Abstract
Humans possess three major isoforms of the apolipoprotein E (ApoE) gene encoded by three alleles: ApoE ε2 (ApoE2), ApoE ε3 (ApoE3), and ApoE ε4 (ApoE4). It is established that the three ApoE isoforms confer differential susceptibility to Alzheimer’s disease (AD); however, an in-depth molecular understanding of the underlying mechanisms is currently unavailable. In this study, we examined the cortical proteome differences among the three ApoE isoforms using 6-month-old female, human ApoE2, ApoE3, and ApoE4 gene-targeted replacement mice and two-dimensional proteomic analyses. The results reveal that the three ApoE brains differ primarily in two areas: cellular bioenergetics and synaptic transmission. Of particular significance, we show for the first time that the three ApoE brains differentially express a key component of the catalytic domain of the V-type H+-ATPase (Atp6v), a proton pump that mediates the concentration of neurotransmitters into synaptic vesicles and thus is crucial in synaptic transmission. Specifically, our data demonstrate that ApoE2 brain exhibits significantly higher levels of the B subunit of Atp6v (Atp6v1B2) when compared to both ApoE3 and ApoE4 brains, with ApoE4 brain exhibiting the lowest expression. Our additional analyses show that Atp6v1B2 is significantly impacted by aging and AD pathology and the data suggest that Atp6v1B2 deficiency could play a role in the progressive loss of synaptic integrity during early development of AD. Collectively, our findings indicate that human ApoE isoforms differentially modulate regulatory mechanisms of bioenergetic and synaptic function in female brain. A more efficient and robust status in both areas could serve as a potential mechanism contributing to the neuroprotective and cognition-favoring properties associated with the ApoE2 genotype.
Keywords: Alzheimer’s disease, ApoE2, ApoE3, ApoE4, cellular bioenergetics, synaptic transmission, V-type H+-ATPase
Introduction
Alzheimer’s disease (AD) currently affects 35 million people worldwide, including 5.4 million Americans [1]. At present, the underlying cause of AD is unknown and an effective treatment is yet to be established. The past 15 years of research have yielded over 200 clinical trials aimed at the treatment of AD; all of which have failed to succeed [2, 3]. The unanticipated challenges associated with the development of a successful treatment combined with the estimated rapid increases in AD prevalence stress the importance of identifying the underlying AD risk-mechanisms that would allow prevention, risk reduction, and early intervention in the preclinical stage of AD [4].
Apolipoprotein E (ApoE) is peripherally synthesized and secreted by the liver where it regulates the transport of cholesterol, lipoproteins, and fat-soluble vitamins [5, 6]. In the brain, ApoE is synthesized and secreted by astrocytes and functions to transport cholesterol into neurons via interaction with ApoE receptors [7]. The human ApoE gene exists as three major isoforms encoded by three polymorphic alleles: ε2, ε3, and ε4. The three translated ApoE proteins differ sequentially by two amino acid substitutions at residues 112 and 158: ApoE2 (Cys112, Cys158), ApoE3 (Cys112, Arg158), and ApoE4 (Arg112, Arg158) [8, 9]. These amino acid differences affect the 3D structure of ApoE and are suggested to alter the ability of ApoE to bind lipids [10], LDL receptor [11], and Aβ [12, 13]. Apart from structural differences, the population distribution and associated AD risk of ApoE alleles also differ significantly. ApoE3, the most common ApoE isoform, is present in 75% of the population and is considered to be risk-neutral [14]. ApoE2 is relatively rare, with only 5% incidence, and is considered to be a neuroprotective variant [15]. In contrast, ApoE4, occurring in 20% of the population, is present in nearly 50% of AD patients [16].
As the greatest genetic risk factor for AD [16], ApoE4 is clinically associated with an increased rate and severity of cognitive decline [17–19], a younger age of onset [20], and altered response to AD treatments [6]. On the molecular level, ApoE4 has been shown to reduce brain glucose utilization [21], increase neuronal inflammation [22], and is associated with increased Aβ dyshomeostasis [23, 24]. Moreover, multiple studies have demonstrated that ApoE4-associated AD risk is significantly more pronounced in the female population [25–27]. For example, a recent longitudinal study conducted in a cohort of 8,084 elderly individuals (healthy controls: n=5,496; MCI cases: n=2,588) demonstrated that the risk of clinical conversion from healthy aging to MCI or from MCI to AD conferred by the ApoE4 allele is significantly greater in women than in men [28]; a finding that corresponds with several earlier reports [29, 30]. Contrarily, possession of the ApoE2 allele has been associated with significantly decreased susceptibility for AD and improved cognitive function in the aging brain particularly in the male population [31]. A 3-year follow-up study conducted in 917 non-demented elderly subjects revealed that subjects possessing the ε2/ε2 or ε2/ε3 genotype maintained consistent verbal learning performance while the learning ability of subjects with alternative ApoE genotypes deteriorated [32]. An additional 8-year cohort study involving 669 elderly individuals determined that subjects who possessed one or more copies of the ε2 allele exhibited improved episodic memory, while episodic memory declined sharply in subjects possessing one ε4 allele [33].
While there exists an abundance of research demonstrating the neurodegenerative impact of the ApoE4 status, far less is known about the mechanisms by which ApoE2 exhibits neuroprotection. To address this research gap, our laboratory has initiated a series of novel studies designed to identify the molecular differences that separate ApoE2 brain from ApoE3 and ApoE4 brains, which could contribute to the neuroprotective properties of ApoE2. We have recently shown that the three human ApoE isoforms differentially modulate signaling pathways involved in glucose uptake and metabolism; compared to ApoE3 and ApoE4 brains, the ApoE2 brain exhibits the most bioenergetically robust profile, providing a possible mechanism whereby ApoE2 delegates neuroprotection [34]. In the current study, we continued our examination of the three ApoE isoforms in female, human ApoE2, ApoE3, and ApoE4 gene-targeted replacement mice. Using two-dimensional (2D) proteomics coupled with liquid chromatography mass spectrometry analyses (LC-MS/MS), we detected differential expression patterns of enzymes involved in bioenergetic and synaptic processes among the three ApoE brains. Of particular significance, we found for the first time that ApoE2 brain expresses a significantly higher level of a key component of the V-type H+-ATPase (V-ATPase; Atp6v), a proton pump that mediates neurotransmitter accumulation in synaptic vesicles and thus is essential in synaptic transmission. An extension of our proteomic analyses revealed that the expression of the B subunit of Atp6v (Atp6v1B2) is predominantly localized to the membrane of synaptic vesicles and is significantly impacted by aging and AD pathology. These data provide another possible mechanistic explanation for the neuroprotective properties associated with the ApoE2 genotype and present a possible molecular entity that could be targeted to sustain synaptic function thus preventing or reducing the risk of AD in particularly high-risk population of female ApoE4 carriers.
Materials and Methods
Animals
The use of animals was approved by the Institutional Animal Care and Use Committee at the University of Kansas and followed NIH guidelines for the care and use of laboratory animals. This study was carried out in human ApoE2, ApoE3, and ApoE4 gene-targeted replacement (hApoE2-TR, hApoE3-TR, and hApoE4-TR) mouse models. These mouse lines were created by gene targeting and carry one of the three human ApoE alleles in place of the endogenous murine ApoE gene while retaining the endogenous regulatory sequences required for modulating hApoE expression [35]. These mice share a C57BL/6J genetic background and express the hApoE protein at physiological levels; thus, they provide a complete in vivo system that allows direct measurement and comparison of hApoE isoform-specific effects. The following experiments were conducted on cortical tissues collected from 6-month-old hApoE2-TR, hApoE3-TR, and hApoE4-TR female mice (n=3 for proteomic analyses; n=5 for immunoblot analyses). For aging and pathology studies, cortical tissues were collected from female 129/C57BL/6 and female triple transgenic AD (3xTg-AD) mice at ages of 6, 9, 12 and 15 months (n=4–5 for each age group). The 3xTg-AD mouse model harbors three human AD-relevant generic alternations: APPswe, PS1M146V and tauP301L [36]. On the day of sacrifice, brain tissues were immediately dissected on ice; brains were separated into the left and right hemispheres and hippocampal and non-cortical tissues were removed. The remaining left and right cortices were cut into two pieces yielding a total of 4 cortical samples (left – upper, left – lower, right – upper, and right – lower) which were immediately flash frozen in dry ice. In the studies presented here, one piece of isolated cortical tissue (left – upper) from each animal was processed for protein extraction as indicated.
Protein extraction, isoelectric focusing, and 2D-PAGE
Cortical tissues from 6-month-old female hApoE2-TR, hApoE3-TR, and hApoE4-TR mice were homogenized using the Bullet Blender 24 Homogenizer (Next Advance, Averill Park, NY) in sample lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, 40 mM Tris pH 8–9, 100 mM dithiothreitol (DTT), 0.2% BioLyte 3–10 ampholyte) supplemented with protease and phosphatase inhibitors with 100 μL 0.5 mm glass beads at speed 8 for 3 min at 4°C followed by centrifugation at 12,000 rpm for 8 min at 4°C. Homogenates were transferred to a new microcentrifuge tube and protein concentration was determined via BCA assay (Thermo Scientific, Grand Island, NY). 150 μg of homogenized cortical protein sample was precipitated using cold 100% trichloroacetic acid (TCA) to obtain a concentration of 15% (v/v) TCA in solution and incubated on ice for 15 minutes. Samples were centrifuged at 14,000 rpm for 10 min at 4 °C. The resulting pellets were washed four times by re-suspension in a 1:1 (v/v) cold ethanol: ethyl acetate solution. Following the final wash, pellets were completely dried at RT and re-hydrated in 200 μL rehydration buffer (8 M urea, 2 M thiourea, 20 mM DTT, 2.0% (w/v) CHAPS, 0.2% Biolytes, trace amount Bromophenol Blue) for 2 hr at RT followed by vortexing for 15 seconds at 100% power. Total samples were added to each lane of the IEF tray and an 11 cm pH 3–10 ReadyStrip IPG Strip (Bio-Rad, Hercules, CA) was placed on top. Samples were run for 45 min, paused and overlaid with 2 mL of mineral oil to prevent evaporation during the IEF process. Strips were actively rehydrated at 20 °C for 18 h at 50 V, focused at a constant temperature of 20 °C beginning at 300 V linear for 2 h, 500 V linear for 2 h, 1000 V linear for 2 h, 8000 V linear for 8 h, and finishing at 8000 V rapid for 10 h. Following completion of IEF, IPG strips were stored at −80 °C until required for second dimension analysis. Prior to gel electrophoresis, IEF strips were allowed to acclimate to RT for approximately 30 min followed by incubation for 10 min in the dark in 4 mL equilibration buffer A (1.5 M Tris-HCl, pH 8.8, 6 M urea, 1% (w/v) SDS, 30% v/v glycerol, 0.5% DTT) followed by 10 min incubation in equilibration buffer B (1.5 M Tris-HCl, pH 8.8, 6 M urea, 1% (w/v) SDS, 30% v/v glycerol, 4.5% Iodoacetamide (IAA)). Equilibrated IPG strips were immersed in TGS running buffer to remove any residual equilibration buffer and then placed onto 8–16% linear gradient pre-cast Criterion Tris-HCl polyacrylamide gels (Bio-Rad). Fresh agarose was overlaid and allowed to set on the strips to ensure complete adhesion to the 2D gel. Gels were resolved for 10 min at 80 V followed by a constant voltage of 200V for 50 min.
SYPRO Ruby staining and gel image analysis
Following electrophoresis, gels were placed in fixing solution (7% acetic acid v/v, 10% methanol v/v in water) and incubated for 60 min at RT with continuous rocking. Gels were then incubated in 50 mL SYPRO Ruby fluorescent gel stain (Bio-Rad) overnight at RT with constant rocking. Following overnight incubation, SYPRO Ruby stain was removed and the gels were washed and stored in 50 mL ddH20. SYPRO Ruby-stained gels were scanned using a Typhoon Trio Scanner (Amersham Biosciences, Pittsburg, PA) set at normal sensitivity with excitation and emission wavelengths of 470 and 618 nm and saved as TIFF files. PDQuest 2D analysis software (Bio-Rad) was then utilized to adjust the size and orientation of the images via cropping and rotating tools followed by spot alignment across gels from different genotypes. For this set of experiments, which were carried out in triplicate, one reference gel was generated and automatic gel comparison to the reference gel was performed. Protein spots were standardized to the total density detected in each individual gel image. From this analysis, a total of 270 protein spots (101 spots in ApoE2 vs ApoE3 and 169 spots in ApoE2 vs ApoE4) were identified. Using the Student’s t-test, 37 spots (18 spots in ApoE2 vs ApoE3 and 19 spots in ApoE2 vs ApoE4) were determined to be statistically significant (p<0.05).
Spot excision, trypsin digestion, LC-MS/MS and database search
Of the 37 spots identified, 20 spots were selected for protein identification using LC-MS/MS; and these spots remained significant from the additional one-way ANOVA with Student-Newman-Keuls post-hoc test. These spots were excised from 2D gels using a UV transilluminator and sterilized micropipette tips and transferred to sterile microcentrifuge tubes in deionized water. Excised spots were then de-stained by incubation with 200 mM NH4HCO3/50% (v/v) acetonitrile at 37°C. Following incubation, solutions were removed from the tubes and gel plugs were allowed to dry under constant airflow in a sterile hood. Gel pieces were then reduced by incubation with dithiothreitol (DTT) at 60°C for 30 min and alkylated by incubation with IAA at RT for 30 min. Trypsin (Promega, Madison, WI) was added to the gel plugs in 200 mM NH4HCO3/10% (v/v) acetonitrile and incubated overnight at 37°C. Digested protein samples were analyzed by LC-MS/MS using a NanoAcquity chromatographic system coupled to an LTQ-FT mass spectrometer. Peptides were separated on a reverse-phase C18 column, 15 cm, 300 μm i.d. (Thermo Acclaim PepMap300, 300 Å, 5 μm). A gradient was developed from 1 to 40% B in 50 min, ramped to 95% B in 4 min, and held at 95% B for 5 min at a flow rate of 10 μL/min with solvents A (99.9% H2O, 0.1% formic acid) and B (99.9% acetonitrile, 0.1% formic acid). The electrospray ionization (ESI) source was operated with spray voltage of 2.8 kV, a tube lens offset of 200 V and a capillary temperature of 200 °C. All other source parameters were optimized for maximum sensitivity of the Met-Arg-Phe-Ala (MRFA) tetrapeptide MH+ ion at m/z 524.26. The instrument was calibrated using an automatic routine based on a standard calibration solution (caffeine, MRFA peptide, and a commercially available mixture of fluorinated phosphazines (Ultramark 1621). The data-dependent acquisition method for the mass spectrometer (version LTQ-FT 2.5.5) was set up using Xcalibur software (Thermo Scientific) and full MS survey scans were acquired at a resolution of 50 000 with an automatic gain control (ACG) target of 5 × 105. The five most abundant ions were fragmented in the linear ion trap by collision-induced dissociation with an AGC target of 2 × 103 or maximum ion time of 300 ms and the ion selection threshold was 500 counts. Analysis of MS/MS spectra for peptide and protein identification was performed by protein database searching using Mascot (Matrix Science, Boston, MA, and Version 2.3) and X! Tandem (The Global Proteome Machine Organization, Version 2010.12.01.1). The programs were set up to search the UniProt-Sprot database assuming trypsin as the digestion enzyme. The fragment ion mass tolerance was 0.20 Da and the parent ion tolerance was 20 ppm. Scaffold software was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95% probability as specified by the Peptide Prophet Algorithm.
Immunoblotting
Approximately 30 mg of cortical tissues collected from female hApoE mice, 129/C57BL/6 mice and 3xTg-AD mice was processed for protein extraction using tissue protein extraction reagent (Thermo Scientific) supplemented with protease and phosphatase inhibitors. Protein concentrations were determined using a BCA protein assay kit (Pierce) according to the manufacturer’s instructions. 20 μg of each total protein sample was loaded and separated by 10% SDS-PAGE. Resolved proteins were transferred to 0.2 μm pore-sized PVDF membranes and blocked with 2% blotting grade blocker (BioRad) in 1× TBST (100 mL 10× TBS (200 mM Tris, 1.5 mM NaCl, pH 7.6), 10 mL 10% Tween-20, 890 mL ddH2O) for 1 hr at RT with continuous rocking. Membranes were incubated with customized dilutions of the primary antibody anti-V-ATPase B2 (1:1000, Santa Cruz, Dallas, TX) at 4°C overnight, followed by incubation with the HRP-conjugated secondary antibody (1:5000, Pierce) for 1 hr at RT. Bands were visualized using chemiluminescence with an ECL detection kit (BioRad) and scanned using the C-Digit Blot Scanner (LI-COR, Lincoln, NE). Following visualization, blots were stripped with restore PLUS Western blot stripping buffer (Thermo Scientific) and re-probed for β-tubulin (Pierce) immunoreactivity as the loading control. Relative intensities of the immunoreactive bands were quantified using the Image Studio Version 4.0 image digitizing software.
Immunocytochemistry
Primary hippocampal neurons were isolated from Day 18 embryonic rat pups as previously described [37] and grown for 15 days on poly-D-lysine treated coverslips (NeuVitro, Vancouver, WA) at a density of 1.5 × 105. Cells were fixed in 4% paraformaldehyde for 15 minutes at room temperature (RT) and permeabilized in PBS + 0.5% Triton X-100 (PBST) for 5 minutes at RT. Cells were incubated for 2 hr at RT with customized dilutions of the primary antibodies: anti-V-ATPase B2 (1:100, Santa Cruz), anti-synaptophysin (1:500, Abcam, Cambridge, United Kingdom), and anti-tau (1:500; Thermo Scientific) in PBST supplemented with 0.5% bovine serum albumin (BSA, Sigma Aldrich). Triple-stained coverslips were then incubated for 1 hr at RT with customized dilutions of secondary antibodies including FITC-conjugated goat-anti-mouse (1:1000, Abcam), preadsorbed goat-anti-rabbit-Cy3 (1:1000, Abcam) and preadsorbed donkey-anti-goat Alexa-Fluor 647 (1:1000, Abcam) in PBST with 0.5% BSA. Coverslips were mounted on standard glass microscopy slides with vectashield mounting medium containing DAPI (Vector Laboratories) and sealed with clear nail polish. Confocal images were acquired using a customized Olympus IX81/spinning disk confocal inverted microscope (Olympus, Yokogawa) equipped with an Olympus 60× 1.45 NA oil immersion objective (Olympus). Images were collected and analyzed using the Slidebook Version 6.0 imaging software (Intelligent Imaging Innovations, Denver, CO) with 20–25 image stacks with a 0.10 μm step size through the cells.
Campbell-Switzer silver staining of Aβ plaques
Mouse hemispheres were sectioned using MultiBrain processing technology at NeuroScience Associates (NSA; Knoxville, TN). Briefly, hemispheres were treated with 20% glycerol and 2% DMSO to prevent freeze-artifacts and subsequently embedded together in a gelatin matrix. After curing, the block of embedded hemispheres was rapidly frozen by immersion in isopentane chilled to −70°C with crushed dry ice and mounted on a freezing stage of an AO 860 sliding microtome. The block was sectioned at 35 μm in the coronal plane through the hippocampus region of the mouse hemispheres (Bregma −0.5 to −4.5 mm). Cut sections were collected sequentially into 12 containers that were filled with antigen preserve solution (50% PBS, 50% ethylene glycol and 1% polyvinyl pyrrolidone, pH 7.0). Campbell-Switzer silver staining that labels Aβ plaques was performed on a serial set of one-of-every-4th (140 μm interval) sections of mouse hemispheres. Briefly, the sections were placed in freshly prepared 2% ammonium hydroxide for 5 minutes. The sections were next placed in a silver-pyridine-carbonate solution for 40 minutes, 1% citric acid for 3 minutes, and 0.5% acetic acid until ready for development. The sections were developed in Physical Developer ABC solution (after Gallyas; containing Na carbonate citric acid, tungstosilicic acid and formaldehyde) with the development time being visually assessed. The development was stopped by briefly placing the sections in 0.5% acetic acid. The stained sections were then mounted on gelatinized glass slides and viewed under a Zeiss Axiovert 200M Marianas digital microscopy workstation equipped with the Slidebook imaging software. Images were stitched together to create a 10× montage using the Fiji Image Stitching Plugin as previously indicated [38].
Statistical Analysis
Statistically significant differences between groups were determined using one-way analysis of variance (ANOVA) followed by Student-Newman-Keuls pairwise multiple comparisons post hoc tests. Statistical significance is indicated as follows: * P<0.05, ** P<0.01.
Results
ApoE2 brain differentially expresses enzymes associated with cellular homeostasis, cellular bioenergetics, and synaptic transmission
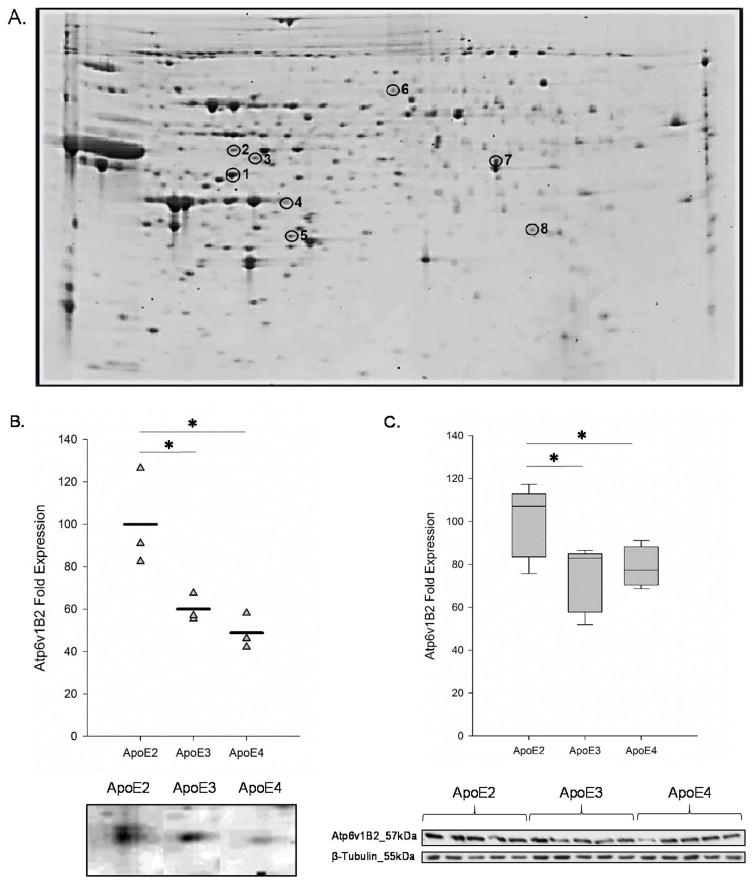
In this study, we sought to determine the cortical proteome differences among the three human ApoE isoforms. Two-dimensional proteomic analysis was performed on 150 μg of cortical total protein derived from 6-month-old, female hApoE2-TR, hApoE3-TR, and hApoE4-TR mice. Comparisons between (a) ApoE2 and ApoE3 and (b) ApoE2 and ApoE4 isoforms were evaluated using PDQuest software. From this analysis, 20 gel spots that were significantly differed in ApoE2 brain when compared to ApoE3 and/or ApoE4 were selected for identification via LC-MS/MS. Of the 20 spots analyzed, 8 proteins were identified with the filter parameters: (a) a percent coverage of greater than 9%, (b) a minimum number of 2 unique peptides, (c) a minimum peptide threshold of 95%, and (d) a minimum protein threshold of 95%. A complete listing of identified proteins with protein accession number, gene name, percent coverage, unique peptide number, and fold change relative to expression levels in ApoE2 brain with the corresponding p-value are indicated in Table 1. Of the 8 proteins identified, 4 proteins exhibited significantly higher levels in ApoE2 brain versus ApoE3, including V-type H+-ATPase subunit 1B2 (Atp6v1b2, 1.67-fold change, p = 0.016), cytosolic non-specific dipeptidase (Cndp2, 1.12-fold change, p = 0.026), dimethylarginine dimethylaminohydrolase (Ddah1, 1.17-fold change, p = 0.021), and Glycine—tRNA ligase (Gars, 2.78-fold change, p = 0.044); while 1 protein, B-type creatine kinase (Ckb, 0.62-fold change, p = 0.033), was found to be higher in ApoE3 brain over ApoE2 brain. Likewise, a comparison between ApoE2 and ApoE4 brain yielded 4 proteins that were expressed at significantly higher levels in ApoE2 brain and 1 protein was found to be higher in ApoE4 brain. Expression levels of cytochrome b-c1 complex subunit 1 (Uqcrc1, 1.35-fold change, p = 0.032), V-type H+-ATPase subunit 1B2 (Atp6v1b2, 2.05-fold change, p = 0.013), cytosolic non-specific dipeptidase (Cndp2, 1.38-fold change, p = 0.007), and glycerol-3-phosphate dehydrogenase 1-like (Gpd1l, 2.7-fold change, p = 0.020) were significantly higher in ApoE2 brain when compared with ApoE4 while the expression of mitochondrial aldehyde dehydrogenase 2 (Aldh2, 0.7-fold change, p = 0.025) was significantly reduced in ApoE2 brain compared to ApoE4 brain.
Table 1.
Summary of identified proteins with significantly different expression levels in ApoE2 brain compared to ApoE3 and/or ApoE4. Fold Change and p-value are given for only those proteins in which there was a significant difference from the ApoE2 group.
| Biological Classification |
Gel ID |
Protein Name | UniProt Number |
Gene Symbol |
Percent Coverage |
Unique Peptides |
ApoE2 vs ApoE3 | ApoE2 vs ApoE4 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Fold Change |
P- value |
Fold Change |
P- value |
|||||||
| Cellular Bioenergetics | 1 | Cytochrome b-c1 complex subunit 1, mitochondrial | Q9CZ13 | Uqcrc1 | 32% | 18 | 1.35 | 0.032 | ||
| 4 | Creatine kinase, B-type | Q04447 | Ckb | 29% | 10 | 0.62 | 0.033 | |||
| 7 | Aldehyde dehydrogenase 2, mitochondrial, isoform CRA_b | Q544B1 | Aldh2 | 12% | 5 | 0.70 | 0.025 | |||
| 8 | Glycerol-3-phosphate dehydrogenase [NAD(+)] | B2RSR7 | Gpd1l | 13% | 4 | 2.70 | 0.020 | |||
| Synaptic Transmission | 2 | V-type proton ATPase subunit B, brain isoform | P62814 | Atp6v1b2 | 17% | 7 | 1.67 | 0.016 | 2.05 | 0.013 |
| 5 | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | Q9CWS0 | Ddah1 | 12% | 2 | 1.17 | 0.021 | |||
| 6 | Glycine—tRNA ligase | Q9CZD3 | Gars | 9.3% | 5 | 2.78 | 0.044 | |||
| Other Proteins | 3 | Cytosolic non-specific dipeptidase | Q9D1A2 | Cndp2 | 28% | 9 | 1.12 | 0.026 | 1.38 | 0.007 |
ApoE2 brain exhibits higher expression of a key catalytic subunit of V-type H+-ATPase
To validate our initial findings from 2D-PAGE, immunoblot analysis was performed on cortical protein samples derived from the same hApoE2-TR, hApoE3-TR, and hApoE4-TR mice (n=5) and normalized to β-tubulin immunoreactivity. Of the 8 proteins identified (Table 1), we were particularly interested in Atp6v1B2 for three reasons: a) the expression levels of Atp6v1B2 were significantly higher in ApoE2 brain compared to both ApoE3 and ApoE4 brains, b) Atp6v plays an essential role in synaptic transmission by mediating the concentration of neurotransmitters into pre-synaptic vesicles, and c) of the 8 subunits that comprise the cytoplasmic domain of Atp6v, the B subunit is essential for ATP hydrolysis and thus successful proton transport [39, 40]. Consistent with findings from our proteomic analysis (Fig. 1B), the expression of Atp6v1B2 was reduced by 20% in ApoE3 brain (0.80-fold change, p = 0.03) and 22% in ApoE4 brain (0.78-fold change, p = 0.04) when compared to ApoE2 brain (Fig. 1C). No significant change in Atp6v1B2 expression was observed between ApoE3 and ApoE4 brains (p = 0.671). These data confirm that ApoE2 brain exhibits a higher expression level of a key component of the catalytic domain of Atp6v when compared to both ApoE3 and ApoE4 brains.
Figure 1. ApoE2 brain expresses significantly higher levels of Atp6v1B2.
(A) A representative 2D gel image of protein spots derived from 6-month-old female hApoE2-TR mouse cortex. Selected proteins are circled on the gel image and protein identification number corresponding to Table 1 is indicated adjacent to protein spot. (B) Proteomic analysis of 150 μg of cortical protein demonstrates that the expression of Atp6v1B2 is significantly higher in ApoE2 brain when compared to both ApoE3 and ApoE4 brains. The data are shown as a vertical point plot with the mean value indicated by a horizontal line; n=3. (C) Immunoblot analysis of cortical tissues harvested from 6-month-old female hApoE2-TR, hApoE3-TR, and hApoE4-TR mice confirmed the proteomic results. The data are shown as a box plot with the bottom bar representing the lowest data point, the middle bar representing the median, and the upper bar representing the highest data point; n=5. *p<0.05 when compared to ApoE2 brain.
Atp6v1B2 is altered by aging and AD pathology in female brain
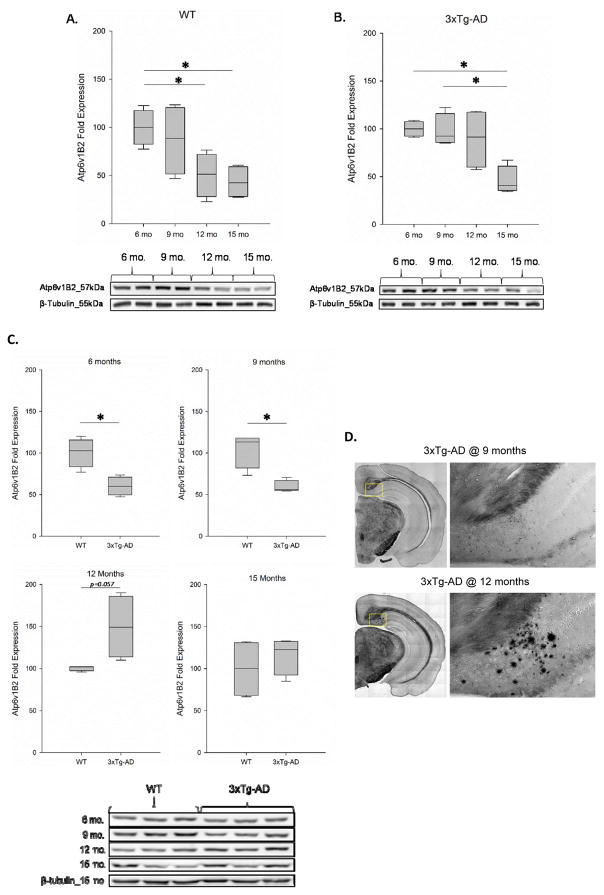
To better understand the function of Atp6v1B2 in relation to brain aging and the development of AD pathology, we further examined the age-related changes in the protein expression of Atp6v1B2 in the cortices of 129/C57BL/6 (WT) and 3xTg-AD female mice at 6, 9, 12 and 15 months of age. In WT mice, we observed a non-significant change in Atp6v1B2 expression at 9 months of age (0.86-fold change, p = 0.470) followed by a significant reduction at both 12 months (0.50-fold change, p = 0.039) and 15 months (0.43-fold change, p = 0.032) of age when compared to 6-month-old animals. Though the downward trend in Atp6v1B2 expression continued following 9 months of age, the change between 9–12 months of age (p = 0.061), 9–15 months of age (p = 0.070) and between 12–15 months of age (p = 0.688) did not reach a level of significance (Fig 2A). In 3xTg-AD mice, similar to our observations in WT mice, we detected a non-significant change in Atp6v1B2 expression at 9 months of age (0.98-fold change, p = 0.42) when compared to 6-month-old animals. However, unlike WT animals, we did not see a significant reduction in Atp6v1B2 expression until 15 months of age (0.41-fold change). The decline in Atp6v1B2 expression at 15 months of age was significant when compared to both 6-month-old animals (p = 0.045) and 9-month-old animals (p = 0.042) and though the downward trend continued between 12–15 months of age, this change did not reach a level of significance (p = 0.226) (Fig. 2B). These data indicate that both aging and AD pathology alter the expression of Atp6v1B2 in female brain. To further examine the role of Atp6v1B2 in AD pathogenesis, we compared the expression of Atp6v1B2 in age-matched female WT and 3xTg-AD mice at 6, 9, 12, and 15 months of age. We observed a significant decrease in Atp6v1B2 expression in 3xTg-AD mice at both 6 months (0.67-fold change, p = 0.024) and 9 months (0.68-fold change, p = 0.021) of age when compared to age-matched WT females, indicating that Atp6v1B2 is reduced in the early development of AD pathology. Contrary to these data, we observed a profound increase in Atp6v1B2 expression in 3xTg-AD mice at 12 months of age when compared to WT controls, however this comparison did not reach a level of significance (1.42-fold change, p = 0.057). No significant change was detected between WT and 3xTg-AD mice at 15 months of age (1.16-fold change, p = 0.249) (Fig 2C). In order to investigate the possible association between the altered trajectory in Atp6v1B2 expression and the stage of AD pathology, 3xTg-AD mouse brain hemispheres were sectioned into 35 μm coronal planes through the hippocampal region and subjected to Campbell-Switzer silver staining to detect Aβ plaques. Our data demonstrate that extraneuronal plaque formation is not visible until 3xTg-AD mice reach approximately 12 months of age indicating that the increase in Atp6v1B2 expression in 12-month-old AD animals is likely a compensatory response to the significantly advanced AD pathology (Fig. 2D).
Figure 2. Aging and AD pathology alter the expression of Atp6v1B2 in female brain.
Immunoblot analysis was performed on cortical protein samples derived from female (A) 129/C57BL/6 and (B) 3xTg-AD mice at ages of 6, 9, 12, and 15 months. n=4–5; *p<0.05. (C) Immunoblot analysis of Atp6v1B2 expression in cortical protein samples derived from age-matched female 129/C57BL/6 and 3xTg-AD mice. n=4–5; *p<0.05. (D) Campbell-Switzer staining of mouse brain hemispherical sections harvested from 9- and 12-month-old 3xTg-AD mice. Campbell-Switzer staining detects the presence of Aβ plaques shown in black.
Atp6v1B2 is localized to synaptic vesicles in primary hippocampal neurons
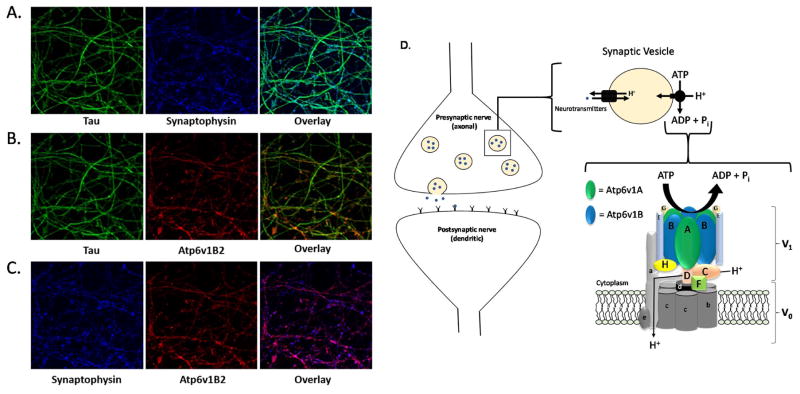
As Atp6v1B2 expression is significantly impacted by aging and the progression of AD pathology, and it is previously reported that Atp6v is critically involved in synaptic transmission, we next examined the distribution of Atp6v1B2 within neurons. In light of the fact that mouse and rat Atp6v1B2 share 100% sequence homology, we chose to utilize rat primary neurons as a model for this experiment. Hippocampal neurons were isolated from day 18 embryonic rat pups and stained with the axonal marker tau, the synaptic vesicle membrane marker, synaptophysin, and Atp6v1B2. Axons were imaged using three-dimensional 60× oil immersion confocal microscopy with three-dimensional image stacks in 0.10 μm planes. As expected, our analysis demonstrates a high level of overlap between synaptophysin and tau confirming the presence of synaptic vesicles within axons (Fig. 3A). Additionally, we detect a high degree of co-localization between Atp6v1B2 and tau indicating that Atp6v1B2 is also expressed within the axon (Fig. 3B). Moreover, our data indicate that synaptophysin and Atp6v1B2 co-localize within the axons of primary hippocampal neurons indicating the presence of Atp6v1B2 on the vesicular membrane (Fig. 3C). Together, these imaging data confirm that Atp6v1B2 is predominantly localized on the membrane of synaptic vesicles in the axons of neurons and are consistent with previous reports that implicate Atp6v1B2 in synaptic transmission (Fig. 3D).
Figure 3. Atp6v1B2 is localized to the vesicular membrane of synaptic vesicles.
(A–C) Primary hippocampal neurons were fixed on day 15 and stained for tau (green), synaptophysin (blue) and Atp6v1B2 (red) immunoreactivity. Images were visualized using 60× oil immersion confocal microscopy and acquired in three-dimensional stacks with 0.10 μm planes. (D–F) A schematic representation of Atp6v on the membrane of synaptic vesicles in presynaptic (axonal) nerve terminals. The 8-subunit V1 complex generates energy via ATP hydrolysis which allows the 5-subunit V0 complex to transport protons across the vesicular membrane. Simultaneously, the neurotransmitter transporters utilize this electrochemical gradient to concentrate neurotransmitters into the synaptic vesicles.
Discussion
Human ApoE genetic isoforms (ApoE2, ApoE3, and ApoE4) play a differential role in the development of late-onset AD, with ApoE4 currently recognized as the most potent genetic risk factor. Existing literature has predominantly focused on the identification of ApoE4-mediated AD-risk mechanisms via comparisons between ApoE3 and ApoE4 brains and between ApoE4 carriers and non-carriers with relatively few studies including the ApoE2 genotype. To address this gap, our laboratory has initiated a series of studies aimed to define the molecular differences that could contribute to the differential impact of the three human ApoE isoforms in the development of AD; in particular, the molecular bases that separate ApoE2 brain from ApoE3 and ApoE4 brains. The goal of this study was to examine the cortical proteome differences in 6-month-old female hApoE2-TR, hApoE3-TR, and hApoE4-TR mice in an effort to determine the mechanisms by which ApoE2 delegates neuroprotection. Our data indicate that three ApoE brains differentially express enzymes primarily involved in cellular bioenergetics and synaptic transmission and provide a potential molecular rationale for the positive influences associated with the ApoE2 genotype.
ApoE2 brain exhibits a more robust bioenergetic profile than ApoE3/ApoE4 brains
Energy hypometabolism occurs early in the preclinical development of AD and several reports have demonstrated that carriers of the ApoE4 allele exhibit deficiencies in key neuronal bioenergetic processes [41–44]. Using 6-month-old female hApoE-TR mice, our laboratory has recently provided the first evidence that the three human ApoE isoforms differentially regulate hippocampal signaling pathways involved in bioenergetics. Of importance, our data indicated significantly more robust Igf1-mediated signaling activity and downstream glucose uptake and metabolism in ApoE2 brain compared to ApoE3 and ApoE4 brains [34]. In the current study, we have identified 4 enzymes involved in cellular bioenergetics that are significantly altered in ApoE2 brain when compared to ApoE3 and/or ApoE4 brains. Expression levels of mitochondrial cytochrome b-c1 complex subunit 1 (Uqcrc1) and glycerol-3-phosphate dehydrogenase 1-like (Gpd1l) were both significantly increased in ApoE2 brain when compared to ApoE3 or ApoE4 brains. Uqcrc1 is one of the 11 subunits that comprise the respiratory chain protein ubiquinol cytochrome c reductase, or Complex III, and is therefore crucial in the production of ATP. Gpd1l is a regulatory enzyme implicated in the modulation of cardiac sodium channels. While Gpd1l has not been previously implicated in the progression of any neurodegenerative disorders, it has been demonstrated that Uqcrc1 expression is significantly reduced in synaptosomal fractions of ApoE4 mice when compared to ApoE3 mice [45] - a finding that corresponds with our data. Contrary to these data, aldehyde dehydrogenase 2 (Aldh2) and B-type creatine kinase (Ckb) were both significantly down-regulated in ApoE2 brain when compared to ApoE3 or ApoE4 brains. While the down-regulation of cellular bioenergetic enzymes in ApoE2 brain was not expected, a possible explanation could be found by examining the function of these enzymes within the cell. Aldh2, an enzyme found in the mitochondrial matrix, functions as a protectant from oxidative stress [46] specifically through the detoxification of toxic aldehydes, like 4-hydroxynonenal [47], while Ckb primarily functions to create the secondary energy source phosphocreatine [48]. Therefore, it can be postulated that under normal resting conditions, an environment that a) contains less oxidative stress and b) is more glucose metabolically robust (i.e. ApoE2 brain) may not require the increased input of detoxifying enzymes and the increased generation of a secondary energy source thereby exhibiting reduced expression of enzymes such as Aldh2 and Ckb. Despite these findings, our data overall do further confirm the differential modulation of bioenergetic processes by human ApoE isoforms and bioenergetic robustness associated with ApoE2.
ApoE2 brain exhibits a more robust synaptic profile than ApoE3/ApoE4 brains
Cognitive impairment, a hallmark symptom of AD, could be the clinical consequence of a series of long-term molecular changes including a reduction in the expression of the synaptic proteome [49, 50] and a reduction in synaptic density [51], plasticity and transmission [52, 53]. It is well established that the ApoE4 genotype is clinically associated with accelerated cognitive decline and earlier incidence of MCI. However, the neurophysiological phenotypes of the ApoE4 genotype are only recently being characterized on the molecular level. One study indicates that co-culture of ApoE4-expressing glial cells with wild-type neurons induces a loss of mature dendritic spines by negatively impacting the expression of glutamate receptors [54]. Dumanis et al. demonstrates significantly reduced cortical dendritic spine density in aged ApoE4-TR mice compared to age-matched ApoE2-TR and ApoE3-TR mice [55]. Young adult (7-month-old) ApoE4-TR mice display reduced excitatory synaptic transmission, dendritic arborization, and spine density in the absence of AD-related neuropathological hallmarks. This same study demonstrates significantly reduced synaptic transmission in 1-month-old ApoE4-TR mice when compared to age-matched ApoE3-TR mice [52]. Additionally, the study demonstrates that the synaptic deficits associated with the ApoE4 genotype in young (1-month) mice were significantly attenuated in ApoE2/E4 heterozygous mice [52]. Collectively, these data indicate that the ApoE4 genotype negatively whereas ApoE2 positively impacts synaptic function.
In support of these findings, our proteomic analysis identified three proteins that were a) significantly increased in expression in ApoE2 brain compared to ApoE3 and/or ApoE4 brain and b) are involved in some aspect of synaptic transmission. The first two proteins identified were found to be significantly upregulated in ApoE2 brain over ApoE3 brain. Dimethylarginine dimethylaminohydrolase (Ddah1) is a regulatory enzyme that mediates the N(G),N(G) asymmetric dimethyl-L-arginine (ADMA)-nitric oxide (NO) system and therefore regulates the levels of NO within the brain [56]. Glycine—tRNA ligase (Gars) is a ubiquitously expressed enzyme that catalyzes the synthesis of glycyl tRNA, a molecule that is required to insert glycine residues into proteins during translation [57]. Alternatively, Gars mutations have been implicated in the development of axonal neuropathies [58, 59] indicating the importance of Gars in the nervous system. While neither Ddah1 nor Gars expression have been previously implicated in AD pathogenesis, our data demonstrate significantly higher expression of both proteins in ApoE2 brain indicating that upregulation of Ddah1 and Gars may contribute to the positive impact on synaptic function associated with the ApoE2 genotype.
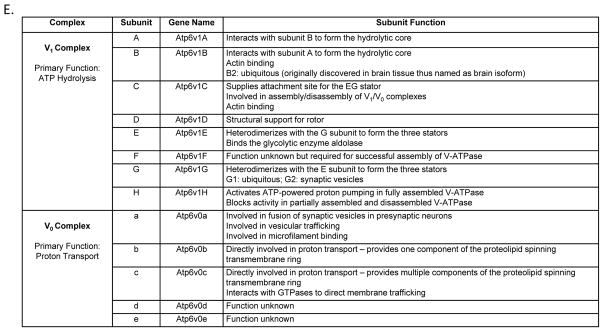
The third synaptic protein identified, and the primary focus of our follow-up studies, is a key component of the V-ATPase (Atp6v): an intricate proton pump that has the highest expression in the brain and accounts for approximately 20% of the total protein resided on the membrane of presynaptic vesicles [60]. Atp6v is composed of a total of 13 subunits separated into two functionally distinct complexes (Fig. 3D–F). The 5-subunit membrane-embedded V0 complex facilitates the movement of protons across a membrane driven by energy generated via ATP hydrolysis by the 8-subunit cytoplasmic V1 complex [40]. Of the 8 subunits that comprise the V1 complex, subunits A (Atp6v1A) and B (Atp6v1B), which are arranged in a heterohexameric rotor pattern composed of 3 alternating A and B subunits, bind ATP via catalytic residues located in Atp6v1A [61] and with the structural support of Atp6v1B perpetuate the hydrolysis of ATP to ADP at the interface between Atp6v1A and Atp6v1B subunits [62, 63]; thus this heterohexameric complex is considered to be the catalytic core of Atp6v [40, 64, 65]. On the membrane of synaptic vesicles, Atp6v works in coordination with neurotransmitter transporters, such as acetylcholine, glutamate and GABA transporters, to facilitate neurotransmitter concentration into synaptic vesicles (Fig. 3D) [66, 67]. Inhibition of Atp6v activity via bafilomycin A1 treatment has been shown to a) significantly deplete glutamate and GABA accumulation in synaptic vesicles, b) substantially reduce glutamatergic and GABAergic synaptic transmission, and c) reduce the frequency and amplitude of miniature postsynaptic currents in rat primary hippocampal neurons [68], connecting Atp6v dysfunction to impaired synaptic transmission. Our data indicate that ApoE brains of three isoforms differentially express Atp6v1B2, with ApoE2 brain exhibiting the highest expression and ApoE4 exhibiting the lowest. As Atp6v1B serves as one of the two essential building blocks that comprise the catalytic core of Atp6v, significantly increased Atp6v1B2 expression in ApoE2 brain over both ApoE3 and ApoE4 brains suggests that the ε2 allele of the ApoE gene may confer a positive impact on the function of Atp6v in the brain. This promising implication, however, needs to be extensively investigated in future studies.
Pertaining to Atp6v1B2 expression in AD, the current literature contains 3 separate proteomic studies that demonstrate significantly increased expression of Atp6v1B2 in 14–16-month-old triple transgenic mice and in 9-month-old APP/PS-1 transgenic mice [69–71]. Contrary to these data, a more recent gene array study performed in normally aging F344 rats demonstrates significantly reduced expression of Atp6v1B2 in 12–23-month-old rats [72]. Taken together, these data indicate that Atp6v1B2 expression is reduced with normal aging but increased in aged AD animals. To further clarify the impact of aging and AD progression on the expression of Atp6v1B2, we examined Atp6v1B2 protein levels in female WT and 3xTg-AD mice at 6, 9, 12, and 15 months of age. Consistent with the previously discussed study by Kadish et al., our data indicate that Atp6v1B2 expression is significantly reduced with aging in WT mice beginning at 12 months of age. These data correspond with the well-established aging-mediated decline in synaptic transmission [31, 36, 72] and suggest that reduced Atp6v1B2 expression could be an underlying contributor to age-associated cognitive decline. As well as being impacted by age, we also find that Atp6v1B2 expression is significantly reduced in response to the progression of AD pathology with the most severe decline occurring at 15 months of age in 3xTg-AD mice. A further comparison between age-matched female WT and 3xTg-AD mice indicates a significant reduction in Atp6v1B2 expression in AD mice compared to WT mice at 6 and 9 months of age and a nearly significant increase in Atp6v1B2 expression at 12 months of age with no significant change at 15 months of age. These data suggest that Atp6v1B2 deficiency could contribute to the gradual loss of synaptic integrity during early disease progression while the increase observed in 12-month-old animals could be a compensatory response to the significantly advanced AD pathology including the onset of Aβ plaque formation and cognitive deficit associated with the animal model at this age.
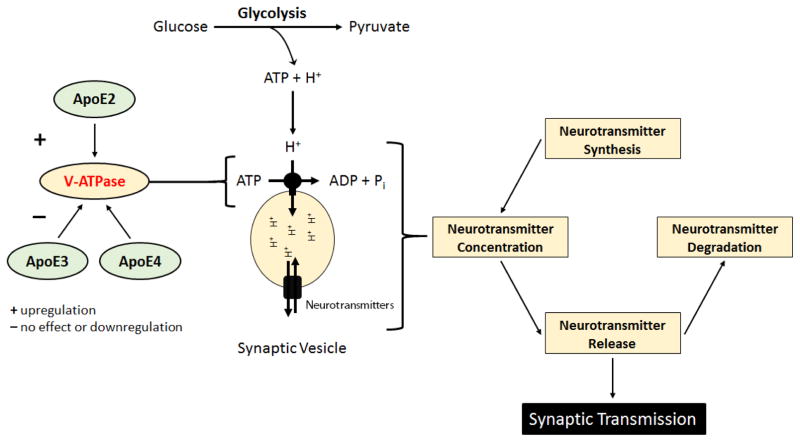
In addition to being regulated by ApoE status, aging, and the progression of AD pathology, emerging evidence suggests that Atp6v function is integrally linked to cellular bioenergetics, specifically glycolysis. It has been previously reported that the glycolytic enzyme phosphofructokinase-1 interacts with the a-subunit of the V0 complex (Atp6v0a) [73]. Additionally, several reports have indicated that aldolase, another glycolytic enzyme, interacts with multiple Atp6v subunits including Atp6v1B, Atp6v1E, and Atp6v0a [74]. These studies also demonstrated that treatment with extracellular glucose significantly increased the interaction between aldolase and Atp6v subunits and that disruption of the aldolase-Atp6v1B interaction results in the disassembly of Atp6v [74, 75]. Furthermore, Nakamura and colleagues demonstrated that Atp6v function is completely inhibited upon treatment with the glycolytic inhibitor, 2-deoxy-D-glucose. This same study indicated that treatment with the mitochondrial electron transport chain inhibitor, antimycin, only partially reduced Atp6v function while the PI3K inhibitor, wortmannin, completely abolished glucose-mediated activation of Atpv6 activity [76]. In addition to these data, Sautin and colleagues demonstrated that stimulation with extracellular glucose initiates the rapid assembly of the V1 and V0 domains while glucose deprivation induces the reversible disassembly of the two protein complexes [77]. Taken together, these studies indicate that the glycolytic pathway directly regulates the assembly, disassembly, and function of Atp6v. As we have previously demonstrated that ApoE2 brain exhibits a more bioenergetically robust profile, and the glycolytic pathway appears to directly impact Atp6v function, we hypothesize that the enhanced synaptic function observed in ApoE2 brain is, in part, due to an ApoE2-mediated upregulation of Atp6v subunit expression. Moreover, we hypothesize that the increased glucose uptake and metabolism previously observed in ApoE2 brain provides a direct source of ATP and protons for Atp6v and thus facilitates increased Atp6v activity thereby increasing neurotransmitter concentration and ultimately synaptic function (Fig. 4).
Figure 4. A representative schematic of our proposed hypothesis.
In order for synaptic transmission to occur, neurotransmitters must first be synthesized, concentrated in synaptic vesicles, and released across the synaptic cleft. V-ATPase (Atp6v), a multi-subunit proton pump, is the primary mediator of ATP-dependent concentration of neurotransmitters into synaptic vesicles and is thus vital for healthy synaptic transmission. Our data indicate that human ApoE isoforms differentially modulate the expression of Atp6v1B2, a key component of the catalytic core of Atp6v, with ApoE2 brain exhibiting the highest expression and ApoE4 brain exhibiting the lowest. Moreover, several studies have indicated that the glycolytic pathway of glucose metabolism is a direct regulator of Atp6v function. Based on these findings, we hypothesize that ApoE2 upregulates Atp6v in the brain and that the increased glucose uptake and metabolism previously observed in ApoE2 brain provides a direct source of ATP and protons for Atp6v thus facilitating increased Atp6v activity. Moreover, we hypothesize that the increased activity of Atp6v results in significantly increased neurotransmitter concentration and thus enhanced synaptic function.
Conclusions
In this study, we examined the cortical proteome differences among the three human ApoE isoforms, with the primary focus on the differences presented in ApoE2 brain compared to ApoE3 and ApoE4 brains. The data indicate that the three ApoE brains are significantly different in two major areas – bioenergetically and synaptically – with ApoE2 brain exhibiting a much more robust profile in both areas. Of particular significance, our data demonstrate for the first time that ApoE isoforms differentially modulate a key component of the catalytic domain of Atp6v, a complex that is crucial for synaptic transmission, with ApoE2 brain exhibiting the highest expression and ApoE4 brain exhibiting the lowest. An extension of these studies indicates that the expression of Atp6v1B2 is significantly reduced before the onset of plaque formation in 3xTg-AD mice compared to WT animals suggesting that Atp6v1B2 deficiency could contribute to the gradual loss of synaptic integrity during early disease progression. These findings warrant further in-depth investigations to explore the potential role of Atp6v as an underlying mechanism responsible for the clinically proven neuroprotective and cognition-favoring properties associated with the ApoE2 genotype.
Acknowledgments
We would like to acknowledge the University of Kansas Mass Spectrometry Core for their assistance in post-2D protein identification and analyses. This work was supported by grants from the Alzheimer’s Association (IIRG-10-172459), the NIH-funded Institutional Development Award (P20GM103418), the NIH-funded University of Kansas Alzheimer’s Disease Center (P30AG035982), and the University of Kansas general research and start-up funds to LZ. The authors declare no conflict of interest.
References
- 1.Alzheimer’s A. 2015 Alzheimer’s disease facts and figures. Alzheimers Dement. 2015;11:332–384. doi: 10.1016/j.jalz.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 2.McBride R. FierceBiotech. 2012 http://www.fiercebiotech.com/story/pharma-counts-just-3-alzheimers-drug-wins-13-years-101-losses/2012-09-14.
- 3.Schnabel J. The Dana Foundation. 2013:1. http://www.dana.org/News/Why_Do_All_the_Large_Alzheimer_s_Drug_Trials_Fail_/
- 4.Mullard A. Sting of Alzheimer’s failures offset by upcoming prevention trials. Nat Rev Drug Discov. 2012;11:657–660. doi: 10.1038/nrd3842. [DOI] [PubMed] [Google Scholar]
- 5.Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 6.Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10:333–344. doi: 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weisgraber KH. Apolipoprotein E: structure-function relationships. Adv Protein Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 9.Zhong N, Weisgraber KH. Understanding the association of apolipoprotein E4 with Alzheimer disease: clues from its structure. J Biol Chem. 2009;284:6027–6031. doi: 10.1074/jbc.R800009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka M, Vedhachalam C, Sakamoto T, Dhanasekaran P, Phillips MC, Lund-Katz S, Saito H. Effect of carboxyl-terminal truncation on structure and lipid interaction of human apolipoprotein E4. Biochemistry. 2006;45:4240–4247. doi: 10.1021/bi060023b. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen D, Dhanasekaran P, Nickel M, Nakatani R, Saito H, Phillips MC, Lund-Katz S. Molecular basis for the differences in lipid and lipoprotein binding properties of human apolipoproteins E3 and E4. Biochemistry. 2010;49:10881–10889. doi: 10.1021/bi1017655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Li Q, Wang J. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc Natl Acad Sci U S A. 2011;108:14813–14818. doi: 10.1073/pnas.1106420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frieden C, Garai K. Structural differences between apoE3 and apoE4 may be useful in developing therapeutic agents for Alzheimer’s disease. Proc Natl Acad Sci U S A. 2012;109:8913–8918. doi: 10.1073/pnas.1207022109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Utermann G, Langenbeck U, Beisiegel U, Weber W. Genetics of the apolipoprotein E system in man. Am J Hum Genet. 1980;32:339–347. [PMC free article] [PubMed] [Google Scholar]
- 15.Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC, Jr, Rimmler JB, Locke PA, Conneally PM, Schmader KE, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7:180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- 16.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 17.Risacher SL, Kim S, Shen L, Nho K, Foroud T, Green RC, Petersen RC, Jack CR, Jr, Aisen PS, Koeppe RA, Jagust WJ, Shaw LM, Trojanowski JQ, Weiner MW, Saykin AJ Alzheimer’s Disease Neuroimaging Initiative d. The role of apolipoprotein E (APOE) genotype in early mild cognitive impairment (E-MCI) Front Aging Neurosci. 2013;5:11. doi: 10.3389/fnagi.2013.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farlow MR, He Y, Tekin S, Xu J, Lane R, Charles HC. Impact of APOE in mild cognitive impairment. Neurology. 2004;63:1898–1901. doi: 10.1212/01.wnl.0000144279.21502.b7. [DOI] [PubMed] [Google Scholar]
- 19.Mayeux R, Small SA, Tang M, Tycko B, Stern Y. Memory performance in healthy elderly without Alzheimer’s disease: effects of time and apolipoprotein-E. Neurobiol Aging. 2001;22:683–689. doi: 10.1016/s0197-4580(01)00223-8. [DOI] [PubMed] [Google Scholar]
- 20.Blacker D, Haines JL, Rodes L, Terwedow H, Go RC, Harrell LE, Perry RT, Bassett SS, Chase G, Meyers D, Albert MS, Tanzi R. ApoE-4 and age at onset of Alzheimer’s disease: the NIMH genetics initiative. Neurology. 1997;48:139–147. doi: 10.1212/wnl.48.1.139. [DOI] [PubMed] [Google Scholar]
- 21.Langbaum JB, Chen K, Lee W, Reschke C, Bandy D, Fleisher AS, Alexander GE, Foster NL, Weiner MW, Koeppe RA, Jagust WJ, Reiman EM Alzheimer’s Disease Neuroimaging I. Categorical and correlational analyses of baseline fluorodeoxyglucose positron emission tomography images from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Neuroimage. 2009;45:1107–1116. doi: 10.1016/j.neuroimage.2008.12.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynch JR, Tang W, Wang H, Vitek MP, Bennett ER, Sullivan PM, Warner DS, Laskowitz DT. APOE genotype and an ApoE-mimetic peptide modify the systemic and central nervous system inflammatory response. J Biol Chem. 2003;278:48529–48533. doi: 10.1074/jbc.M306923200. [DOI] [PubMed] [Google Scholar]
- 23.Drzezga A, Grimmer T, Henriksen G, Muhlau M, Perneczky R, Miederer I, Praus C, Sorg C, Wohlschlager A, Riemenschneider M, Wester HJ, Foerstl H, Schwaiger M, Kurz A. Effect of APOE genotype on amyloid plaque load and gray matter volume in Alzheimer disease. Neurology. 2009;72:1487–1494. doi: 10.1212/WNL.0b013e3181a2e8d0. [DOI] [PubMed] [Google Scholar]
- 24.Fleisher AS, Chen K, Liu X, Roontiva A, Thiyyagura P, Ayutyanont N, Joshi AD, Clark CM, Mintun MA, Pontecorvo MJ, Doraiswamy PM, Johnson KA, Skovronsky DM, Reiman EM. Using positron emission tomography and florbetapir F18 to image cortical amyloid in patients with mild cognitive impairment or dementia due to Alzheimer disease. Arch Neurol. 2011;68:1404–1411. doi: 10.1001/archneurol.2011.150. [DOI] [PubMed] [Google Scholar]
- 25.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 26.Bretsky PM, Buckwalter JG, Seeman TE, Miller CA, Poirier J, Schellenberg GD, Finch CE, Henderson VW. Evidence for an interaction between apolipoprotein E genotype, gender, and Alzheimer disease. Alzheimer Dis Assoc Disord. 1999;13:216–221. doi: 10.1097/00002093-199910000-00007. [DOI] [PubMed] [Google Scholar]
- 27.Payami H, Zareparsi S, Montee KR, Sexton GJ, Kaye JA, Bird TD, Yu CE, Wijsman EM, Heston LL, Litt M, Schellenberg GD. Gender difference in apolipoprotein E-associated risk for familial Alzheimer disease: a possible clue to the higher incidence of Alzheimer disease in women. Am J Hum Genet. 1996;58:803–811. [PMC free article] [PubMed] [Google Scholar]
- 28.Altmann A, Tian L, Henderson VW, Greicius MD Alzheimer’s Disease Neuroimaging Initiative I. Sex modifies the APOE-related risk of developing Alzheimer disease. Ann Neurol. 2014;75:563–573. doi: 10.1002/ana.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fleisher A, Grundman M, Jack CR, Jr, Petersen RC, Taylor C, Kim HT, Schiller DH, Bagwell V, Sencakova D, Weiner MF, DeCarli C, DeKosky ST, van Dyck CH, Thal LJ Alzheimer’s Disease Cooperative S. Sex, apolipoprotein E epsilon 4 status, and hippocampal volume in mild cognitive impairment. Arch Neurol. 2005;62:953–957. doi: 10.1001/archneur.62.6.953. [DOI] [PubMed] [Google Scholar]
- 30.Payami H, Montee KR, Kaye JA, Bird TD, Yu CE, Wijsman EM, Schellenberg GD. Alzheimer’s disease, apolipoprotein E4, and gender. JAMA. 1994;271:1316–1317. [PubMed] [Google Scholar]
- 31.Johnson JK, McCleary R, Oshita MH, Cotman CW. Initiation and propagation stages of beta-amyloid are associated with distinctive apolipoprotein E, age, and gender profiles. Brain Res. 1998;798:18–24. doi: 10.1016/s0006-8993(98)00363-1. [DOI] [PubMed] [Google Scholar]
- 32.Helkala EL, Koivisto K, Hanninen T, Vanhanen M, Kervinen K, Kuusisto J, Mykkanen L, Kesaniemi YA, Laakso M, Riekkinen P., Sr Memory functions in human subjects with different apolipoprotein E phenotypes during a 3-year population-based follow-up study. Neurosci Lett. 1996;204:177–180. doi: 10.1016/0304-3940(96)12348-x. [DOI] [PubMed] [Google Scholar]
- 33.Wilson RS, Bienias JL, Berry-Kravis E, Evans DA, Bennett DA. The apolipoprotein E epsilon 2 allele and decline in episodic memory. J Neurol Neurosurg Psychiatry. 2002;73:672–677. doi: 10.1136/jnnp.73.6.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keeney JT, Ibrahimi S, Zhao L. Human ApoE Isoforms Differentially Modulate Glucose and Amyloid Metabolic Pathways in Female Brain: Evidence of the Mechanism of Neuroprotection by ApoE2 and Implications for Alzheimer’s Disease Prevention and Early Intervention. J Alzheimers Dis. 2015;48:411–424. doi: 10.3233/JAD-150348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan PM, Mezdour H, Aratani Y, Knouff C, Najib J, Reddick RL, Quarfordt SH, Maeda N. Targeted replacement of the mouse apolipoprotein E gene with the common human APOE3 allele enhances diet-induced hypercholesterolemia and atherosclerosis. J Biol Chem. 1997;272:17972–17980. doi: 10.1074/jbc.272.29.17972. [DOI] [PubMed] [Google Scholar]
- 36.Oddo S, Caccamo A, Shepherd JD, Murphy MP, Golde TE, Kayed R, Metherate R, Mattson MP, Akbari Y, LaFerla FM. Triple-transgenic model of Alzheimer’s disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron. 2003;39:409–421. doi: 10.1016/s0896-6273(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 37.Zhao L, Wu TW, Brinton RD. Estrogen receptor subtypes alpha and beta contribute to neuroprotection and increased Bcl-2 expression in primary hippocampal neurons. Brain Res. 2004;1010:22–34. doi: 10.1016/j.brainres.2004.02.066. [DOI] [PubMed] [Google Scholar]
- 38.Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coughenour HD, Spaulding RS, Thompson CM. The synaptic vesicle proteome: a comparative study in membrane protein identification. Proteomics. 2004;4:3141–3155. doi: 10.1002/pmic.200300817. [DOI] [PubMed] [Google Scholar]
- 40.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 41.McGeer EG, Peppard RP, McGeer PL, Tuokko H, Crockett D, Parks R, Akiyama H, Calne DB, Beattie BL, Harrop R. 18Fluorodeoxyglucose positron emission tomography studies in presumed Alzheimer cases, including 13 serial scans. Can J Neurol Sci. 1990;17:1–11. doi: 10.1017/s0317167100029930. [DOI] [PubMed] [Google Scholar]
- 42.Smith GS, de Leon MJ, George AE, Kluger A, Volkow ND, McRae T, Golomb J, Ferris SH, Reisberg B, Ciaravino J, et al. Topography of cross-sectional and longitudinal glucose metabolic deficits in Alzheimer’s disease. Pathophysiologic implications. Arch Neurol. 1992;49:1142–1150. doi: 10.1001/archneur.1992.00530350056020. [DOI] [PubMed] [Google Scholar]
- 43.Mielke R, Herholz K, Grond M, Kessler J, Heiss WD. Clinical deterioration in probable Alzheimer’s disease correlates with progressive metabolic impairment of association areas. Dementia. 1994;5:36–41. doi: 10.1159/000106692. [DOI] [PubMed] [Google Scholar]
- 44.Drzezga A, Riemenschneider M, Strassner B, Grimmer T, Peller M, Knoll A, Wagenpfeil S, Minoshima S, Schwaiger M, Kurz A. Cerebral glucose metabolism in patients with AD and different APOE genotypes. Neurology. 2005;64:102–107. doi: 10.1212/01.WNL.0000148478.39691.D3. [DOI] [PubMed] [Google Scholar]
- 45.Shi L, Du X, Zhou H, Tao C, Liu Y, Meng F, Wu G, Xiong Y, Xia C, Wang Y, Bi G, Zhou JN. Cumulative effects of the ApoE genotype and gender on the synaptic proteome and oxidative stress in the mouse brain. Int J Neuropsychopharmacol. 2014;17:1863–1879. doi: 10.1017/S1461145714000601. [DOI] [PubMed] [Google Scholar]
- 46.Ohta S, Ohsawa I, Kamino K, Ando F, Shimokata H. Mitochondrial ALDH2 deficiency as an oxidative stress. Ann N Y Acad Sci. 2004;1011:36–44. doi: 10.1007/978-3-662-41088-2_4. [DOI] [PubMed] [Google Scholar]
- 47.Yoval-Sanchez B, Rodriguez-Zavala JS. Differences in susceptibility to inactivation of human aldehyde dehydrogenases by lipid peroxidation byproducts. Chem Res Toxicol. 2012;25:722–729. doi: 10.1021/tx2005184. [DOI] [PubMed] [Google Scholar]
- 48.Schlattner U, Tokarska-Schlattner M, Wallimann T. Mitochondrial creatine kinase in human health and disease. Biochim Biophys Acta. 2006;1762:164–180. doi: 10.1016/j.bbadis.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 49.Counts SE, Nadeem M, Lad SP, Wuu J, Mufson EJ. Differential expression of synaptic proteins in the frontal and temporal cortex of elderly subjects with mild cognitive impairment. J Neuropathol Exp Neurol. 2006;65:592–601. doi: 10.1097/00005072-200606000-00007. [DOI] [PubMed] [Google Scholar]
- 50.Xu PT, Li YJ, Qin XJ, Scherzer CR, Xu H, Schmechel DE, Hulette CM, Ervin J, Gullans SR, Haines J, Pericak-Vance MA, Gilbert JR. Differences in apolipoprotein E3/3 and E4/4 allele-specific gene expression in hippocampus in Alzheimer disease. Neurobiol Dis. 2006;21:256–275. doi: 10.1016/j.nbd.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 51.Scheff SW, Price DA, Schmitt FA, DeKosky ST, Mufson EJ. Synaptic alterations in CA1 in mild Alzheimer disease and mild cognitive impairment. Neurology. 2007;68:1501–1508. doi: 10.1212/01.wnl.0000260698.46517.8f. [DOI] [PubMed] [Google Scholar]
- 52.Klein RC, Mace BE, Moore SD, Sullivan PM. Progressive loss of synaptic integrity in human apolipoprotein E4 targeted replacement mice and attenuation by apolipoprotein E2. Neuroscience. 2010;171:1265–1272. doi: 10.1016/j.neuroscience.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dumanis SB, DiBattista AM, Miessau M, Moussa CE, Rebeck GW. APOE genotype affects the pre-synaptic compartment of glutamatergic nerve terminals. J Neurochem. 2013;124:4–14. doi: 10.1111/j.1471-4159.2012.07908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nwabuisi-Heath E, Rebeck GW, Ladu MJ, Yu C. ApoE4 delays dendritic spine formation during neuron development and accelerates loss of mature spines in vitro. ASN Neuro. 2014;6:e00134. doi: 10.1042/AN20130043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dumanis SB, Tesoriero JA, Babus LW, Nguyen MT, Trotter JH, Ladu MJ, Weeber EJ, Turner RS, Xu B, Rebeck GW, Hoe HS. ApoE4 decreases spine density and dendritic complexity in cortical neurons in vivo. J Neurosci. 2009;29:15317–15322. doi: 10.1523/JNEUROSCI.4026-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tran CT, Leiper JM, Vallance P. The DDAH/ADMA/NOS pathway. Atheroscler Suppl. 2003;4:33–40. doi: 10.1016/s1567-5688(03)00032-1. [DOI] [PubMed] [Google Scholar]
- 57.Freist W, Logan DT, Gauss DH. Glycyl-tRNA synthetase. Biol Chem Hoppe Seyler. 1996;377:343–356. [PubMed] [Google Scholar]
- 58.Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin SQ, Jordanova A, Kremensky I, Christodoulou K, Middleton LT, Sivakumar K, Ionasescu V, Funalot B, Vance JM, Goldfarb LG, Fischbeck KH, Green ED. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldfarb LG, Sivakumar K. GARS-Associated Axonal Neuropathy. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K, editors. GeneReviews(R) Seattle (WA): 1993. [Google Scholar]
- 60.Moriyama Y, Futai M. H(+)-ATPase, a primary pump for accumulation of neurotransmitters, is a major constituent of brain synaptic vesicles. Biochem Biophys Res Commun. 1990;173:443–448. doi: 10.1016/s0006-291x(05)81078-2. [DOI] [PubMed] [Google Scholar]
- 61.MacLeod KJ, Vasilyeva E, Baleja JD, Forgac M. Mutational analysis of the nucleotide binding sites of the yeast vacuolar proton-translocating ATPase. J Biol Chem. 1998;273:150–156. doi: 10.1074/jbc.273.1.150. [DOI] [PubMed] [Google Scholar]
- 62.Liu Q, Kane PM, Newman PR, Forgac M. Site-directed mutagenesis of the yeast V-ATPase B subunit (Vma2p) J Biol Chem. 1996;271:2018–2022. doi: 10.1074/jbc.271.4.2018. [DOI] [PubMed] [Google Scholar]
- 63.Liu Q, Leng XH, Newman PR, Vasilyeva E, Kane PM, Forgac M. Site-directed mutagenesis of the yeast V-ATPase A subunit. J Biol Chem. 1997;272:11750–11756. doi: 10.1074/jbc.272.18.11750. [DOI] [PubMed] [Google Scholar]
- 64.Zhao L, Yao J, Mao Z, Chen S, Wang Y, Brinton RD. 17beta-Estradiol regulates insulin-degrading enzyme expression via an ERbeta/PI3-K pathway in hippocampus: relevance to Alzheimer’s prevention. Neurobiol Aging. 2011;32:1949–1963. doi: 10.1016/j.neurobiolaging.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holliday LS. Hindawi Publishing Corporation New Journal of Science. 2014. p. 21. [Google Scholar]
- 66.Beyenbach KW, Wieczorek H. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J Exp Biol. 2006;209:577–589. doi: 10.1242/jeb.02014. [DOI] [PubMed] [Google Scholar]
- 67.Moriyama Y, Maeda M, Futai M. The role of V-ATPase in neuronal and endocrine systems. J Exp Biol. 1992;172:171–178. doi: 10.1242/jeb.172.1.171. [DOI] [PubMed] [Google Scholar]
- 68.Zhou Q, Petersen CC, Nicoll RA. Effects of reduced vesicular filling on synaptic transmission in rat hippocampal neurones. J Physiol. 2000;525(Pt 1):195–206. doi: 10.1111/j.1469-7793.2000.t01-1-00195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martin B, Brenneman R, Becker KG, Gucek M, Cole RN, Maudsley S. iTRAQ analysis of complex proteome alterations in 3xTgAD Alzheimer’s mice: understanding the interface between physiology and disease. PLoS One. 2008;3:e2750. doi: 10.1371/journal.pone.0002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ciavardelli D, Silvestri E, Del Viscovo A, Bomba M, De Gregorio D, Moreno M, Di Ilio C, Goglia F, Canzoniero LM, Sensi SL. Alterations of brain and cerebellar proteomes linked to Abeta and tau pathology in a female triple-transgenic murine model of Alzheimer’s disease. Cell Death Dis. 2010;1:e90. doi: 10.1038/cddis.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson RA, Joshi G, Huang Q, Sultana R, Baker AS, Cai J, Pierce W, St Clair DK, Markesbery WR, Butterfield DA. Proteomic analysis of brain proteins in APP/PS-1 human double mutant knock-in mice with increasing amyloid beta-peptide deposition: insights into the effects of in vivo treatment with N-acetylcysteine as a potential therapeutic intervention in mild cognitive impairment and Alzheimer’s disease. Proteomics. 2011;11:4243–4256. doi: 10.1002/pmic.201000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kadish I, Thibault O, Blalock EM, Chen KC, Gant JC, Porter NM, Landfield PW. Hippocampal and cognitive aging across the lifespan: a bioenergetic shift precedes and increased cholesterol trafficking parallels memory impairment. J Neurosci. 2009;29:1805–1816. doi: 10.1523/JNEUROSCI.4599-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Su Y, Zhou A, Al-Lamki RS, Karet FE. The a-subunit of the V-type H+-ATPase interacts with phosphofructokinase-1 in humans. J Biol Chem. 2003;278:20013–20018. doi: 10.1074/jbc.M210077200. [DOI] [PubMed] [Google Scholar]
- 74.Lu M, Sautin YY, Holliday LS, Gluck SL. The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. J Biol Chem. 2004;279:8732–8739. doi: 10.1074/jbc.M303871200. [DOI] [PubMed] [Google Scholar]
- 75.Lu M, Ammar D, Ives H, Albrecht F, Gluck SL. Physical interaction between aldolase and vacuolar H+-ATPase is essential for the assembly and activity of the proton pump. J Biol Chem. 2007;282:24495–24503. doi: 10.1074/jbc.M702598200. [DOI] [PubMed] [Google Scholar]
- 76.Nakamura S. Glucose activates H(+)-ATPase in kidney epithelial cells. Am J Physiol Cell Physiol. 2004;287:C97–105. doi: 10.1152/ajpcell.00469.2003. [DOI] [PubMed] [Google Scholar]
- 77.Sautin YY, Lu M, Gaugler A, Zhang L, Gluck SL. Phosphatidylinositol 3-kinase-mediated effects of glucose on vacuolar H+-ATPase assembly, translocation, and acidification of intracellular compartments in renal epithelial cells. Mol Cell Biol. 2005;25:575–589. doi: 10.1128/MCB.25.2.575-589.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]