SUMMARY
To understand the cellular basis of behavior, it is necessary to know the cell types that exist in the nervous system and their contributions to function. Spinal networks are essential for sensory processing and motor behavior and provide a powerful system for identifying the cellular correlates of behavior. Here, we used massively parallel single nucleus RNA sequencing (snRNA-seq) to create an atlas of the adult mouse lumbar spinal cord. We identified and molecularly characterized 43 neuronal populations. Next, we leveraged the snRNA-seq approach to provide unbiased identification of neuronal populations that were active following a sensory and a motor behavior, using a transcriptional signature of neuronal activity. This approach can be used in the future to link single nucleus gene expression data with dynamic biological responses to behavior, injury, and disease.
In Brief
Sathyamurthy et al. use massively parallel single nucleus RNA-seq to probe spinal cord cell types and present an atlas of 43 neuronal populations. By using this approach after a sensory and a motor behavior, they were able to detect and molecularly identify activated neurons associated with each function.

INTRODUCTION
To understand how networks of cells mediate behavior, it is necessary to classify the various cell types of the brain, spinal cord, and peripheral nervous system and to know which populations of cells are involved in specific functions. Gene expression-based definitions of cell identity have been a foundation of spinal cord biology for the past 30 years. In particular, the use of post-natal genetic markers to control defined classes of spinal cord neurons has enabled the functional characterization of many cell types and has advanced our understanding of how these populations contribute to normal sensory-motor behavior (Abraira et al., 2017; Azim et al., 2014; Bikoff et al., 2016; Bourane et al., 2015; Dougherty et al., 2013; Duan et al., 2014; Hilde et al., 2016; Koch et al., 2017b; Mishra and Hoon, 2013; Peirs et al., 2015; Satoh et al., 2016; Sun et al., 2009). However, there are three important limitations to this approach. First, there is no census of neuronal cell types in the adult spinal cord. The lack of such a resource limits the application and interpretation of genetic manipulations, and it is not known how previously described cell types relate to one another. Second, the unique gene expression profiles that endow cell types with their functional repertoires are not known. Third, we lack an unbiased approach to identify the set of spinal cord cell types associated with a given neural function, such as motor behavior or the response to a sensory stimulus.
Pioneering work using massively parallel single-cell sequencing has established that a cell’s transcriptional program is a powerful strategy for defining cell type (Campbell et al., 2017; Chen et al., 2017; Jaitin et al., 2014; Lake et al., 2016; Li et al., 2016; Macosko et al., 2015; Shin et al., 2015; Tasic et al., 2016; Usoskin et al., 2015; Villani et al., 2017). Furthermore, single-cell RNA sequencing has been adapted to provide unbiased detection of immediate-early gene expression in molecularly defined cell types following seizure, acute anxiety, or sensory experience in the striatum and visual cortex (Hrvatin et al., 2018; Wu et al., 2017).
We sought to develop an approach that simultaneously provides a single-cell gene expression census of the cell types of the adult spinal cord and the ability to overlay a map of the transcriptional signature of neuronal activity following behavior. To characterize the gene expression and cell-type composition of the adult mouse spinal cord, we used massively parallel single nucleus RNA-seq (snRNA-seq). We created a catalog of spinal cord neuronal cell types, characterizing 43 classes of neurons. Analysis of the genes expressed in each cell type provided a powerful resource for understanding the mechanistic basis of functional neuronal heterogeneity. This work also revealed distinct organizing principles for molecular heterogeneity between neuronal populations in the dorsal and ventral horns. To provide unbiased characterization of the classes of spinal neurons that were associated with defined behaviors, we performed this technique immediately following a painful sensory stimulation or a locomotor behavior. This approach could be used to reveal comprehensive single nucleus response maps for a range of behaviors and disease states, establishing an unprecedented link between single nucleus gene expression and circuit- and system-level function within the spinal cord.
RESULTS
snRNA-Seq Identification of Major Spinal Cord Cell Types
To adapt massively parallel RNA sequencing approaches to the spinal cord, we opted to perform single nucleus, rather than single cell, analysis for three key reasons: single nucleus transcriptional profiling accurately permits cell-type analysis, avoids experimental artifacts from transcriptional changes induced in intact cells during the tissue dissociation process, and can be performed easily from whole tissue, including tissue that is difficult to dissociate (such as the spinal cord), frozen material, and human biobank material (Grindberg et al., 2013; Habib et al., 2017; Lake et al., 2016, 2017; Matevossian and Akbarian, 2008).
To establish an snRNA-seq strategy for the adult spinal cord, we used a detergent-based protocol, which allowed rapid and thorough nuclear release and transfer of the material to cold temperatures, thereby minimizing gene expression changes (Figure 1A; Figure S1A). Nuclei were easily isolated from adult mouse spinal cord and frozen adult human spinal cord. We next sought to modify Drop-Seq (Macosko et al., 2015), a droplet-based approach for massively parallel single-cell RNA capture, cDNA synthesis, and sequencing, to allow this technique to be used for single nuclei. We found that simply increasing the concentration of detergent in the Drop-Seq lysis buffer improved nuclear lysis and generated smaller droplets than standard Drop-Seq (mean 0.48 ± 0.06 nL SEM) (Figures S1B–S1D). To determine whether this approach enables single nucleus droplet encapsulation, nuclei from human spinal cord were pooled with nuclei from mouse spinal cord, and we examined how many beads contained both human and mouse transcripts (Figure 1B). We found that 2% of droplets with a mouse nucleus also contained a human nucleus (4/196 mouse nuclei), which represents a calculated doublet rate of 4.1%. Thus, single nuclei can be obtained from difficult-to-dissociate and frozen human spinal cord tissue and can be processed through Drop-Seq with a simple buffer modification.
Figure 1. Massively Parallel snRNA-Seq Was Used to Define Cell Types in the Adult Mouse Spinal Cord.
(A) Summary of experimental strategy.
(B) Barnyard plot of pooled human and mouse spinal cord nuclei showing beads that were associated with human transcripts, mouse transcripts, both human and mouse transcripts (mixed), or those that could not be determined (undetermined).
(C) tSNE visualization plot of 17,354 spinal cord nuclei, colored according to seven major SC3-defined clusters: neurons, oligodendrocytes (oligos), meningeal and Schwann cells, astrocytes, vascular cells, oligodendrocyte precursor cells (OPCs), and microglia.
(D) Heatmap of normalized mean expression for key marker genes for each major SC3-defined cluster.
Using this approach, we sequenced and analyzed 17,354 nuclei from adult mouse lumbar spinal cord. We found seven major clusters that corresponded to the following cell types, based on marker expression: neurons (52% of total nuclei), oligodendrocytes (16% of total nuclei), a mixed population of meningeal and Schwann cells (14% of total nuclei), astrocytes (9% of total nuclei), vascular cells (5% of total nuclei), oligodendrocyte precursor cells (1% of total nuclei), and microglia (1% of total nuclei) (Figure 1D; Figure S1E; Table S1). The diversity of the RNA transcript yield that we obtained, reflected in the number of genes per nucleus, varied among cell types (Figure S1F).
Census of Adult Spinal Cord Neuronal Populations
To identify and characterize neuronal classes within the adult mouse spinal cord, 4,280 neuronal nuclei were analyzed and partitioned into 43 clusters (Figure 2A; Figure S2A; Table S2). We first characterized the neurotransmitter status, a core feature of neuronal identity, of each cluster by analyzing excitatory, inhibitory, and cholinergic marker expression. We found that 53% of neuronal nuclei were in 23 predominantly excitatory clusters (including 2 cholinergic clusters), 45% were in 18 predominantly inhibitory clusters, and 2.5% were in 2 clusters with both excitatory and inhibitory markers (Figure S3A). Within this latter group, only rare individual neuronal nuclei co-expressed excitatory and inhibitory markers (n = 2/109), which may reflect doublets and cannot account for the substantial fractions of excitatory and inhibitory marker-expressing nuclei in these clusters. Therefore, these two clusters contained separate excitatory and inhibitory populations that share overall similar gene expression.
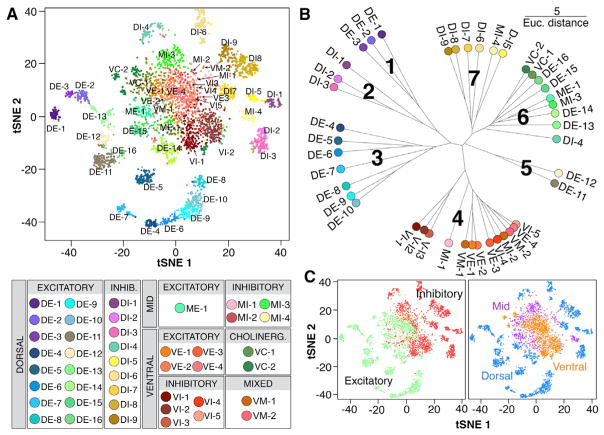
Figure 2. Massively Parallel snRNA-Seq Identified 43 Neuronal Populations in the Adult Spinal Cord.
(A) tSNE visualization plot and cluster key of 4,280 spinal cord neuronal nuclei, colored according to membership in 43 SC3-defined clusters. Cluster names were assigned based on cluster location (D, dorsal; M, mid; and V, ventral) and neuro-transmitter status (E, excitatory; I, inhibitory; M, mixed; and C, cholinergic), as shown in the key below the plot.
(B) Unrooted dendrogram depicting cluster relationships based on mean gene expression for each cluster. Units shown are Euclidean distance.
(C) tSNE visualization plots of spinal cord neuronal nuclei, colored to depict neurotransmitter status (green, excitatory; red, inhibitory; yellow, mixed; cholinergic clusters were also predominantly excitatory and are green) or location (blue, dorsal; orange, ventral; purple, deep dorsal, intermediate zone, or mid).
See also Figures S2 and S3 and Tables S2 and S3.
Next, we analyzed relationships between clusters by performing Euclidean-based hierarchical clustering on the mean expression of each gene in each cluster (Figure 2B). A dendrogram presentation of these relationships revealed seven major groupings, five of which shared a common neurotransmitter status (groups 1, 2, 3, 5, and 7) (Figure 2B). To determine what other parameters are major organizing features of neuronal populations, we compared all genes that significantly contributed to defining any cluster with public gene expression databases (Gong et al., 2003; Lein et al., 2007). This allowed us to probe the cluster distribution of previously known marker genes (Figure S3B) and to determine the spatial location of each cluster. We found that 55% of neuronal nuclei were in 25 dorsal clusters, 34% were in 13 ventral clusters, and 11% were in 5 clusters in the deep dorsal horn or intermediate zone. Four of seven major dendrogram groupings each had a common regional location within the spinal cord (groups 1, 2, 3, and 5) (Figure 2B). To further analyze neurotransmitter status and spatial location across clusters, each population was colored by these features and plotted by t-distributed stochastic neighbor embedding (tSNE) analysis (Figure 2C). The structure of the tSNE distribution of neurons according to these parameters supported the importance of location and neuro-transmitter status as defining features of spinal cord cell types. Accordingly, clusters were named by their spatial location (D, dorsal; V, ventral; or M, deep dorsal, intermediate, or “mid” cord) and their neurotransmitter status (E, excitatory; I, inhibitory; M, mixed; C, cholinergic) (Figure 2).
A major difference in cluster organization was observed between dorsal and ventral clusters. tSNE visualization revealed that dorsal clusters form an outer ring of discrete groups while ventral clusters overlapped one another in the center of the plot, with deep dorsal and intermediate clusters between (Figure 2C). Similarly, dorsal clusters were generally present as homogeneous blocks in a cell consensus matrix and had high SC3 silhouette width consensus values (a measure that represents the diagonality of the matrix), while ventral clusters showed inter-relatedness with other ventral clusters and had low silhouette width consensus values (Figure S2). At a molecular level, markers for ventral clusters were often shared across ventral populations (as explained later). These differences were not based on a failure to segregate ventral neurons due to low molecular information content, because the number of genes per nucleus was 2,465 ± 202 in ventral neurons and 1,346 ± 33 in dorsal neurons (mean ± SEM). This analysis suggests a general principle that the dorsal horn of the spinal cord contains more molecularly distinct populations while the ventral horn displays overlapping gene expression patterns.
We next sought to determine what categories of genes drive neuronal diversity. Gene ontology (GO) term analysis of the top genes associated with each cluster was performed. We found that neurotransmitter receptors and ion channels, transcription factors, and cyclic AMP (cAMP) signal transduction components are significantly over-represented among the top genes that contribute to defining these clusters (Figure 3A). Many genes within these molecular function families were enriched or specifically expressed in particular populations or related groups (Figures S3C–S3E).
Figure 3. Gene Expression that Defined Spinal Cord Neuronal Populations.
(A) Summarized GO terms that were significantly enriched (>1.3 enrichment score) among the top genes associated with each cluster.
(B) Validation co-labeling for pairs of cluster-defining genes using immuno-fluorescence (DE-4 and DE-7) or fluorescent in situ hybridization (DE-5, DI-1, and DI-4). Images taken at 20×, with the full image (scale bar, 200 μm) and magnification (scale bar, 100 μm) shown in the left and right panels of each pair, respectively.
To identify candidate marker genes for the 43 clusters, and to characterize their gene expression profiles, we further analyzed all genes that significantly contributed to defining each cluster. Many clusters were partially defined by previously established markers (Figure S3B) (Abraira et al., 2017; Bikoff et al., 2016; Koch et al., 2017a; Lu et al., 2015; Todd, 2017), and in most of these clusters, we identified new key genes (Figures 3B and 4). In addition, we identified previously unrecognized cell populations (Figure 4). Table S3 is a searchable database of the mean gene expression and the percent cluster membership for each gene. This provides the opportunity to probe the molecular identity of each population or to search across clusters for a gene of interest. A summary of each group of clusters with highlighted findings follows.
Figure 4. Summary of 43 Spinal Cord Neuronal Populations.
For each population, the cluster name, a putative cell-type assignment, and key marker genes are shown. Previously undescribed cell types and markers are shown in bold. The expression of the marker genes across clusters are shown as a heatmap of normalized mean gene expression. See also Table S3.
Group 1 was composed of the dorsal excitatory (DE) clusters DE-1–DE-3. These clusters shared expression of the transcription factor Ebf2 (together with DE-4–DE-7) and the γ-aminobutyric acid (GABA) receptor Gabrg3. They were peptidergic, expressing the enzyme Pam and the genes for neuropeptides Grp and/or Sst. DE-1 expressed the mu opioid receptor Oprm1, DE-2 expressed the peptide receptor Npy1r, and DE-3 expressed Ntrk2/TrkB.
Group 2 was composed of the dorsal inhibitory (DI) clusters DI-1–DI-3. These clusters shared expression of the peptide receptor Sstr2, as well as the glutamate receptor Grik2 and the potassium channel Kcnc2. DI-1 expressed Calb2/calretinin, and DI-2 and DI-3 were peptidergic, expressing the enzyme Pam and the genes for Gal (DI-2) and Pnoc/nociception (DI-3), as well as Nos1 (DI-3).
Group 3 was composed of the DE clusters DE-4–DE-10. There were two subgroups. DE-4–DE-7 expressed Ebf2 (together with DE-1–DE-3) and Calb1/calbindin and were peptidergic, expressing Pam, as well as Sst/SOM (DE-4 and DE-5), Tac2/NeurokininA (DE-5), Calca/CGRP (DE-5), Nts/neurotensin (DE-6), Penk/enkephalin (DE-6), and Cck (DE-7). DE-4 also expressed Prkcg/PKCγ. DE-8–DE-10 expressed the transcription factor Maf and were not peptidergic. DE-8 expressed Cbln2, DE-9 expressed Adarb2, and DE-10 expressed enriched levels of Slc17a8/vGlut3.
Group 4 was composed of the ventral inhibitory (VI) clusters VI-1–VI-5, ventral excitatory (VE) clusters VE-1–VE-4, ventral mixed (VM) clusters VM-1 and VM-2, and mid inhibitory (MI) deep dorsal clusters MI-1 and MI-2. Overall, this group shared expression of the transcription factors Esrrg/ERRγ and Foxp2, as well as the sodium channel Scn1a, which has been shown to correlate positively with maximum firing rate (Tripathy et al., 2017). Several ventral clusters (VI-1, VI-4, VI-5, VE-3, and VE-4) and MI-2 also shared expression of the peri-neuronal net components Acan and Bcan and the link protein Hapln1. This is consistent with the observation that peri-neuronal nets have been shown to surround many previously unidentified ventral neurons (Galtrey et al., 2008). Cluster VI-5 was also distinguished by being enriched for nearly all genes associated with the mammalian target of rapamycin (mTOR) complexes mTORC1 and mTORC2 that play key roles in cell metabolism and survival (Laplante and Sabatini, 2012), including Mtor/mTOR, Rptor/RAPTOR, Mlst8/GβL, Deptor, Rictor, and Mapkap1/SIN1, as well as the mTOR pathway regulators Rheb, Tsc1, and Tsc2. Surprisingly, many embryonic and early postnatal ventral cell fate markers were found within these clusters and may provide a link between the embryonic lineage-defined identity and the adult populations described here (Figure S3F) (Alvarez et al., 2005; Bikoff et al., 2016; Catela et al., 2015; Lu et al., 2015; Perry et al., 2015; Seredick et al., 2014). As examples, VE-1–VE-3 were enriched for embryonic lineage V2a markers, including Vsx2/Chx10, Shox2, Lhx3, and Lhx4, and cluster VI-4 was enriched for the embryonic lineage dI6 markers Wt1 and Dmrt3 and the embryonic lineage V1-subtype markers Chrna2 (Renshaw), Pvalb (1a inhibitory interneurons), and Esrrb/Nr3b2. However, many of these embryonically expressed genes had very low expression levels in these clusters, and even for the genes with stronger expression, it is not certain whether the same cells continue to express these genes from embryonic through adult stages.
Group 5 was composed of the DE clusters DE-11 and DE-12, which shared expression of the transcription factor Sox5 and the potassium channel Kcnd3. DE-11 was peptidergic, expressing Pam, Penk, and Tac1/substance P, while DE-12 was not peptidergic but expressed the peptide receptor Grpr.
Group 6 was composed of a diverse collection of clusters: DI-4, DE-13–DE-16, MI-3, mid excitatory (ME) cluster ME-1, and ventral cholinergic (VC) clusters VC-1 and VC-2. Clusters DI-4 and DE-13–DE-16 are peptidergic, expressing Npy (DI-4), Calca/CGRP (DE-13), Cck and Tac1/substance P (DE-14), Pdyn/dynorphin (DE-15), and Penk/enkephalin (DE-16). VC-1 expressed the embryonic lineage V0c marker Pitx2. VC-2 expressed markers of spinal motoneurons, including Prph/peripherin, Isl1, Map1b, Nrg1, and Slit3, as well as Nkain1, which has been shown to correlate positively with input resistance (Tripathy et al., 2017).
Group 7 was composed of the DI clusters DI-5–DI-9 and the deep dorsal cluster MI-4 that shared expression of the glutamate receptor Grik2. These clusters expressed previously described DI transcription factors Gbx1 (DI-5), Lhx1 (DI-7), and Rorb (DI-9). DI-5 also expressed the estrogen receptor Esr1, DI-6 expressed Cdh3 and Kcnip2, and DI-8 expressed Nrgn/Neurogranin.
Collectively, these 43 clusters establish an atlas of spinal cord neuronal populations and their constituent molecules.
snRNA-Seq Following Behavior Identified Active Neurons
Having characterized the spinal cord neuron populations, we next considered that snRNA-seq could provide an unbiased, cell-type based characterization of neurons that express immediate-early genes following a behavioral paradigm. We found that direct isolation of nuclei did not induce Fos RNA (Figure S1A) but that detectable Fos expression in nuclei could be induced 5 min following a painful sensory stimulus (formalin hindpaw injection) (Figure S4). To determine whether Fos RNA can be detected at the single nucleus level following behavior, snRNA-seq was performed following formalin injection or rotarod locomotion. Fos was expressed in a higher proportion of nuclei following rotarod locomotion (1.6%) or formalin administration (1.9%) compared with baseline (0.48%), and the level of Fos gene expression was significantly increased (0.0082 ± 0.0025 counts per million (cpm) after rotarod, 0.0175 ± 0.0040 cpm after formalin, and 0.0024 ± 0.0009 cpm at baseline; mean ± SEM; p < 0.001, ANOVA, corrected p value). Thus, massively parallel snRNA-seq following behavior detected a transcriptional signature of neuronal activity.
Next, we used the distribution of Fos RNA expression to map neuronal activity across clusters following rotarod locomotion or formalin administration, because these experimental paradigms produce classic patterns of cFOS protein expression (Figure 5A) (Herdegen et al., 1994; Jasmin et al., 1994). During locomotion, each of the major ventral embryonic lineage domains gives rise to neurons that are important for specific features of locomotion, such as flexor and extensor alternation (V1-and V2b-derived cells) and left and right alternation (V2a-derived cells) (Crone et al., 2008; Zhang et al., 2014). In addition, cholinergic interneurons express cFOS protein following locomotion in cat, and these may correlate with V0c neurons (Huang et al., 2000; Zagoraiou et al., 2009). However, the identities of locomotor-associated intermediate and dorsal horn neurons are not well established, with the exceptions of protein kinase C gamma (PKCγ)-expressing neurons in the rat (Neumann et al., 2008) and Rorb-expressing neurons (Koch et al., 2017b). We hypothesized that the application of snRNA-seq following rotarod running would help to reveal the set of adult neurons that are active during locomotion.
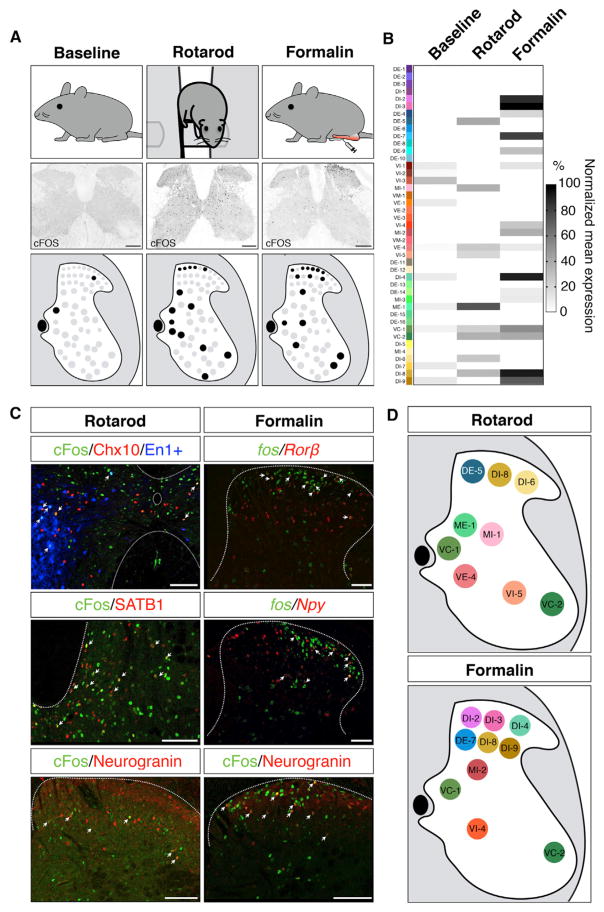
Figure 5. snRNA-Seq Identified Active Neurons Following Behavior.
(A) Characteristic pattern of cFOS expression at baseline, following rotarod locomotion or after formalin injection in the hindpaw, at 60 min following behavior.
(B) Fos RNA expression as detected by snRNA-seq across clusters in baseline, rotarod, and formalin samples, shown as normalized mean gene expression per cluster.
(C) Experimental validation of clusters associated with each behavior, as detected by snRNA-seq. For each cluster, a marker gene was compared with cFOS protein expression by immunofluorescence (En1:Cre;Ai9/Chx10, Satb1, and Neurogranin) or Fos RNA by fluorescent in situ hybridization (Rorb and Npy) (scale bars, 100 μm).
(D) Summary of the set of neuronal populations associated with each behavior, as identified by snRNA-seq.
See also Figure S4.
Following locomotion, Fos RNA was detected in ventral clusters VC-1 (which includes Pitx2-expressing V0c neurons), VC-2 (spinal motoneurons), VE-4 (which includes putative V2a neurons) and VI-5 (which includes putative V1/V2b neurons), thereby confirming that these cell types are associated with locomotion (Figure 5B). We used markers to validate this approach and found that VE-4 (Chx10) and VI-5 cells (En1:Cre;Ai9) express cFOS after rotarod locomotion (Figure 5C). Within the intermediate zone and dorsal horn, clusters ME-1, MI-1, DE-5, DI-6, and DI-8 expressed Fos RNA, but we did not detect Fos RNA in cluster DE-4 that expresses Prkcg/PKCγ (Figure 5B). This may be due to species differences or a technical false negative. Rorb expression is highest in cluster DI-9, which was not detected using this approach, but it is also present in cluster DI-8, which did express Fos after locomotion. Using markers for clusters ME-1 (Satb1) and DI-8 (Nrgn), we confirmed that these newly defined clusters express cFOS protein after locomotion, thereby expanding the known set of neuronal populations that are associated with this core behavior (Figure 5D).
Formalin administration is a well-established pain assay, and it has previously been shown to activate predominantly dorsal horn spinal cord neurons, including those that express Gal, Sstr2, Nos1, Npy, Penk, and Tacr1/Nk1r, creating specific expectations for which clusters should be detected (Herdegen et al., 1994; Hossaini et al., 2010; Lee et al., 1993; Polgár et al., 2013). Following formalin injection, Fos RNA was observed in clusters DI-2 (which includes Gal and Sstr2-expressing neurons), DI-3 (which includes Nos1-expressing neurons), and DI-4 (which includes Npy-expressing neurons), confirming that these neurons express Fos after formalin administration, as well as DI-8, DI-9, DE-7, VI-4, VC-1, VC-2, and MI-2 (Figure 5C). Penk and Tacr1/Nk1r are both distributed across several clusters. We used markers for clusters DI-4 (Npy), DI-8 (Nrgn), and DI-9 (Rorb) to validate these findings and found that these populations express Fos RNA or cFOS protein following formalin administration (Figure 5C). Thus, in both a sensory test and a motor behavior, massively parallel snRNA-seq provided an unbiased definition of cell types that displayed activity-induced transcription and revealed new cell types that are associated with each function.
DISCUSSION
The spinal cord plays essential roles in sensory processing and motor control, but how the cells of the cord function together in networks to mediate behavior is not well understood. Here, we sought to identify spinal cord cell types and their contributions to behavior through single nucleus transcriptional profiling. Massively parallel snRNA-seq was used to analyze more than 17,000 nuclei from the adult mouse spinal cord. We created an atlas of spinal cord neuronal populations, characterizing 43 cell types. By applying snRNA-seq following behavior, we detected transcriptional signatures of neuronal activity and identified neuronal populations associated with a sensory and a motor function.
We have described 43 neuronal populations within the adult mouse lumbar spinal cord, including previously unrecognized cell types. This work establishes a cellular framework for the spinal cord and facilitates the comparison and integration of prior work that generally used single markers to define cell types. We detected clusters that correspond to nearly all previously described adult spinal cord neuronal populations, with the primary exceptions being populations that were previously described by a single marker gene that is expressed more broadly within the spinal cord (such as Penk and Pvalb).
The perspective afforded by massively parallel single nucleus sequencing also revealed an intriguing difference between dorsal and ventral neuronal populations. We found that dorsal neuron types were more distinct from one another, forming discrete clusters, while ventral neuron types were more closely associated with one another. Previously, most analysis of spinal cord cell types emphasized adult molecular markers in the dorsal horn and embryonic lineage domain-defined cell types in the ventral horn. Together with our findings, this may reflect different organizing principles for neuronal identity in the dorsal and ventral spinal cord. It is possible that in the adult spinal cord, cellular identity in the dorsal horn is governed by restricted and ongoing expression of genes for specific cellular functions, whereas in the ventral horn, it is governed by factors defined during development, such as cell location, axon guidance, and genetically programmed synaptic specificity.
This work also reveals the molecular repertoire of each neuronal population, providing a significant extension of our understanding of spinal cord cell types. The searchable database that is included here (Table S3) will allow researchers to probe the complement of genes in cell types of interest and analyze the expression of genes of interest across clusters. This will serve as a powerful tool to advance our understanding of the molecular mechanisms that mediate functional heterogeneity among neuronal populations.
Despite the strengths of this work, three major limitations must be noted. First, we detected a lower number of genes per nucleus than is typically detected from whole cells or nuclei (Grindberg et al., 2013; Habib et al., 2016, 2017; Lacar et al., 2016; Lake et al., 2016; Macosko et al., 2015). This is likely due to several factors, including the lower amount of RNA in the nucleus compared with the whole cell, the trade-off between resolution and scale for low-throughput versus massively parallel approaches, technical differences such as mRNA capture and reverse transcription efficiency, and cell-type differences. Despite this first limitation, we obtained a sufficient number of genes per nucleus to permit successful clustering of neuronal populations. A second major limitation is that this work only characterized the cell types of the lumbar spinal cord. Although the major classes of known cell types are present along the full rostro-caudal axis of the spinal cord, work has revealed that distinct subpopulations may vary at different segmental levels (Francius et al., 2013; Hayashi et al., 2018; Sweeney et al., 2018). Accordingly, future work is necessary to fully characterize spinal cord cell types outside of the lumbar region. A third major limitation is that the use of snRNA-seq for activity profiling can only identify cells that induced a transcriptional response above a detection threshold. As a result, this approach will not detect all neural activity, and negative results must be interpreted with caution.
Massively parallel single nucleus transcriptional profiling has the potential to reveal unprecedented knowledge about cell types, the transcriptional programs of cell types, and gene expression control in an array of in vivo settings in animal models. With this resource in hand, single nucleus transcriptional profiling can be used to probe spinal cord neuronal and non-neuronal responses to disease or injury, to study how the molecular and cellular composition of a tissue changes over time, and to reveal the selective loss of cell types during degeneration or gain of cell types during inflammation. Because nuclei are readily obtained from human patient-derived and archived bio-bank material, snRNA-seq can be applied to study human biology as well (Habib et al., 2017; Lake et al., 2016; Matevossian and Akbarian, 2008). We now have the tools to reach a new level in our understanding of the molecular and cellular mechanisms by which complex tissues mediate basic function, behavior, and disease.
EXPERIMENTAL PROCEDURES
Mice and Behavior
All animal work was performed in accordance with a protocol approved by the National Institute of Neurological Disorders and Stroke Animal Care and Use Committee. Balanced samples of male and female ICR/CD-1 wild-type mice, between 8 and 12 weeks old, were used for all experiments except those shown in Figure 5C, for which En1:Cre;Ai9 mice (Stock No. 007916 × Stock No. 007909, both from The Jackson Laboratory) were used. Formalin injection was done by injecting 30–40 μL of 2% paraformaldehyde into the plantar surface of the hindpaw. Rotarod testing was done with a standard program accelerating from 0 to 40 rotations per minute over 5 min.
Nuclei Preparation
This protocol was adapted from Halder et al. (2016). Animals were euthanized by CO2 inhalation, the lumbar spinal cord was rapidly dissected, and dorsal root ganglia were removed. The cords were dounced in 500 μL of sucrose buffer (0.32 M sucrose, 10 mM HEPES [pH 8.0], 5 mM CaCl2, 3 mM Mg-acetate, 0.1 mM EDTA, 1 mM DTT) with 0.1% Triton X-100 using five strokes with the A pestle (Kontes Dounce Tissue Grinder) followed by five strokes with the B pestle. The lysate was then diluted with 3 mL of sucrose buffer and was centrifuged at 3,200 × g for 10 min. The supernatant was removed, and 3 mL of sucrose buffer was added to the pellet and incubated for 1–2 min; the loosened pellet was then transferred to an Oak Ridge centrifuge tube. The pellet was then homogenized using Ultra-Turrax on setting 1 for 1 min. 12 mL of density buffer (1 M sucrose, 10 mM HEPES [pH 8.0], 3 mM Mg-acetate, 1 mM DTT) was then added carefully below the nuclei layer, and the tube was centrifuged at 3,200 × g for 20 min. The supernatant was then rapidly poured off, and the nuclei on the walls of the tube were collected with 1 mL of PBS with 0.02% BSA and spun at 3,200 × g for 10 min. Nuclei were then resuspended in PBS with 0.02% BSA.
Drop-Seq and Analysis
Each Drop-Seq sample was produced from the lumbar cords of a pair of ICR mice 8–12 weeks old. There were nine independent samples for baseline (four male and five female), five independent samples for formalin treatment (three male and two female), and five independent samples for rotarod behavior (three male and two female).
The Drop-Seq method was performed as previously described (Macosko et al., 2015) except that the following concentrations were used: 225 nuclei/μL, 250 beads/μL, and 0.7% sarkosyl in the lysis buffer; flow rates were adjusted accordingly. Clustering was performed using SC3 consensus clustering (Kiselev et al., 2017). All antibodies and in situ hybridization probes used for validation are listed in Table S4.
Supplementary Material
Highlights.
An atlas of adult mouse spinal cord cell types
A resource of the molecular repertoires of 43 neuronal populations
A simple method using snRNA-seq to identify activated neurons following behavior
Acknowledgments
Human adult spinal cord tissues were procured by the National Disease Research Interchange (NDRI) with support from NIH grant 2 U42 OD011158 and the assistance of Dr. Christopher Grunseich and Dr. Kenneth Fischbeck. We thank Dr. Arnab Barik for expertise and assistance with in situ hybridization, Dr. Alexander Chesler for sharing reagents, and Dr. Yuesheng Li for expertise and assistance with sequencing. The Lbx1 antibody was a gift of Dr. Carmen Birchmeier. This research was supported by NIH Intramural Research Program funding from the NINDS (1 ZIA NS003153 02 to A.S., K.R.J., L.L., K.J.E.M., C.I.D., A.R.R., T.B.B., and A.J.L.) and the NIDCD (1 ZIA DC000059 18 to M.C.K. and M.W.K.).
Footnotes
DATA AND SOFTWARE AVAILABILITY
The accession number for the raw sequencing data reported in this paper is GEO: GSE103892.
Supplemental Information includes Supplemental Experimental Procedures, four figures, and four tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.02.003.
AUTHOR CONTRIBUTIONS
A.S. and A.J.L. conceived the project. K.R.J. performed all bioinformatic analysis. K.J.E.M., M.C.K., and A.J.L. performed Drop-Seq experiments. A.S., L.L., A.R.R., T.B.B., and C.I.D. performed validation experiments. A.S., K.R.J., K.J.E.M., M.C.K., M.W.K., and A.J.L. analyzed experiments. A.S., K.R.J., and A.J.L. wrote the manuscript.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Abraira VE, Kuehn ED, Chirila AM, Springel MW, Toliver AA, Zimmerman AL, Orefice LL, Boyle KA, Bai L, Song BJ, et al. The cellular and synaptic architecture of the mechanosensory dorsal horn. Cell. 2017;168:295–310. e19. doi: 10.1016/j.cell.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez FJ, Jonas PC, Sapir T, Hartley R, Berrocal MC, Geiman EJ, Todd AJ, Goulding M. Postnatal phenotype and localization of spinal cord V1 derived interneurons. J Comp Neurol. 2005;493:177–192. doi: 10.1002/cne.20711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azim E, Jiang J, Alstermark B, Jessell TM. Skilled reaching relies on a V2a propriospinal internal copy circuit. Nature. 2014;508:357–363. doi: 10.1038/nature13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikoff JB, Gabitto MI, Rivard AF, Drobac E, Machado TA, Miri A, Brenner-Morton S, Famojure E, Diaz C, Alvarez FJ, et al. Spinal inhibitory interneuron diversity delineates variant motor microcircuits. Cell. 2016;165:207–219. doi: 10.1016/j.cell.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourane S, Grossmann KS, Britz O, Dalet A, Del Barrio MG, Stam FJ, Garcia-Campmany L, Koch S, Goulding M. Identification of a spinal circuit for light touch and fine motor control. Cell. 2015;160:503–515. doi: 10.1016/j.cell.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JN, Macosko EZ, Fenselau H, Pers TH, Lyubetskaya A, Tenen D, Goldman M, Verstegen AMJ, Resch JM, McCarroll SA, et al. A molecular census of arcuate hypothalamus and median eminence cell types. Nat Neurosci. 2017;20:484–496. doi: 10.1038/nn.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catela C, Shin MM, Dasen JS. Assembly and function of spinal circuits for motor control. Annu Rev Cell Dev Biol. 2015;31:669–698. doi: 10.1146/annurev-cellbio-100814-125155. [DOI] [PubMed] [Google Scholar]
- Chen R, Wu X, Jiang L, Zhang Y. Single-cell RNA-seq reveals hypothalamic cell diversity. Cell Rep. 2017;18:3227–3241. doi: 10.1016/j.celrep.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crone SA, Quinlan KA, Zagoraiou L, Droho S, Restrepo CE, Lundfald L, Endo T, Setlak J, Jessell TM, Kiehn O, Sharma K. Genetic ablation of V2a ipsilateral interneurons disrupts left-right locomotor coordination in mammalian spinal cord. Neuron. 2008;60:70–83. doi: 10.1016/j.neuron.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Dougherty KJ, Zagoraiou L, Satoh D, Rozani I, Doobar S, Arber S, Jessell TM, Kiehn O. Locomotor rhythm generation linked to the output of spinal shox2 excitatory interneurons. Neuron. 2013;80:920–933. doi: 10.1016/j.neuron.2013.08.015. [DOI] [PubMed] [Google Scholar]
- Duan B, Cheng L, Bourane S, Britz O, Padilla C, Garcia-Campmany L, Krashes M, Knowlton W, Velasquez T, Ren X, et al. Identification of spinal circuits transmitting and gating mechanical pain. Cell. 2014;159:1417–1432. doi: 10.1016/j.cell.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francius C, Harris A, Rucchin V, Hendricks TJ, Stam FJ, Barber M, Kurek D, Grosveld FG, Pierani A, Goulding M, et al. Identification of multiple subsets of ventral interneurons and differential distribution along the rosstrocaudal axis of the developing spinal cord. PLoS One. 2013;8:e70325. doi: 10.1371/journal.pone.0070325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtrey CM, Kwok JCF, Carulli D, Rhodes KE, Fawcett JW. Distribution and synthesis of extracellular matrix proteoglycans, hyaluronan, link proteins and tenascin-R in the rat spinal cord. Eur J Neurosci. 2008;27:1373–1390. doi: 10.1111/j.1460-9568.2008.06108.x. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Grindberg RV, Yee-Greenbaum JL, McConnell MJ, Novotny M, O’Shaughnessy AL, Lambert GM, Araúzo-Bravo MJ, Lee J, Fishman M, Robbins GE, et al. RNA-sequencing from single nuclei. Proc Natl Acad Sci USA. 2013;110:19802–19807. doi: 10.1073/pnas.1319700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, Hession C, Zhang F, Regev A. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353:925–928. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14:955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder R, Hennion M, Vidal RO, Shomroni O, Rahman RU, Rajput A, Centeno TP, van Bebber F, Capece V, Garcia Vizcaino JC, et al. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat Neurosci. 2016;19:102–110. doi: 10.1038/nn.4194. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Hinckley CA, Driscoll SP, Moore NJ, Levine AJ, Hilde KL, Sharma K, Pfaff SL. Graded arrays of spinal and supraspinal V2a interneuron subtypes underlie forelimb and hindlimb motor control. Neuron. 2018 doi: 10.1016/j.neuron.2018.01.023. Published online February 1, 2018 https://doi.org/10.1016/j.neuron.2018.01.023. [DOI] [PMC free article] [PubMed]
- Herdegen T, Rüdiger S, Mayer B, Bravo R, Zimmermann M. Expression of nitric oxide synthase and colocalisation with Jun, Fos and Krox transcription factors in spinal cord neurons following noxious stimulation of the rat hindpaw. Brain Res Mol Brain Res. 1994;22:245–258. doi: 10.1016/0169-328x(94)90053-1. [DOI] [PubMed] [Google Scholar]
- Hilde KL, Levine AJ, Hinckley CA, Hayashi M, Montgomery JM, Gullo M, Driscoll SP, Grosschedl R, Kohwi Y, Kohwi-Shigematsu T, Pfaff SL. Satb2 is required for the development of a spinal exteroceptive microcircuit that modulates limb position. Neuron. 2016;91:763–776. doi: 10.1016/j.neuron.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossaini M, Duraku LS, Saraç C, Jongen JLM, Holstege JC. Differential distribution of activated spinal neurons containing glycine and/or GABA and expressing c-fos in acute and chronic pain models. Pain. 2010;151:356–365. doi: 10.1016/j.pain.2010.07.023. [DOI] [PubMed] [Google Scholar]
- Hrvatin S, Hochbaum DR, Nagy MA, Cicconet M, Robertson K, Cheadle L, Zilionis R, Ratner A, Borges-Monroy R, Klein AM, et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat Neurosci. 2018;21:120–129. doi: 10.1038/s41593-017-0029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A, Noga BR, Carr PA, Fedirchuk B, Jordan LM. Spinal cholinergic neurons activated during locomotion: localization and electro-physiological characterization. J Neurophysiol. 2000;83:3537–3547. doi: 10.1152/jn.2000.83.6.3537. [DOI] [PubMed] [Google Scholar]
- Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasmin L, Gogas KR, Ahlgren SC, Levine JD, Basbaum AI. Walking evokes a distinctive pattern of Fos-like immunoreactivity in the caudal brainstem and spinal cord of the rat. Neuroscience. 1994;58:275–286. doi: 10.1016/0306-4522(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Kiselev VY, Kirschner K, Schaub MT, Andrews T, Yiu A, Chandra T, Natarajan KN, Reik W, Barahona M, Green AR, Hemberg M. SC3: consensus clustering of single cell RNA-seq data. Nat Methods. 2017;14:483–486. doi: 10.1038/nmeth.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch SC, Acton D, Goulding M. Spinal circuits for touch, pain, and itch. Annu Rev Physiol. 2017a doi: 10.1146/annurev-physiol-022516-034303. Published online September 27, 2017 https://doi.org/10.1146/annurev-physiol-022516-034303. [DOI] [PMC free article] [PubMed]
- Koch SC, Del Barrio MG, Dalet A, Gatto G, Günther T, Zhang J, Seidler B, Saur D, Schüle R, Goulding M. RORβ spinal interneurons gate sensory transmission during locomotion to secure a fluid walking gait. Neuron. 2017b;96:1419–1431. e5. doi: 10.1016/j.neuron.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacar B, Linker SB, Jaeger BN, Krishnaswami SR, Barron JJ, Kelder MJ, Parylak SL, Paquola AC, Venepally P, Novotny M, et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun. 2016;7:1–12. doi: 10.1038/ncomms11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung HL, Chen S, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Codeluppi S, Yung YC, Gao D, Chun J, Kharchenko PV, Linnarsson S, Zhang K. A comparative strategy for single-nucleus and single-cell transcriptomes confirms accuracy in predicted cell-type expression from nuclear RNA. Sci Rep. 2017;7:6031. doi: 10.1038/s41598-017-04426-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Price RH, Williams FG, Mayer B, Beitz AJ. Nitric oxide synthase is found in some spinothalamic neurons and in neuronal processes that appose spinal neurons that express Fos induced by noxious stimulation. Brain Res. 1993;608:324–333. doi: 10.1016/0006-8993(93)91474-7. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Li CL, Li KC, Wu D, Chen Y, Luo H, Zhao JR, Wang SS, Sun MM, Lu YJ, Zhong YQ, et al. Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res. 2016;26:83–102. doi: 10.1038/cr.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu DC, Niu T, Alaynick WA. Molecular and cellular development of spinal cord locomotor circuitry. Front Mol Neurosci. 2015;8:25. doi: 10.3389/fnmol.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matevossian A, Akbarian S. Neuronal nuclei isolation from human postmortem brain tissue. J Vis Exp. 2008;20:e914. doi: 10.3791/914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SK, Hoon MA. The cells and circuitry for itch responses in mice. Science. 2013;340:968–971. doi: 10.1126/science.1233765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann S, Braz JM, Skinner K, Llewellyn-Smith IJ, Basbaum AI. Innocuous, not noxious, input activates PKCgamma interneurons of the spinal dorsal horn via myelinated afferent fibers. J Neurosci. 2008;28:7936–7944. doi: 10.1523/JNEUROSCI.1259-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirs C, Williams SPG, Zhao X, Walsh CE, Gedeon JY, Cagle NE, Goldring AC, Hioki H, Liu Z, Marell PS, Seal RP. Dorsal horn circuits for persistent mechanical pain. Neuron. 2015;87:797–812. doi: 10.1016/j.neuron.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S, Gezelius H, Larhammar M, Hilscher MM, Lamotte d’Incamps B, Leao KE, Kullander K. Firing properties of Renshaw cells defined by Chrna2 are modulated by hyperpolarizing and small conductance ion currents Ih and ISK. Eur J Neurosci. 2015;41:889–900. doi: 10.1111/ejn.12852. [DOI] [PubMed] [Google Scholar]
- Polgár E, Sardella TCP, Tiong SYX, Locke S, Watanabe M, Todd AJ. Functional differences between neurochemically defined populations of inhibitory interneurons in the rat spinal dorsal horn. Pain. 2013;154:2606–2615. doi: 10.1016/j.pain.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh D, Pudenz C, Arber S. Context-dependent gait choice elicited by EphA4 mutation in Lbx1 spinal interneurons. Neuron. 2016;89:1046–1058. doi: 10.1016/j.neuron.2016.01.033. [DOI] [PubMed] [Google Scholar]
- Seredick S, Hutchinson SA, Van Ryswyk L, Talbot JC, Eisen JS. Lhx3 and Lhx4 suppress Kolmer-Agduhr interneuron characteristics within zebrafish axial motoneurons. Development. 2014;141:3900–3909. doi: 10.1242/dev.105718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL, Song H. Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YG, Zhao ZQ, Meng XL, Yin J, Liu XY, Chen ZF. Cellular basis of itch sensation. Science. 2009;325:1531–1534. doi: 10.1126/science.1174868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney LB, Bikoff JB, Gabitto MI, Brenner-Morton S, Baek M, Yang JH, Tabak EG, Dasen JS, Kintner CR, Jessell TM. Origin and segmental diversity of spinal inhibitory interneurons. Neuron. 2018;97:341–355. e3. doi: 10.1016/j.neuron.2017.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd AJ. Identifying functional populations among the interneurons in laminae I–III of the spinal dorsal horn. Mol Pain. 2017;13:1744806917693003. doi: 10.1177/1744806917693003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathy SJ, Toker L, Li B, Crichlow CL, Tebaykin D, Mancarci BO, Pavlidis P. Transcriptomic correlates of neuron electrophysiological diversity. PLoS Comput Biol. 2017;13:e1005814. doi: 10.1371/journal.pcbi.1005814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YE, Pan L, Zuo Y, Li X, Hong W. Detecting activated cell populations using single-cell RNA-seq. Neuron. 2017;96:313–329. e6. doi: 10.1016/j.neuron.2017.09.026. [DOI] [PubMed] [Google Scholar]
- Zagoraiou L, Akay T, Martin JF, Brownstone RM, Jessell TM, Miles GB. A cluster of cholinergic premotor interneurons modulates mouse locomotor activity. Neuron. 2009;64:645–662. doi: 10.1016/j.neuron.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lanuza GM, Britz O, Wang Z, Siembab VC, Zhang Y, Velasquez T, Alvarez FJ, Frank E, Goulding M. V1 and v2b interneurons secure the alternating flexor-extensor motor activity mice require for limbed locomotion. Neuron. 2014;82:138–150. doi: 10.1016/j.neuron.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.