Summary
The mechanism of Zika virus (ZIKV)–induced microcephaly is not well understood. Studies suggest antibody-dependent enhancement (ADE) induced by dengue antibodies. ADE wasn’t observed in vivo in patients who had a dengue virus infection followed by a secondary ZIKV infection.
Keywords: ZIKV, DENV, ADE, cytokines
Abstract
Background
The pathogenesis of severe dengue disease involves immune components as biomarkers. The mechanism by which some dengue virus (DENV)–infected individuals progress to severe disease is poorly understood. Most studies on the pathogenesis of severe dengue disease focus on the process of antibody-dependent enhancement (ADE) as a primary risk factor. With the circulation of Zika virus (ZIKV) in DENV-endemic areas, many people infected by ZIKV were likely exposed to DENV. The influence of such exposure on Zika disease outcomes remains unknown.
Methods
We investigated whether patients previously exposed to DENV exhibited higher viremia when exposed to a subsequent, heterologous dengue or Zika infection than those patients not previously exposed to dengue. We measured viral loads and cytokine profile during patients’ acute infections.
Results
Neither dengue nor Zika viremia was higher in patients with prior DENV infection, although the power to detect such a difference was only adequate in the ZIKV analysis. Of the 10 cytokines measured, only 1 significant difference was detected: Levels of interleukin 1β (IL-1β) were lower in dengue-infected patients who had experienced a previous dengue infection than patients infected with dengue for the first time. However, power to detect differences between groups was low. In Zika-infected patients, levels of IL-1β showed a significant, positive correlation with viral load.
Conclusions
No signs of ADE were observed in vivo in patients with acute ZIKV infection who had prior exposure to DENV.
The development of severe dengue disease is associated with serial infection with dengue viruses (DENVs) of different serotypes; secondary dengue infection is a significant risk factor in >97% of severe cases [1, 2]. The pathogenesis of severe dengue is thought to be largely due to immune mechanisms in which antibody enhancement and T-cell immunopathology are likely to play key roles [3]. Once stimulated, components of the host immune response, which includes cells, cytokines, complements, and other cellular mediators, may serve as biomarkers of severe disease [4–7]. The mechanism by which only a few DENV-infected individuals progress to severe dengue disease is poorly understood. The processes of plasma leakage, shock, and hemorrhagic manifestations are initiated through the enhancement of infection by DENV with the help of opsonizing antibodies and result in an altered immune response that triggers memory T-cell activation and the release of cytokines and chemical mediators. These processes have been found to be a risk factor in secondary infection [6, 7].
The recent spread of Zika virus (ZIKV) in dengue-endemic areas has raised questions about the immunopathogenesis of ZIKV infection in patients who have been previously infected by DENV [2]. Dejnirattisai et al report that immunity to DENV may drive greater ZIKV replication and may have direct implications for disease pathogenesis and future vaccination programs for ZIKV and DENV [2]. On the other hand, other researchers have reported that antibodies presented in sera from dengue patients were found to be highly cross-reactive to ZIKV in that they produced binding and neutralization [8]. Subsequent results showed that a subset of antibodies targeting a conformational epitope from dengue patients is capable of strongly neutralizing ZIKV [9]. It is important to note that the homology between flaviviruses and cross-protection has also been observed in some contexts. For example, memory T-cell responses from patients previously infected with DENV produced potent protection through cellular immune response against Japanese encephalitis virus infection [10].
In this context, our aim was to investigate whether patients in a highly endemic area who had been previously exposed to dengue could exhibit higher viremia when exposed to a subsequent infection by a heterologous flavivirus. To do so, we compared the viral loads in these patients to determine whether the prior infection influenced virus replication. We also analyzed cytokine profiles during the subsequent acute infection.
MATERIALS AND METHODS
Clinical Samples
We selected 65 clinical samples collected from January to July 2016 from patients who exhibited acute febrile disease for ≤5 days and were attended at the at the emergency facility of the reference hospital in the city of São José do Rio Preto, São Paulo, Brazil, during a ZIKV outbreak. The blood samples were collected, and the viral RNA was extracted for the diagnosis of DENV serotypes 1–4 [11], ZIKV [12], Chikungunya virus (CHIKV) [13], or a lack of infection. After the molecular results, the 65 analyzed patients were randomly selected to continue the study. This study is part of an ongoing arbovirus surveillance program approved by the local research ethics committee (number 02078812.8.0000.5415), and all samples were taken from storage at –80°C when the study was performed.
Polymerase Chain Reaction Assays for Dengue Virus 1–4, Zika Virus, and Chikungunya Virus
To determine the presence of DENV, Flavivirus genus-specific primers targeting NS5 regions were first used in a reverse-transcription polymerase chain reaction (RT-PCR) assay. Next, DENV1–4 species-specific primers were used, because these viruses are known to circulate in the region. A multiplex-nested PCR assay was used to identify DENV1–4 in the samples [11]. To detect ZIKV and CHIKV, a 1-step quantitative, real-time, fluorescent probe–based RT-PCR assay was performed using primers targeting the envelope region in the case of ZIKV and the NsP1 region in the case of CHIKV, as described previously [12, 13]. The viral RNA samples used in the PCR assays were extracted from 140 µL using the QIAamp Viral RNA Mini kit (Qiagen) according to the manufacturer’s instructions.
Analysis of Viral Load by Quantitative Reverse-Transcription Polymerase Chain Reaction (Dengue Virus 2 and Zika Virus)
To determine the ZIKV viral load, we used the previously described primers known as ZIKV 1086 (5ʹ CCGCTGCCCAACACAAG 3ʹ) and ZIKV 1162c (5ʹ CCACTAACGTTCTTTTGCAGACAT 3ʹ) [12]. A 1-step reaction was performed using the SuperScript III Platinum SYBR Green One-Step quantitative reverse-transcription polymerase chain reaction kit (Life Technologies). For each sample, reactions were prepared with a final volume of 15 μL containing 10 μL 2X SYBR Green Reaction Mix, 0.4 μL SuperScript III RT/Platinum Taq Mix, 0.3 μL of each primer at 10 µM, ROX Dye to a final concentration of 50 nM, 5 µL of RNA, and water to complete the volume. Thermal cycling consisted of an initial step of 42°C for 3 minutes and 95°C for 5 minutes, followed by 40 cycles of 95°C for 15 seconds, 60°C for 1 minute, and a final step of 40°C for 1 minute. Finally, a melting curve was applied to the amplicons. The standard curve was performed using ZIKV containing 109–105 plaque-forming units (PFU)/mL.
To determine the DENV 2 viral load, a SYBR Green 2-step protocol was applied as previously described [14] and using Den F (5ʹ-TTAGAGGAGACCCCTCCC-3ʹ) and Den R2 (5ʹ-GAGACAGCAGGATCTCTGG-3ʹ). The standard curve was performed with DENV 2 PE 3808 containing 106–101 PFU/mL.
All of the reactions were performed in the QuantStudio 3 Real-Time PCR System using a MicroAmp Fast Optical 96-well reaction plate (Applied Biosystems). The runs were analyzed in the QuantStudio Design and Analysis Software, version 1.4. The viral loads are presented as genomic equivalent per milliliter.
Dengue Virus Immunoglobulin G Enzyme-Linked Immunosorbent Assay
The patients’ sera were tested for the presence of DENV immunoglobulin G (IgG) using the human anti–DENV IgG enzyme-linked immunosorbent assay (ELISA) kit (Abcam, Cambridge, United Kingdom) according to the manufacturer’s instructions. The assay did not discriminate between the DENV serotypes. The plate was read at 450 nm using a Spectramax Plus ELISA reader (Molecular Devices).
Analysis of Serum Cytokine Levels
Serum cytokine levels were determined through the use of a multiplex bead analysis in a Human Cytokine/Chemokine Magnetic Bead Panel (Millipore, Watford, United Kingdom). Levels of the following 10 molecules were measured according to the manufacturer’s instructions: interferon gamma (IFN-γ), interleukin (IL) 1β, IL-2, IL-4, IL-6, IL-8, IL-9, IL-10, IL-13, and IL-17. Finally, the beads were washed and analyzed in a Luminex IS-100 system (Luminex Corp, Texas). Standard curves of known concentrations of recombinant human cytokines were used to convert fluorescence units into cytokine concentration units (pg/mL). The data were stored and analyzed using GraphPad Prism software version 5.0.
Statistical Analysis
All statistical analyses were carried out using JMP software version 10. Age was normally distributed and analyzed using parametric statistics, whereas number of days between symptoms and medical assessment, cytokine level, and viral load data were not normal and no transformation attempted rendered them normal, so these data were analyzed using nonparametric statistics.
RESULTS
Molecular Results and Dengue Virus Immunoglobulin G Seroprevalence
Among the 65 patients with acute febrile illness who were studied herein, 20 were found to be positive for DENV 2 and 45 were found to be positive for ZIKV according to viral genome detection. An ELISA for specific IgG was performed to determine whether the patients had had a previous exposure to DENV. Among the 20 DENV 2-infected patients, 14 (70%) were IgG positive (DENV 2/IgG+); Among the 45 ZIKV-positive patients, 35 (78%) were IgG+ (ZIKV/IgG+). There was no significant difference in the proportion of patients previously exposed to DENV among the DENV-infected and ZIKV-infected groups (n = 66; P = .54, Fisher exact test).
Patient Age and Lag to Medical Assessment
Among the DENV-infected patients, individuals with evidence of a prior DENV infection were significantly older than those who lacked anti-DENV IgG (mean ± standard error [SE] age, 41.6 ± 3.4 years and 16.0 ± 5.7 years, respectively; t test: degree of freedom (df) = 17, t = 3.87, P = .001) (Supplementary Table 1). In contrast, among the ZIKV-infected patients, those who were DENV/IgG+ were slightly but not significantly older than those who were DENV/IgG– (mean ± SE age, 42.4 ± 2.8 years and 32.3 ± 5.3 years, respectively; t test: df = 43 t = 1.69, P = .10). Overall, the mean ± SE age of patients infected with DENV and ZIKV did not differ (34.9 ± 3.9 vs 40.2 ± 2.5 years, respectively; df = 62, t = 1.13, P = .26). The lag between symptom onset and medical assessment did not differ significantly between DENV/IgG+ and DENV/IgG– patients for either the DENV-infected group or the ZIKV-infected group (Mann-Whitney U test, P > .5 for both comparisons); across all data, this lag ranged from 0 to 7 days.
Viral Load
We analyzed patients’ viral loads to assess whether a prior dengue infection could influence viral replication during a second infection by another flavivirus. Viral load did not correlate with patient age for either DENV- or ZIKV-infected patients (Spearman rank correlation, P > .77 for both comparisons), nor was a significant correlation detected when DENV IgG+ and IgG– patients were analyzed separately within each virus-infected group (P > .2 for all comparisons). Neither was there a significant correlation between viral load and lag between symptom onset and medical treatment in DENV- or ZIKV-infected patients (Spearman rank correlation, P > .15 for both comparisons), although in the DENV-infected patients there was a trend of decreasing viral load as lag increased.
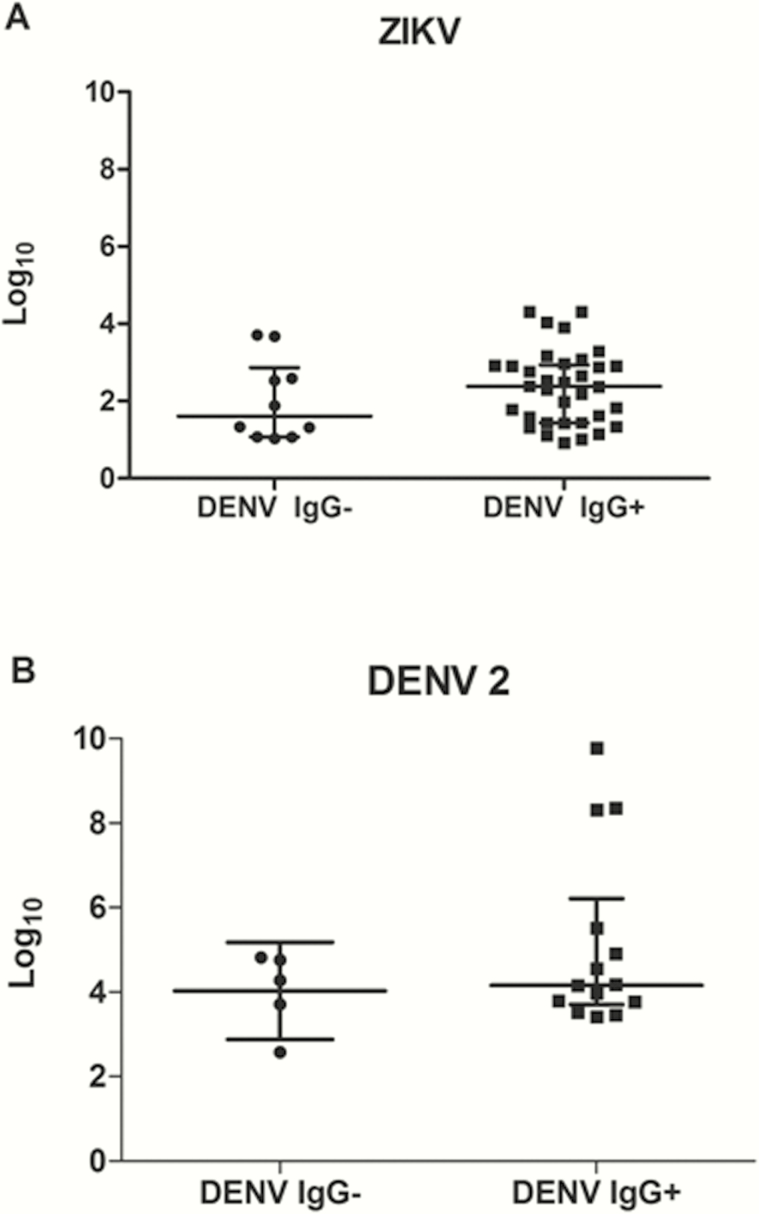
There was no significant difference in viral load between the DENV IgG+ and IgG– patients infected by DENV 2 (n = 19; P = .57, Mann-Whitney U test). Given the sample size and standard deviation of the data, the smallest difference detectable in this analysis (ie, the least significant value) is 2.08 log10 PFU/mL. Put another way, this analysis had high power to detect differences of ≥100-fold in viral load between the 2 samples. In the existing data, the DENV/IgG+ patients had only a 10-fold higher viremia, on average, than the DENV/IgG– patients. To have 90% power to detect a difference of this magnitude would have required 154 patients, equally distributed between the IgG+ and IgG– categories.
When DENV 2-infected patients were considered individually, 3 DENV 2/IgG+ patients (2056, 2287, and 2425) had viral loads (1 × 108 to 1 × 109 genomic equivalent/mL) that were up to 2 logs higher than the other patients. This might represent the antibody-dependent enhancement (ADE) of DENV viral load, despite the fact that only 1 of these patients had severe manifestations of dengue (Supplementary Table 2).
There was also no significant difference in viral load between the DENV IgG+ and IgG– patients infected by ZIKV (n = 44; P = .25, Mann-Whitney U test). The least significant value for this analysis was 0.70 log10 PFU/mL; thus, the analysis had high power (90%) to detect a 10-fold difference in viral load between IgG+ and IgG– patients. Moreover, no IgG+ patients had dramatically higher levels of ZIKV than the IgG– patients (Figure 1).
Figure 1.
Viral load in patients with Zika virus (ZIKV) and dengue virus (DENV) serotype 2 during primary and secondary dengue infections. A, Viral load quantified in ZIKV-positive patients with primary (immunoglobulin G–negative [IgG–]) DENV infection (n = 10) and secondary (immunoglobulin G–positive [IgG+]) DENV infection (n = 35). The Mann-Whitney U test demonstrated no significant difference in viral load. B, Viral load quantified in DENV 2+ patients with primary (IgG–) DENV infection (n = 6) and secondary (IgG+) DENV infection (n = 14). The Mann-Whitney U test demonstrated no significant difference in viral load. Medians with interquartile range are shown.
Cytokine Levels
A commercial panel consisting of IFN-γ, IL-1β, IL-2, IL-4, IL-6, IL-8, IL-9, IL-10, IL-13, and IL-17 was used to analyze the levels of circulating cytokines in patients exposed to a second infection. These cytokines are involved in the “cytokine storm” phenomenon that is known to affect patients with severe dengue and which may be considered a marker of severe disease [7] (Supplementary Table 2).
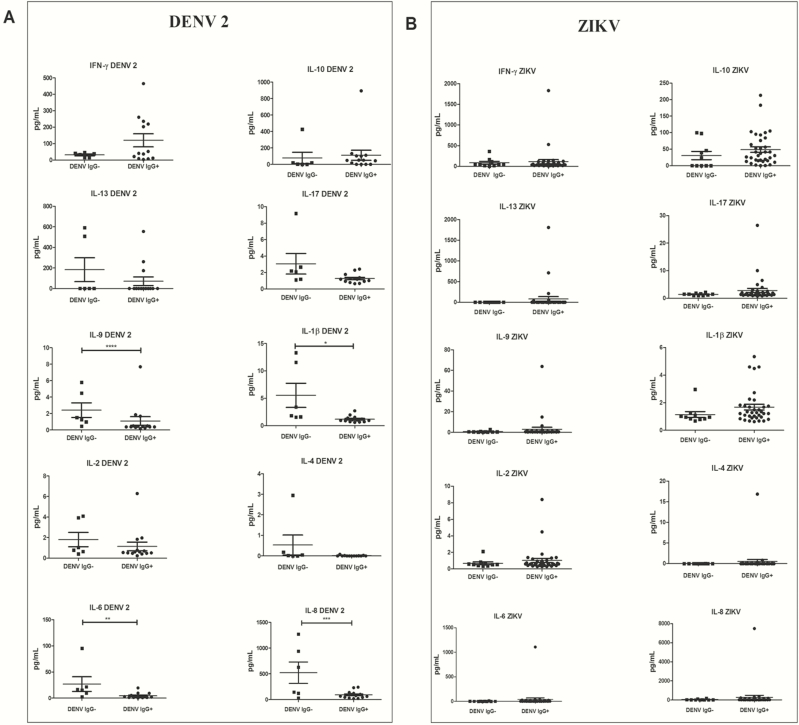
We compared levels of 10 cytokines between IgG+ and IgG– patients in the DENV-infected cohort (Figure 2A). To correct for multiple testing, the threshold for significance (α) was adjusted to .005 (.05/10). Only 1 cytokine, IL-1β, was significantly different between the 2 groups (n = 19; P = .004, Mann-Whitney U test), and this cytokine was lower in IgG+ patients. However, these analyses had very low power, ranging from 6% to 12% power to detect a deviation of ≥25% from the larger of the mean cytokine values in the 2 groups. Generally, DENV 2/IgG+ patients presented higher levels of INF-γ and IL-10 and lower levels of the other 8 cytokines tested. Individually, the DENV 2/IgG+ patients were found to have high levels of some cytokines at rates very similar to those of the DENV 2/IgG– patients (1 patient each for IL-1β, IL-9, and IL-2). Levels of 2 cytokines, IL-1β and IL-6, showed a significant, negative correlation with age (P < .004 for both comparisons, Spearman rank correlation).
Figure 2.
Cytokine (interleukin [IL]; interferon [IFN]) response in patients with dengue virus (DENV) serotype 2 and Zika virus (ZIKV) during primary and secondary dengue infections. A, Cytokine levels measured in DENV 2-positive patients with primary (immunoglobulin G–negative [IgG–]) DENV infection (n = 06) and secondary (immunoglobulin G–positive [IgG+]) DENV infection (n = 14). The Mann-Whitney U test demonstrated a significant P value in the cases of IL-1β (*P = .0039), IL-6 (**P = .0133), IL-8 (***P = .0477), and IL-9 (****P = .0391). B, Cytokine levels measured in ZIKV-positive patients with primary (IgG–) DENV infection (n = 10) and secondary (IgG+) DENV infection (n = 35). The Mann-Whitney U test demonstrated no significant P value.
In the ZIKV-infected cohort, none of the 10 cytokines differed significantly between IgG+ and IgG– patients (Figure 2B). Again, the power of these analyses was low, ranging from 5% to 20% power to detect a deviation of ≥25% from the larger of the mean cytokine values in the 2 groups. When patients were considered individually, a few ZIKV/IgG+ individuals were found to exhibit substantially higher levels of IL-4, IL-6, IL-8, IL-9, IL-13, and IL-17 (1 patient), of IFN-γ, IL-10, and IL-2 (2 patients), and of IL-1β (4 patients) compared with the levels found in ZIKV/IgG– individuals. None of the cytokine levels showed a significant correlation with age.
Cytokines Versus Viral Load
Viral load did not show a significant correlation with any cytokine level in the DENV 2/IgG+ patients or the DENV 2/IgG– patients. However, viral load did show a significant positive correlation with IL-1β (n = 34; P = .03, Spearman rank correlation) in the ZIKV/IgG+ patients and a marginally significant correlation (n = 10; P = .05) in the ZIKV/IgG– patients. ZIKV load did not show a significant correlation with any other cytokine level (Supplementary Table 2 and Supplementary Figure 1).
DISCUSSION
Infection with any DENV serotype (or with any flavivirus) is known to result in long-term homotypic protection, whereas in the case of heterotypic infection, only short-term immunity is produced [15]. Moreover, secondary infections by heterologous DENV serotypes can lead to ADE through the cross-reactivity among anti-DENV antibodies that enhance uptake by Fc receptor–bearing cells [2]. Previous studies using animal models have shown that ADE produces a higher DENV viral load [16, 17]. Thus, it is worrisome that some vaccine strategies against DENV or even ZIKV could produce ADE in a population that has been previously exposed to several flaviviruses, which is the case of many Brazilian residents.
ADE has been proposed in ZIKV-infected individuals who have had previous DENV infection [2, 8, 18]. Both viruses belong to the same genus and exhibit a homology of 50% in the amino acid sequence of the envelope protein, the major antigenic target [19]. However, ADE is only one component of a combination of immunological mechanisms that drive the pathogenesis of severe dengue [20]. In this study, we investigated a cohort of patients who had been previously exposed to DENV via natural infection and who had experienced secondary infection by DENV 2 or ZIKV. We analyzed viral load and levels of proinflammatory and anti-inflammatory cytokines and assessed possible correlations between viral load and cytokine levels in this very original and valuable cohort.
The most significant finding of our study is that ADE was not observed in our in vivo investigation into patients who had been infected by DENV followed by a secondary infection by ZIKV. Moreover, none of the patients in this cohort exhibited a severe course of the disease, and all recovered after the recommended supportive therapy for infections caused by arboviruses.
Our analysis, which had high power to detect a 10-fold difference in virus titer, found no difference in viral load between ZIKV-infected patients who had been exposed previously to DENV and those who had not. We also detected no apparent enhancement of DENV replication in patients who had or had not been previously infected with DENV; however, this latter analysis had low power to detect biologically realistic differences in virus titer.
A cytokine storm is an overexuberant immunological response that can be activated during a secondary DENV infection. It may worsen pathology through processes such as vascular permeability, plasma leakage, and fever [20, 21]. Among the cytokines associated with this phenomenon, the increase of IL-6, IL-8, IL-10, IFN-γ, IFN-α, and vascular endothelial growth factor, combined with tumor necrosis factor α, are considered poor prognostic markers of the occurrence of severe dengue as they indicate a poor outcome [20, 22]. Furthermore, some of these cytokines are important for overcoming the disease. Our data showed that DENV-infected patients with previous exposure to dengue presented significantly lower levels of IL-1β and higher, albeit not significantly so, levels of IFN-γ and IL-10, whereas the same differences were not observed in ZIKV-infected patients who had or had not experienced a previous DENV infection. We did make the intriguing observation that levels of IL-1β in both ZIKV-infected groups were positively correlated with viral load. IL-1β is related to the processes of coagulation and fibrinolysis during a viral infection [23]. Its concentration was shown to be elevated during the early stages of DENV infection without being related to the disease severity [24, 25].
Most studies on severe dengue focus on ADE. With the circulation of ZIKV in DENV-endemic areas, many people infected by ZIKV are likely to have been previously exposed to DENV. In these cases, little is known about the consequences or whether the subsequent infection is worsened by prior flavivirus exposure. We are aware that a study based on cytokine profiles and viral load alone is not the most appropriate strategy for investigating disease progress or severity, nor it is the best choice for tracking patients’ immunological response. However, the use of cytokines may be an important tool in the initial screening of patient response to infection with DENV and ZIKV. Our focus now is to continue to evaluate this cohort to determine whether primary infection by DENV does, in fact, provide protective benefits in secondary infections by Zika and by other dengue serotypes. Our strategies will be to analyze different immunological aspects, such as subclasses of immunoglobulins and different subsets of immune cells, to understand the mechanisms underlying the results that we have observed in the present study.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Author contributions. All authors had full access to all of the data for this study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Financial support. This study was supported by the São Paulo Research Foundation (FAPESP) (grant number 2013/21719-3 to M. L. N. and fellowship grant number 2015/12295-0 to A. C. B. T.); by the Butantan Institute; the Brazilian National Institutes for Science and Technology for Dengue Studies (INCT em Dengue); and the FAPESP Zika Network. M. L. N. is a Brazilian National Research Council (CNPq) fellow. N. V. and K. A. H. were supported by the National Institutes of Health (grant number 1U01AI115577-01).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Guzman MG. Global voices of science. Deciphering dengue: the Cuban experience. Science 2005; 309: 1495–7. [DOI] [PubMed] [Google Scholar]
- 2. Dejnirattisai W, Supasa P, Wongwiwat W et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat Immunol 2016; 17:1102–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Screaton G, Mongkolsapaya J, Yacoub S, Roberts C. New insights into the immunopathology and control of dengue virus infection. Nat Rev Immunol 2015; 15:745–59. [DOI] [PubMed] [Google Scholar]
- 4. Pawitan JA. Dengue virus infection: predictors for severe dengue. Acta Med Indones 2011; 43:129–35. [PubMed] [Google Scholar]
- 5. Green S, Rothman A. Immunopathological mechanisms in dengue and dengue hemorrhagic fever. Curr Opin Infect Dis 2006; 19:429–36. [DOI] [PubMed] [Google Scholar]
- 6. Nielsen DG. The relationship of interacting immunological components in dengue pathogenesis. Virol J 2009; 6:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pang T, Cardosa MJ, Guzman MG. Of cascades and perfect storms: the immunopathogenesis of dengue haemorrhagic fever-dengue shock syndrome (DHF/DSS). Immunol Cell Biol 2007; 85:43–5. [DOI] [PubMed] [Google Scholar]
- 8. Priyamvada L, Quicke KM, Hudson WH et al. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci U S A 2016; 113:7852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barba-Spaeth G, Dejnirattisai W, Rouvinski A et al. Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature 2016; 536:48–53. [DOI] [PubMed] [Google Scholar]
- 10. Turtle L, Bali T, Buxton G et al. Human T cell responses to Japanese encephalitis virus in health and disease. J Exp Med 2016; 213:1331–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Morais Bronzoni RV, Baleotti FG, Ribeiro Nogueira RM, Nunes M, Moraes Figueiredo LT. Duplex reverse transcription-PCR followed by nested PCR assays for detection and identification of Brazilian alphaviruses and flaviviruses. J Clin Microbiol 2005; 43:696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lanciotti RS, Kosoy OL, Laven JJ et al. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 2008; 14:1232–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lanciotti RS, Kosoy OL, Laven JJ et al. Chikungunya virus in US travelers returning from India, 2006. Emerg Infect Dis 2007; 13:764–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chutinimitkul S, Payungporn S, Theamboonlers A, Poovorawan Y. Dengue typing assay based on real-time PCR using SYBR Green I. J Virol Methods 2005; 129:8–15. [DOI] [PubMed] [Google Scholar]
- 15. Montoya M, Gresh L, Mercado JC et al. Symptomatic versus inapparent outcome in repeat dengue virus infections is influenced by the time interval between infections and study year. PLoS Negl Trop Dis 2013; 7:e2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zompi S, Harris E. Animal models of dengue virus infection. Viruses 2012; 4:62–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Milligan GN, Sarathy VV, Infante E et al. A dengue virus type 4 model of disseminated lethal infection in AG129 mice. PLoS One 2015; 10:e0125476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paul LM, Carlin ER, Jenkins MM et al. Dengue virus antibodies enhance Zika virus infection. Clin Transl Immunology 2016;5:e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sirohi D, Chen Z, Sun L et al. The 3.8 a resolution cryo-EM structure of Zika virus. Science 2016; 352:467–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. John DV, Lin YS, Perng GC. Biomarkers of severe dengue disease—a review. J Biomed Sci 2015; 22:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rothman AL. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nature Rev Immunol 2011; 11:532–43. [DOI] [PubMed] [Google Scholar]
- 22. Tsai TT, Chuang YJ, Lin YS et al. Antibody-dependent enhancement infection facilitates dengue virus-regulated signaling of IL-10 production in monocytes. PLoS Negl Trop Dis 2014; 8:e3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Suharti C, van Gorp ECM, Setiati TE, et al. The role of cytokines in activation of coagulation and fibrinolysis in dengue shock syndrome. Thromb Haemost 2002. January; 87(1):42–6. [PubMed] [Google Scholar]
- 24. Bozza Fa, Salluh JI, Japiassu AM, et al. Cytokine profiles as markers of disease severity in sepsis: a multiplex analysis. Crit Care 2007; 11(2):R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hober D, Poli L, Roblin B, et al. Serum levels of tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), and interleukin-1β (IL-1β) in dengue-infected patients. Am J Trop Med Hyg 1993; 48(3):324–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.