introductions of MRSA had occurred. By examining the timing and location of these classes, we deduced that interclass MRSA transmission may have occurred and that successive cohorts of trainees may have been exposed to a common source of MRSA, environmental or otherwise, while at Fort Benning.
Crowding and recent contact with an individual with SSTI are well-established risk factors for infection [3]. Crowding also increases the likelihood of MRSA colonization; household contacts of MRSA index patients readily acquire MRSA, and the number of household members and frequency of interaction are positively correlated with acquisition risk [24]. In military training, up to 50 soldiers occupy a single (“open bay”) barrack where sleeping/living, restroom, and laundry facilities are shared. Genomic data simply confirm what past epidemiologic studies, the majority of them household based, have indicated: close contact and prolonged exposure to individuals with SSTI favors the person-to-person transmission of MRSA.
These findings are consistent with previous genomic studies of MRSA in households [7, 10] and community-based hospitals [8]. However, our population of close-quartered military training cohorts is unique, owing to the “community” size, the uniformity of daily/weekly training activities experienced (and thus, SSTI risk factors encountered), as well as the duration and intensity of MRSA exposure. The incorporation of genomics into the epidemiologic investigation of MRSA SSTI in this population has provided new insight into the relatedness, potential sources, and persistence of MRSA in this setting. These data will be highly informative for SSTI prevention strategies for this high-risk population.
The demonstration of interclass phylogenetic clusters suggests the existence of epidemiologic links that extend beyond training classes themselves. While we did not evaluate the various settings (eg, obstacle courses, recreational facilities) where interclass interaction and MRSA transmission could have occurred, there are at least 2 possibilities. First, colonization and/or infection among training-associated personnel and others with whom the trainees have contact (eg, healthcare personnel) may have served as a reservoir of MRSA. Second, environmental reservoirs (eg, healthcare facilities, wrestling mats, exercise equipment, and other common-touch surfaces contaminated with MRSA) may also have played a role [25, 26]. Household studies have shown that S. aureus contamination of common-touch surfaces is correlated with both SSTI risk and recurrence [27, 28]. As such, environmental cleaning measures, both during and in between training cycles, may need to be used as a SSTI prevention strategy in this setting.
Among recurrent cases, pairwise SNV differences ranged from 1 to 48. There was no correlation between pairwise differences and the interval between episodes. For some cases, the isolates associated with recurrence were genetically distant from the isolates that immediately preceded or proceeded them in the chronology of cases. For 4 cases of recurrent SSTI, <10 SNVs separated the respective pairs, suggesting that the case himself may have served as a source of reinfection (presumably through persistent colonization) [9]. Whether the observed limited diversity of these isolate pairs from recurrent cases reflects a limited intrahost bacterial population is not known.
The genomic epidemiology of recurrent MRSA SSTI has been described in 2 other studies [9, 29]. In a community-based study, isolate pairs differed by a median of 8 (0–37) SNVs [9]. While most pairs indicated a correlation between SNV differences and duration between episodes, 2 pairs separated by 6 months differed by 0–2 SNVs. The second evaluation stemmed from a hospital/clinic-based study of recurrent SSTI (3–6 episodes per patient) in a small, diverse patient population [29]. Genomic characterization revealed a spectrum of diversity, ranging from minimal variation (eg, 6 SNVs among 4 isolates collected over 424 days) to acquisition of an entirely new strain (ie, strain replacement).
In our study, SNVs from recurrent USA300 MRSA isolates resided in the IS6 family transposase gene. Although the function of IS6 transposases specifically have not been described in the literature, these DNA-binding proteins may be critical to the movement of mobile genetic elements in bacterial species [30, 31]. Specific changes to the core genome have important implications for the virulence and persistence of MRSA [10, 32–34]. Whether mutations in transposases are important to S. aureus pathogenicity remains unknown. Further studies of MRSA colonization, infection, and recurrence are needed to advance our current understanding and to dissect the underlying mechanisms of pathogenesis.
We identified a phylogenetic cluster of mupA-positive isolates that spanned contemporaneous classes. The origin of these mupA-positive isolates is unclear. Medical record data revealed several prescriptions for mupirocin; the reasons for these prescriptions are not known. Also, it is not known whether nonstudy participants were prescribed mupirocin. Similarly, the origin of the qacA/B-positive isolate is unclear. Chlorhexidine-based antiseptics (eg, Hibiclens) may still be used on an infrequent basis among military trainees, although a group-randomized trial in this population showed that chlorhexidine was ineffective in preventing SSTIs [6].
This genomic study among military trainees broadens our understanding of MRSA transmission dynamics, specifically in non-healthcare settings (eg, prisons, athletic facilities, childcare centers) where the risk of acquisition and infection is also high [7, 8]. Households play an important role in endemicity of MRSA [9], and many household risk factors for MRSA SSTI (eg, recent exposure to an individual with MRSA, sharing of towels, contamination of common touch surfaces) are likely present, if not amplified, in military barracks. Military trainees endure intense and prolonged exposure to S. aureus, as evidenced by a high prevalence of colonization [35]. MRSA colonization is very likely a critical component in the intra- and interclass transmission chain among military trainees, and USA300 MRSA is highly transmissible when compared to other types [36].
This study has several strengths. First, this was a prospective study among military trainees who, for a 14-week period, had very limited interaction with others outside of their respective classes, even less so with the population surrounding the base. Second, all trainees followed the same schedule and thus likely experienced the same risk factors for S. aureus colonization and infection. Third, the overall study spanned a 2-year period and encompassed the entire training cycle for the classes we evaluated. Fourth, cases accessed a single troop medical clinic and 90% of those who presented for care were enrolled. Thus, the participants are highly representative of the SSTI cases among military trainees at Fort Benning.
There are limitations. First, cases were recruited at the time of clinical presentation for SSTI, meaning we were unable to identify MRSA acquisition and colonization events that preceded infection. Second, the classification of MRSA cases relied on the selection of a single, purified colony from a cultured l specimen, whereas the actual within-host bacterial diversity is likely much greater than what is observed [37]. Third, we were unable to distinguish between isolates that were introduced via incoming trainees vs those that were already in circulation at Fort Benning.
Whole genome sequencing is a valuable epidemiologic tool for studying MRSA in the military training setting, where the continuous cycling of closed cohorts through well-defined, highly structured environments permits comprehensive evaluations of disease transmission. This tool has great potential for the identification and characterization of individual clonal variants of MRSA in this and other congregate settings, including athletic facilities and prisons. Further genomic characterization will improve our understanding of MRSA in the military training environment, from pathogenesis to transmission to environmental persistence, and will yield insight for future prevention strategies.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgment. We are indebted to the study team of clinical research coordinators, laboratory personnel, and data management staff for their dedication to the project.
Disclaimer. The contents of this publication are the sole responsibility of the author(s) and do not necessarily reflect the views, opinions, or policies of Uniformed Services University of the Health Sciences (USUHS); the Department of Defense (DoD); the departments of the Army, Navy, or Air Force; or the Henry M. Jackson Foundation for the Advancement of Military Medicine. Mention of trade names, commercial products, or organizations does not imply endorsement by the US government.
Financial support. This work was supported by a US Department of Defense program project grant (HT9404-12-1-0019 to M. W. E.). Additional support for this work was provided by the Department of Defense Global Emerging Infections Surveillance and Response System (HU0001-10-1-0018 to M. W. E] and the Military Infectious Diseases Research Program (HT9404-12-1-0012 to M. W. E]. The protocol was executed by the Infectious Disease Clinical Research Program, a DoD program executed through the USUHS and funded in part by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health, under an interagency agreement (Y1-AI-5072).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. David MZ, Mennella C, Mansour M, Boyle-Vavra S, Daum RS. Predominance of methicillin-resistant Staphylococcus aureus among pathogens causing skin and soft tissue infections in a large urban jail: risk factors and recurrence rates. J Clin Microbiol 2008; 46:3222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fontanilla JM, Kirkland KB, Talbot EA et al. . Outbreak of skin infections in college football team members due to an unusual strain of community-acquired methicillin-susceptible Staphylococcus aureus. J Clin Microbiol 2010; 48:609–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Campbell KM, Vaughn AF, Russell KL et al. . Risk factors for community-associated methicillin-resistant Staphylococcus aureus infections in an outbreak of disease among military trainees in San Diego, California, in 2002. J Clin Microbiol 2004; 42:4050–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leamer NK, Clemmons NS, Jordan NN, Pacha LA. Update: community-acquired methicillin-resistant Staphylococcus aureus skin and soft tissue infection surveillance among active duty military personnel at Fort Benning GA, 2008–2010. Mil Med 2013; 178:914–20. [DOI] [PubMed] [Google Scholar]
- 5. Morrison-Rodriguez SM, Pacha LA, Patrick JE, Jordan NN. Community-associated methicillin-resistant Staphylococcus aureus infections at an army training installation. Epidemiol Infect 2010; 138:721–9. [DOI] [PubMed] [Google Scholar]
- 6. Ellis MW, Schlett CD, Millar EV et al. . Hygiene strategies to prevent methicillin-resistant Staphylococcus aureus skin and soft tissue infections: a cluster-randomized controlled trial among high-risk military trainees. Clin Infect Dis 2014; 58:1540–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alam MT, Read TD, Petit RA 3rd et al. . Transmission and microevolution of USA300 MRSA in U.S. households: evidence from whole-genome sequencing. MBio 2015; 6:e00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Popovich KJ, Snitkin E, Green SJ et al. . Genomic epidemiology of USA300 methicillin-resistant Staphylococcus aureus in an urban community. Clin Infect Dis 2016; 62:37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Uhlemann AC, Dordel J, Knox JR et al. . Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community. Proc Natl Acad Sci U S A 2014; 111:6738–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uhlemann AC, Kennedy AD, Martens C, Porcella SF, Deleo FR, Lowy FD. Toward an understanding of the evolution of Staphylococcus aureus strain USA300 during colonization in community households. Genome Biol Evol 2012; 4:1275–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schlett CD, Millar EV, Crawford KB et al. . Prevalence of chlorhexidine-resistant methicillin-resistant Staphylococcus aureus following prolonged exposure. Antimicrob Agents Chemother 2014; 58:4404–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biodefense and Emerging Infections Research Resources Repository Available at: beiresources.org. Accessed 15 December 2016.
- 13. FastQC Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 2 November 2016.
- 14. Joshi NA. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33) 2011. Available at https://github.com/najoshi/sickle. Accessed 2 November 2016.
- 15. Bankevich A, Nurk S, Antipov D et al. . SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403–10. [DOI] [PubMed] [Google Scholar]
- 17. BAGA Available at: https://github.com/daveuu/baga. Accessed 2 November 2016.
- 18. Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009; 25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McKenna A, Hanna M, Banks E et al. . The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 2010; 59:307–21. [DOI] [PubMed] [Google Scholar]
- 21. Udo EE, Sarkhoo E. Genetic analysis of high-level mupirocin resistance in the ST80 clone of community-associated methicillin-resistant Staphylococcus aureus. J Med Microbiol 2010; 59:193–9. [DOI] [PubMed] [Google Scholar]
- 22. Centers for Disease C, Prevention. Norwalk-like viral gastroenteritis in U.S. Army trainees—Texas, 1998. MMWR Morb Mortal Wkly Rep 1999; 48:225–7. [PubMed] [Google Scholar]
- 23. Kolavic-Gray SA, Binn LN, Sanchez JL et al. . Large epidemic of adenovirus type 4 infection among military trainees: epidemiological, clinical, and laboratory studies. Clin Infect Dis 2002; 35:808–18. [DOI] [PubMed] [Google Scholar]
- 24. Mollema FP, Richardus JH, Behrendt M et al. . Transmission of methicillin-resistant Staphylococcus aureus to household contacts. J Clin Microbiol 2010; 48:202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mukherjee N, Sulaiman IM, Banerjee P. Characterization of methicillin-resistant Staphylococcus aureus isolates from fitness centers in the Memphis metropolitan area, Tennessee. Am J Infect Control 2016; 44:1681–3. [DOI] [PubMed] [Google Scholar]
- 26. Harrison EM, Ludden C, Brodrick HJ et al. . Transmission of methicillin-resistant Staphylococcus aureus in long-term care facilities and their related healthcare networks. Genome Med 2016; 8:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Knox J, Uhlemann AC, Miller M et al. . Environmental contamination as a risk factor for intra-household Staphylococcus aureus transmission. PLoS One 2012; 7:e49900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Uhlemann AC, Knox J, Miller M et al. . The environment as an unrecognized reservoir for community-associated methicillin resistant Staphylococcus aureus USA300: a case-control study. PLoS One 2011; 6:e22407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Azarian T, Daum RS, Petty LA et al. . Intrahost evolution of methicillin-resistant Staphylococcus aureus USA300 among individuals with reoccurring skin and soft-tissue infections. J Infect Dis 2016; 214:895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aziz RK, Breitbart M, Edwards RA. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res 2010; 38:4207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hennig S, Ziebuhr W. Characterization of the transposase encoded by IS256, the prototype of a major family of bacterial insertion sequence elements. J Bacteriol 2010; 192:4153–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tewhey R, Cannavino CR, Leake JA et al. . Genetic structure of community acquired methicillin-resistant Staphylococcus aureus USA300. BMC Genomics 2012; 13:508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kennedy AD, Otto M, Braughton KR et al. . Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci U S A 2008; 105:1327–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prosperi M, Veras N, Azarian T et al. . Molecular epidemiology of community-associated methicillin-resistant Staphylococcus aureus in the genomic era: a cross-sectional study. Sci Rep 2013; 3:1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Singh J, Johnson RC, Schlett CD et al. . Multi-body-site microbiome and culture profiling of military trainees suffering from skin and soft tissue infections at Fort Benning, Georgia. mSphere 2016;1. doi: 10.1128/mSphere.00232-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miller LG, Eells SJ, Taylor AR et al. . Staphylococcus aureus colonization among household contacts of patients with skin infections: risk factors, strain discordance, and complex ecology. Clin Infect Dis 2012; 54:1523–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Worby CJ, Lipsitch M, Hanage WP. Within-host bacterial diversity hinders accurate reconstruction of transmission networks from genomic distance data. PLoS Comput Biol 2014; 10:e1003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.