Abstract
We report a typhoid fever case with a Salmonella enterica serovar Typhi isolate showing extended spectrum β-lactamase (ESBL) production in the Democratic Republic of the Congo. Whole genome sequencing revealed that the strain carried a plasmid-mediated CTX-M-15 ESBL gene and did not belong to the dominant H58 Salmonella Typhi clade.
Keywords: typhoid fever, Salmonella Typhi, extended-spectrum beta lactamases (ESBL), Democratic Republic of the Congo
Typhoid fever, caused by Salmonella enterica serovar Typhi (further referred to as Salmonella Typhi) is a major health threat in low- and middle-income countries. In the Democratic Republic of the Congo (DRC), Salmonella Typhi is among the leading causes of bloodstream infections [1]. Given the high prevalence of multidrug resistance (MDR), defined as coresistance to ampicillin, trimethoprim/sulfamethoxazole, and chloramphenicol, among Salmonella Typhi, the current treatment options are fluoroquinolones and 3rd generation cephalosporins. The availability and use of antibiotics in the DRC are described in the supplementary material.
On November 24th 2015, a 6-years old boy presented at the health center of Tambu-Tseke in southwestern DRC, with a 3-day history of fever, abdominal pain, and vomiting. Clinical examination was unremarkable, and a malaria rapid diagnostic test was negative. Given his good clinical condition, he was sent home with oral treatment consisting of cefixime, paracetamol, and albendazole. As part of an ongoing epidemic investigation for suspicion of typhoid fever, blood was drawn for culture (BacT/ALERT-PF, bioMérieux, Marcy L’Etoile, France) and shipped to the National Institute of Biomedical Research (INRB) in Kinshasa (DRC). Upon arrival there was growth of Gram-negative rods, which were identified as Salmonella Typhi by standard biochemical reactions. The isolate was shipped to Belgium for further investigation.
Serotyping (Pro-Lab Diagnostics, Richmond Hill, Ontario, Canada) confirmed the isolate as Salmonella Typhi (9,12,[Vi]:d:-). Disk diffusion antibiotic susceptibility testing (CLSI, M100-S26 2016) showed resistance to ampicillin, trimethoprim-sulfamethoxazole, aztreonam, cephalosporins II (cefuroxime), III (cefotaxime, ceftriaxone and ceftazidime) and IV (cefixime) with susceptibility to amoxicillin-clavulanic acid, chloramphenicol, azithromycin, gentamicin, tetracycline, piperacillin-tazobactam, meropenem, and ertapenem. Production of extended spectrum β-lactamase (ESBL) was confirmed by the disk diffusion clavulanate inhibition test using ceftazidime and cefotaxime. The isolate further showed decreased ciprofloxacin susceptibility (DCS, MIC-value 0.38 mg/L) with resistance to pefloxacin and nalidixic acid (5 µg and 30 µg disks, respectively). An in-house Luminex-based assay to identify mutations in quinolone resistance-determining regions (QRDR) and plasmid-mediated quinolone resistance genes (PMQR) showed that the DCS was associated with a single mutation in residue Ser83 of the gyrA gene. This is one of the dominant genetic determinants among Salmonella Typhi showing DCS in DRC [1]. Polymerase chain reaction (PCR) screening for genes encoding conventional ESBL revealed the blaCTX-M gene belonging to CTX-M-1 group G1.
The genome of the isolate was sequenced on Illumina HiSeq 2500 with 2 × 125bp paired-end reads (GATC Biotech, Mulhouse, France). Sequence reads were submitted to the European Nucleotide Archive (ENA) with accession number PRJEB19771. A draft sequence of the genome (named Typhi 10040_15) was assembled into 47 contigs using de novo assembly. Presence of acquired antibiotic resistance genes was assessed by Short Read Sequence Typing 2 (SRST2) in a curated ARG-annot database (ARGannot.r1) [2], and mutations in the QRDRs were determined by Basic Local Alignment Search Tool – Nucleotide (BLASTN) search of the Salmonella Typhi CT18 QRDR reference sequences (GI 16758993 gyrA, gyrB, parC, and parE) against the Typhi10040_15 assemblies. Resistance genes against sulfonamides (sulI), trimethoprim (dfrA7), and ampicillin (TEM-1D) were detected, the Ser83Phe substitution in gyrA was confirmed, and the ESBL gene was further identified as CTX-M-15. The aac6-iaa gene, which is associated with resistance against aminoglycosides, was present. However, 10 single-nucleotide polymorphisms (SNPs) compared to the reference sequence (AF144880) were detected. This may explain the aminoglycoside susceptible phenotype of the strain. We identified 49 putative IS sites in the contigs, of which 10 could be annotated using the semiautomatic pipeline of ISsaga (Supplementary table 1). The CTX-M-15 gene was encoded downstream of the mobile insertion site element ISEcp1. ISEcp1 is a member of the IS1380 family that may enable mobilization of CTX-M-15 genes. Presence of the repA replicon gene K02380 in the CTX-M-15 containing contig indicates that this gene is located on a type Y plasmid. The IncHI1-plasmid replicon, which is associated with MDR in Salmonella Typhi [3], was not identified using SRST2 and the PlasmidFinder database. BLASTN analysis of the assembled plasmid region against NCBI databases showed highest similarity (85% coverage and 97% identity) with the Klebsiella pneumoniae plasmid pKP12226, which also harbors a CTX-M-15 gene. This suggests that the isolate has acquired the CTX-M-15 containing plasmid from other Enterobacteriaceae, for which CTX-M-15 is frequently reported [4]. In 2015, Kariuki et al. reported a novel IncHI2 plasmid (pKST313) carrying the CTX-M-15 gene in Salmonella Typhimurium isolates from Kenya [5], but there is no similarity other than the CTX-M-15 region between pKST313 and the CTX-M-15 contig in Typhi10040_15 (Supplementary Figure 1).
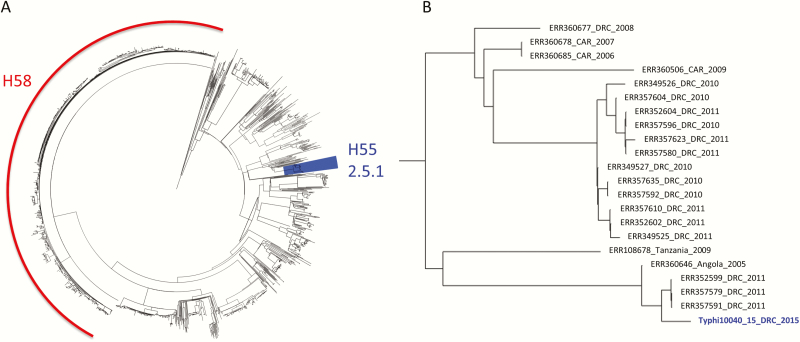
A phylogenetic analysis of the Typhi 10040_15 genome was undertaken using a global collection of 1831 Salmonella Typhi sequences [6]. All sequences were mapped against the Salmonella Typhi CT18 reference genome with SMALT (WTSI, Hinxton, UK), and a maximum likelihood phylogenetic tree was built with Gubbins (Figure 1). The Typhi 10040_15 isolate was most closely related to isolates recovered from DRC in 2011 and belongs to the Salmonella Typhi H55 clade (genotype 2.5.1) [7].
Figure 1.
A, The extended spectrum β-lactamase producing Typhi 10040_15 isolate within the global phylogenetic context of 1831 published Salmonella Typhi genomes (4). The H58 clade is indicated in red and the H55 clade (genotype 2.5.1), containing Typhi 10040_15, is highlighted in blue. B, Detailed view of the branch with Typhi 10040_15. Strains are annotated with the accession number of the sequencing data, the country of isolation and the year of isolation. Abbreviations: CAR, Central African Republic; DRC, The Democratic Republic of the Congo.
In contrast to other Salmonella serovars, ESBL production has been rarely reported in Salmonella Typhi, and most cases originate from Asia [8–12]. Only one report of ESBL-producing Salmonella Typhi in Africa has been published. In 2015, Akinyemi reported CTX-M-1 ESBL producing Salmonella Typhi in hospitalized patients in Lagos, Nigeria [13]. Unlike the Nigeria outbreak, the present Typhi 10040_15 isolate showed ESBL production in combination with DCS. The finding of ESBL production in Salmonella Typhi in DRC is of huge concern, as it adds to the high proportion of multidrug resistance and decreased ciprofloxacin susceptibility among Salmonella Typhi isolates in DRC (37.8% and 37.2%, respectively, for the period 2011–2014) [1]. Further surveillance as well as judicious use of azithromycin, which is considered as an orally available reserve antibiotic for typhoid fever, are imperative.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We thank Prof. Youri Glupczynski and Dr Pierre Bogaerts from the Belgian National Reference Centre for Antimicrobial Resistance of Gram-Negative Resistant Bacteria at CHU-UCL (Namur, Belgium) for their help in characterizing the ESBL phenotype and genotype of the strain, and for critical reading of the manuscript. We also thank Ms Tessa de Block for the technical assistance in this work and the International Typhoid Consortium for providing Salmonella Typhi genome sequences [5].
Financial support. This work was funded by the Belgian Directorate of Development Cooperation (DGD) through Project 2.01 of the Third Framework Agreement between the Belgian DGD and the Institute of Tropical Medicine, Belgium; the Flemish Ministry of Sciences (EWI, SOFI project IDIS); and the Wellcome Trust to G. D. and V. K. W.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Kalonji LM, Post A, Phoba MF et al. Invasive Salmonella infections at multiple surveillance sites in the Democratic Republic of the Congo, 2011–2014. Clin Infect Dis 2015; 61:S346–53. [DOI] [PubMed] [Google Scholar]
- 2. Inouye M, Dashnow H, Raven LA et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 2014; 6:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Holt KE, Phan MD, Baker S et al. Emergence of a globally dominant IncHI1 plasmid type associated with multiple drug resistant typhoid. PLoS Negl Trop Dis 2011; 5:e1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Woerther PL, Burdet C, Chachaty E, Andremont A. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 2013; 26:744–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kariuki S, Okoro C, Kiiru J et al. Ceftriaxone-resistant Salmonella enterica serotype typhimurium sequence type 313 from Kenyan patients is associated with the blaCTX-M-15 gene on a novel IncHI2 plasmid. Antimicrob Agents Chemother 2015; 59:3133–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wong VK, Baker S, Pickard DJ et al. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet 2015; 47:632–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wong VK, Baker S, Connor TR et al. ; International Typhoid Consortium. An extended genotyping framework for Salmonella enterica serovar Typhi, the cause of human typhoid. Nat Commun 2016; 7:12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Al Naiemi N, Zwart B, Rijnsburger MC et al. Extended-spectrum-beta-lactamase production in a Salmonella enterica serotype Typhi strain from the Philippines. J Clin Microbiol 2008; 46:2794–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rotimi VO, Jamal W, Pal T, Sovenned A, Albert MJ. Emergence of CTX-M-15 type extended-spectrum beta-lactamase-producing Salmonella spp. in Kuwait and the United Arab Emirates. J Med Microbiol 2008; 57:881–6. [DOI] [PubMed] [Google Scholar]
- 10. Pfeifer Y, Matten J, Rabsch W. Salmonella enterica serovar Typhi with CTX-M beta-lactamase, Germany. Emerg Infect Dis 2009; 15:1533–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morita M, Takai N, Terajima J et al. Plasmid-mediated resistance to cephalosporins in Salmonella enterica serovar Typhi. Antimicrob Agents Chemother 2010; 54:3991–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ahmed D, Hoque A, Mazumder R et al. Salmonella enterica serovar Typhi strain producing extended-spectrum β-lactamases in Dhaka, Bangladesh. J Med Microbiol 2012; 61:1032–3. [DOI] [PubMed] [Google Scholar]
- 13. Akinyemi KO, Iwalokun BA, Alafe OO, Mudashiru SA, Fakorede C. bla CTX-M-I group extended spectrum beta lactamase-producing Salmonella Typhi from hospitalized patients in Lagos, Nigeria. Infect Drug Resist 2015; 8:99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.