Abstract
To investigate the prognostic impact of MET copy number (MET-CN) in patients with non-small cell lung cancer (NSCLC), we retrospectively reviewed clinical and pathologic data of NSCLC patients whose tumors were assessed for MET-CN using fluorescence in situ hybridization (FISH). We correlated MET-CN status with patient overall survival (OS) and optimized MET-FISH reporting criteria. The study group included 384 patients with NSCLC of which 88% were adenocarcinoma and 55.7% of patients had distant metastases. There were 170 patients with stages I-III and 214 patients with stage IV disease. Based on the MET-CN and MET/CEP7 ratio the patients were classified into 3 categories: MET-amplification (METamp): MET/CEP7 ≥ 2 or MET-CN ≥ 5; MET-CN-gain (METcng): MET-CN ≥ 4 to < 5; and MET-negative (METneg): MET-CN < 4. METamp was associated with high fatality (P=.036) and stage IV tumors (P=.038). In patients with stages I-III NSCLC, patients in the METamp category had the shortest OS (P=.015) and more often developed distant metastases within 1 year (P=.004). In patients with stage IV tumors, METamp did not further impact the OS. Patients in the METcng category had the longest OS (P=.053). Multivariate analysis confirmed METamp to be an independent high-risk factor (HR 3.26; P=.026) and predicted earlier progression to distant metastasis (HR 4.86; P=.001). In conclusion, we suggest that the MET-FISH criteria presented optimizes risk stratification by defining 3 categories of NSCLC patients. METamp is an independent risk factor predicting early distant metastasis and patients with METcng could represent a lower-risk group.
Keywords: MET gene amplification, MET FISH reporting criteria, non-small cell lung cancer, distant metastasis
INTRODUCTION
Chromosomal aneuploidy or somatic copy number alterations (SCNAs) are frequently observed in malignant neoplasms including lung cancers and have been proposed to drive tumorigenesis or treatment resistance [1, 2]. SCNAs such as amplification of the mesenchymal-epithelial transition factor gene (MET) which resides on chromosome 7q31, can be detected conveniently by using fluorescence in situ hybridization (FISH). MET amplification (METamp) has been associated with shorter overall survival (OS) in patients with non-small cell lung cancer (NSCLC) [3–6]. Therefore, MET is considered a potentially targetable oncogenic driver [7, 8]. Recent studies have shown that amplified MET is a clinically valid therapeutic target for MET inhibitors or MET-tyrosine kinase inhibitors (e.g. crizotinib) approved recently for the treatment of patients with NSCLC [9].
The frequency of METamp as detected by FISH in NSCLC is variable in the literature, ranging from 3% to 10%. This range in frequency is likely attributable to a lack of standardization of FISH techniques, different cutoffs for defining MET positivity, and/or patient selection criteria across studies [6, 10–16]. For example, in a study of 213 Asian patients with NSCLC, Okuda et al. [16] used a cutoff of 3 or more copies of MET per cell to define MET-FISH positivity. They reported that patients with MET-positive tumors had a shorter OS than did patients with a normal MET-FISH result. In contrast, Cappuzzo et al. [6] used a cutoff of 5 copies per cell to designate MET-FISH positivity because patients with 5 or more copies of MET showed a worse clinical outcomes than did those with fewer than 5 copies. In another study involving 141 patients with stage I lung adenocarcinoma, a mean MET copy number (MET-CN) ≥ 3.4 per cell was regarded as a positive result and was associated with a poorer prognosis [3]. In a recent study, Noonan and colleagues [17] proposed using a combination of criteria to establish MET-FISH positivity: a MET/CEP7 (centromeric probe of chromosome 7) ratio of ≥ 1.8 or a MET-CN ≥ 5. The variations among these studies indicate that the criteria used to define MET-FISH positivity are inconsistent and that researchers need to reach a consensus on a positive cutoff point above which MET-CN has a clinical impact on outcomes in patients with NSCLC [6, 10–16].
Although preclinical studies have shown that tumor cells with METamp display significantly increased sensitivity to MET inhibitors or MET tyrosine kinase inhibitors (MET-TKIs) [18–20], several clinical trials of MET inhibitors have failed in patients with METamp as determined by FISH [21–23]. One potential reason for the failure of these trials may be that MET-CN status cannot be assessed consistently or accurately because no standard MET-FISH reporting criteria is available. In addition, most clinical studies related to MET-CN status were conducted in NSCLC patients who had undergone surgical resection; very limited data are available on patients with unresectable stage IV disease [3, 6, 16]. Although one recent study population included about 60% stage IV patients, no detailed clinical data were provided [17].
To improve patient selection criteria for trials of targeted MET inhibitors, we conducted a comprehensive retrospective review of MET-FISH data of NSCLC patients evaluated during a 6-year period. We evaluated and validated MET-FISH reporting criteria proposed in the literature and correlated MET-FISH results with clinical outcome and disease progression in NSCLC patients who presented without distant metastasis. We also used these data to suggest more optimized MET-FISH reporting criteria to facilitate accurate determination of clinically relevant MET-CN status and improve risk stratification of patients with NSCLC.
RESULTS
Clinicopathologic data
The study group included 384 NSCLC specimens tested for MET-CN. The patient cohort consisted of 184 (47.9%) men and 200 (52.1%) women, with a median age of 64 years (range, 31–89 years). More than three quarters of the patients (305, 79.4%) were white. Adenocarcinoma accounted for 338 (88%) cases and squamous cell carcinoma accounted for 40 (10.4%) cases. The cohort consisted of 42 (10.9%) patients with stage I NSCLC, 42 (10.9%) with stage II disease, 86 (22.4%) with stage III disease, and 214 (55.7%) with stage IV disease. All patients received the standard-of-care therapy according to their disease stage at diagnosis, including but not limited to surgery, chemotherapy, radiation therapy, or epidermal growth factor receptor (EGFR) inhibitor therapy when applicable. No patients were treated with MET inhibitors. Table 1 provides detailed clinicopathological and demographic data summarized by disease stage. Stages I, II, III were grouped together because most patients who do not have distant metastasis are considered clinically eligible for surgery, whereas patients with stage IV disease are not candidates for surgery.
Table 1. Patient demographic and clinicopathologic characteristics.
| Characteristic | Total | Stages I-III | Stage IV | P value |
|---|---|---|---|---|
| (n =384) | (n =170) | (n =214) | ||
| Median age, years (range) | 64 (31–89) | 65 (32–88) | 62 (31–89) | .25 |
| Sex, n (%) | .751 | |||
| Male | 184 (47.9) | 83 (48.8) | 101 (47.2) | |
| Female | 200 (52.1) | 87 (51.2) | 113 (52.8) | |
| Race/ethnicity, n (%) | .463 | |||
| White | 305 (79.4) | 138 (81.2) | 167 (78.0) | |
| Black | 25 (6.5) | 11 (6.5) | 14 (6.5) | |
| Hispanic | 24 (6.3) | 12 (7.1) | 12 (5.6) | |
| Asian | 23 (6.0) | 6 (3.5) | 17 (7.9) | |
| Unknown | 7 (1.8) | 3 (1.8) | 4 (1.9) | |
| Histologya, n (%) | .002* | |||
| Adenocarcinoma | 338 (88.0) | 139 (81.8) | 199 (93.0) | |
| Squamous cell carcinoma | 40 (10.4) | 26 (15.3) | 14 (6.5) | |
| Other | 6 (1.6) | 5 (2.9) | 1 (0.5) |
a Other histology included 5 patients with unclassified non-small cell lung cancer and 1 patient with adenosquamous carcinoma.
*Indicates statistically significant result (P <.05).
Optimization of MET-FISH reporting criteria
Patients were first stratified into 5 groups according to MET-CN and MET/CEP7 ratio (Groups 1-4 MET/CEP7 ratio < 2.0) and: Group 1 characterized by MET-CN < 3 (273 patients, 71.1%); Group 2 with MET-CN ≥ 3 to < 4 (65 patients, 16.9%); Group 3 characterized by MET-CN ≥ 4 to < 5 (17 patients, 4.4%); Group 4 with MET-CN ≥ 5 (8 patients, 2.1%); and Group 5 in which the MET/CEP7 ratio ≥ 2.0 or signal clusters seen in > 10% of tumor cells (21 patients, 5.5%) (Supplementary Table 1) [24].
When we tested for associations between these groups and OS in patients with stages I to III disease, we found that patients in Groups 1 and 2 had a similar median OS (48.9 months and 59.1 months, respectively; P =.321). Groups 4 and 5 also had similar OS (28.1 months vs. 23.6 months, respectively; P =.782). Patients in Groups 1 and 2 had markedly longer median OS than did patients in Groups 4 and 5. Interestingly, patients in Group 3 had significantly longer median OS than patients in any other group (134.4 months; P =.03) (Figure 1A and Supplementary Table 2).
Figure 1. Kaplan-Meier curves comparing overall survival (OS) by MET FISH status.
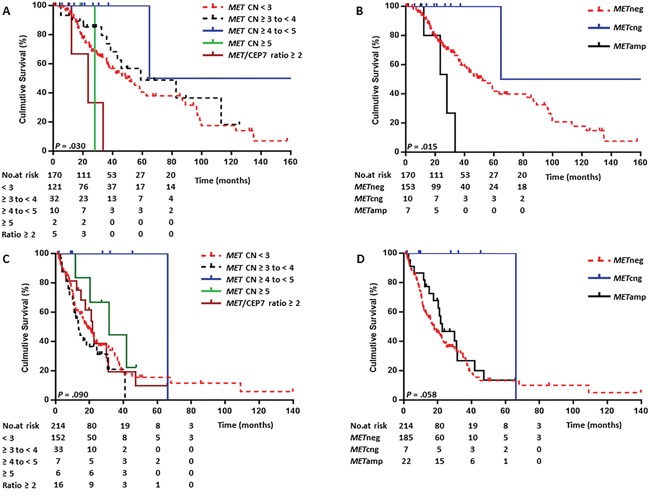
(A) Comparison of OS in patients with stage I-III NSCLC stratified into 5 MET-CN groups. (B) Comparison of OS in the stage I-III NSCLC patients using the optimized MET-FISH reporting criteria. (C) Comparison of OS in patients with stage IV NSCLC stratified into 5 MET-CN groups. (D) Comparison of OS in patients with stage IV NSCLC using the optimized MET-FISH reporting criteria.
Because our OS data for patients with stages I-III disease were consistent with those of reported previously in a large study [6], we next re-categorized the data using a MET-CN cutoff of 5 copies and a MET/CEP7 ratio cutoff of 2.0. These optimized MET-FISH reporting categories were as follows:
– METamp: (A) MET/CEP7 ratio ≥ 2.0, or (B) MET-CN ≥ 5, or (C) MET/CEP7 ratio < 2.0 but MET-CN ≥ 20 or MET signal clusters in more than10% of tumor cells.
– MET-CN-gain (METcng): MET/CEP7 ratio < 2.0 and MET-CN ≥ 4 to < 5.
– METneg: MET/CEP7 ratio < 2.0 and MET-CN < 4.
We reclassified all cases into these 3 categories for further analyses of OS and risk stratification (Figure 1B and Supplementary Table 2).
METamp is highly associated with stage IV NSCLC
The mean MET-CN in the overall study cohort was 3.1 copies per cell (range, 1.5 - 20.5). The mean MET/CEP7 ratio was 1.15 (range, 0.5 - 8.5). The mean MET-CN and mean MET/CEP7 ratio were significantly higher in patients with stage IV NSCLC than in patients with stages I-III disease (P =.042 and P =.016, respectively) (Supplementary Table 3). Using the optimized MET-FISH reporting criteria, 29 of 384 (7.6%) patients were categorized as having METamp.
Among the 170 patients with stage I-III disease, 153 (90%) were in the METneg group, 10 (5.9%) in the METcng group, and 7 (4.1%) in the METamp group. Of the 214 patients with stage IV disease, 185 (86.4%) were in the METneg group and 7 (3.3%) in the METcng group. We determined that 22 (10.3%) patients with stage IV NSCLC had METamp, a significantly higher rate than that found in patients with stages I-III disease (P =.038), indicating that METamp was highly associated with advanced disease stage (Supplementary Table 3 and Table 2).
Table 2. Demographic and clinical characteristics of 384 patients in the three optimized MET categories.
| Characteristic | Total (n = 384) |
METneg (n = 338) |
METcng (n = 17) |
METamp (n = 29) |
P value |
|---|---|---|---|---|---|
| Median age at diagnosis (years) | 63 | 64 | 62 | 61 | .338 |
| Sex, n (%) | .549 | ||||
| Male | 184 (47.9) | 165 (48.8) | 8 (47.1) | 11 (37.9) | |
| Female | 200 (52.1) | 173 (51.2) | 9 (52.9) | 18 (62.1) | |
| Race/ethnicity, n (%) | 1.000 | ||||
| White | 305 (80.9) | 267 (80.7) | 14 (82.4) | 24 (82.8) | |
| Non-whitea | 72 (19.1) | 64 (19.3) | 3 (17.6) | 5 (17.2) | |
| Histology, n (%) | .364 | ||||
| Adenocarcinoma | 338 (88.0) | 296 (87.6) | 17 (100.0) | 25 (86.2) | |
| Non-adenocarcinomab | 46 (12.0) | 42 (12.4) | 0 (0.0) | 4 (13.8) | |
| Stage, n (%) | .064 | ||||
| I | 42 (10.9) | 37 (10.9) | 4 (23.5) | 1 (3.4) | |
| II | 42 (10.9) | 39 (11.5) | 0 (0.0) | 3 (10.3) | |
| III | 86 (22.4) | 77 (22.8) | 6 (35.3) | 3 (10.3) | |
| IV | 214 (55.7) | 185 (54.7) | 7 (41.2) | 22 (75.9) | |
| Distant metastasisc (%) | .038* | ||||
| No | 170 (44.3) | 153 (45.3) | 10 (58.8) | 7 (24.1) | |
| Yes | 214 (55.7) | 185 (54.7) | 7 (41.2) | 22 (75.9) |
Abbreviations: MET, mesenchymal-epithelial transition factor; FISH, fluorescence in situ hybridization; METneg, MET-negative; METcng, MET copy number gain; METamp, MET amplification.
aNon-white included 25 black, 24 Hispanic, and 23 Asian patients. Seven patients with unknown race were excluded from the analysis.
bNon-adenocarcinoma included 40 patients with squamous cell carcinoma, 5 patients with unclassified non-small cell lung cancer, and 1 patient with adenosquamous carcinoma.
cDistant metastasis included bone, brain, liver, adrenal, contralateral lobe, tumor with pleural nodules, or malignant pleural effusion.
* Indicates statistically significant result (P <.05, Fisher exact test).
MET-FISH and MET-IHC correlations
MET overexpression as assessed by immunohistochemistry (IHC) was available in 202 of 384 (52.6%) patients, including 92 of 170 (54.1%) patients with stages I-III and 110 of 214 (51.4%) patients with stage IV disease. MET overexpression was found in 51 (55.4%) patients in stages I-III and 73 (66.4%) patients with stage IV (P =.14), indicating that MET overexpression is not associated significantly with disease stage (Supplementary Table 5). In addition, when we correlated MET expression with MET-CN, 20 of 122 METneg cases, 3 of 8 METcng cases, and 5 of 22 METamp cases were MET-IHC negative (P =.28) (Supplementary Table 5). The overall concordance between the MET-FISH and the MET-IHC was 44.8% (P =.11) (Supplementary Table 5).
METamp is associated with shorter OS in patients with stages I-III NSCLC
The median follow-up duration was 19.5 months and the median OS was 36.4 months for the overall study cohort. OS was not associated with patient age (P =.18), sex (P =.58), or race/ethnicity (P =.45). As expected, longer OS was highly associated with early-stage (I-III) disease (P <.001) and with adenocarcinoma (P <.001) (Supplementary Table 4).
Under the optimized MET-FISH reporting criteria, the median OS durations of early-stage patients with METamp, METcng, and METneg were 28.1, 134.4, and 51.6 months, respectively. Patients with METamp had significantly shorter OS than did patients with METneg (P =.036), and patients with METcng had longer OS than did patients with METneg, although the difference was not statistically significant (P =.109) (Figure 1B, Table 3)
Table 3. Univariate and multivariate analyses of overall survival and progression to distant metastasis for patients with stages I-III NSCLC.
| Variable | Overall Survival | Progression to Distant Metastasisa | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | ||||||||||
| HR | 95% CI | P value | HR | 95% CI | P value | HR | 95% CI | P value | HR | 95% CI | P value | ||
| Age | > 64 years | - | - | - | - | - | - | - | - | - | - | - | - |
| ≤ 64 years | 0.65 | 0.41-1.03 | .066 | 0.75 | 0.46-1.22 | .243 | 1.24 | 0.79-1.96 | .343 | 1.24 | 0.78-1.98 | .372 | |
| Sex | Female | - | - | - | - | - | - | - | - | - | - | - | - |
| Male | 1.39 | 0.88-2.18 | .159 | 1.03 | 0.64-1.67 | .89 | 1.34 | 0.86-2.11 | .197 | 1.19 | 0.75-1.90 | .467 | |
| Raceb | Non-white | - | - | - | - | - | - | - | - | - | - | - | - |
| White | 2.05 | 0.98-4.29 | .055 | 2.05 | 0.93-4.48 | .074 | 1.2 | 0.64-2.22 | .572 | 1.43 | 0.75-2.76 | .281 | |
| Histologyc | Non-ADC | - | - | - | - | - | - | - | - | - | - | - | - |
| ADC | 0.34 | 0.20-0.57 | <.001* | 0.52 | 0.30-0.90 | .019* | 0.85 | 0.47-1.55 | .593 | 0.97 | 0.52-1.81 | .929 | |
| Stage | I | - | - | - | - | - | - | - | - | - | - | - | - |
| II | 1.57 | 0.78-3.17 | .206 | 1.38 | 0.67-2.84 | .377 | 0.95 | 0.47-1.92 | .88 | 0.92 | 0.45-1.87 | .807 | |
| III | 3.28 | 1.77-6.10 | <.001* | 3.33 | 1.73-6.42 | <.001* | 2.21 | 1.25-3.91 | .007* | 2.25 | 1.24-4.09 | .008* | |
| MET FISH | METneg | - | - | - | - | - | - | - | - | - | - | - | - |
| METcng | 0.2 | 0.03-1.43 | .109 | 0.24 | 0.03-1.79 | .165 | 0.93 | 0.34-2.55 | .883 | 0.93 | 0.33-2.60 | .894 | |
| METamp | 3.01 | 1.08-8.42 | .036* | 3.26 | 1.15-9.23 | .026* | 4.25 | 1.67-10.81 | .002* | 4.86 | 1.85-12.75 | .001* | |
Abbreviations: HR, hazard ratio; CI, confidence interval; MET, mesenchymal-epithelial transition factor; FISH, fluorescence in situ hybridization; METneg, MET-negative; METcng, MET copy number gain; METamp, MET amplification; hyphens (-) indicate the reference category. ADC: Adenocarcinoma.
aDistant metastasis included bone, brain, liver, adrenal, contralateral lobe, tumor with pleural nodules, or malignant pleural effusion.
bNon-white included 11 black, 12 Hispanic, and 6 Asian patients. Three patients with unknown race were not included in the analysis.
cThe adenocarcinoma category also included bronchoalveolar histology. Non-adenocarcinoma included 26 patients with squamous cell carcinoma, 4 patients with unclassified non-small cell lung cancer, and 1 patient with adenosquamous carcinoma.
* Indicates statistically significant result (P <.05).
In patients with stage IV tumors, MET-CN status showed no statistically significant association with OS (Figures 1C and 1D, Table 4). However, patients with METcng did have substantially longer OS than did patients with METneg (66.2 vs. 17.5 months, P =.053). This pattern was similar to that observed in patients with stages I-III disease.
Table 4. Univariate and multivariate analyses of overall survival for patients with stage IV NSCLC.
| Variable | Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | ||
| Age | > 64 years | - | - | - | - | - | - |
| ≤ 64 years | 0.77 | 0.54-1.09 | .141 | 0.81 | 0.56-1.15 | .239 | |
| Sex | Male | - | - | - | - | - | - |
| Female | 0.88 | 0.62-1.25 | .485 | 0.83 | 0.58-1.19 | .315 | |
| Racea | Non-white | - | - | - | - | - | - |
| White | 0.95 | 0.62-1.45 | .803 | 0.98 | 0.64-1.52 | .938 | |
| Histologyb | Non-adenocarcinoma | - | - | - | - | - | - |
| Adenocarcinoma | 0.62 | 0.34-1.12 | .115 | 0.59 | 0.32-1.09 | .095 | |
| MET-FISH | METneg | - | - | - | - | - | - |
| METcng | 0.14 | 0.02-1.02 | .053 | 0.17 | 0.02-1.20 | .075 | |
| METamp | 0.79 | 0.46-1.36 | .374 | 0.79 | 0.46-1.36 | .400 | |
Abbreviations: HR, hazard ratio; CI, confidence interval; MET, mesenchymal-epithelial transition factor; FISH, fluorescence in situ hybridization; METneg, MET-negative; METcng, MET copy number gain; METamp, MET amplification; hyphens (-) indicate the reference category.
a Non-white included 14 black, 12 Hispanic, and 17 Asian patients. Four patients with unknown race were not included in the analysis.
b The adenocarcinoma category also included bronchoalveolar histology. Non-adenocarcinoma included 14 patients with squamous cell carcinoma and 1 patient with unclassified non-small cell lung cancer.
METamp is associated with earlier distant metastases in patients with stages I-III NSCLC
We assessed the relationship between MET-CN and disease metastases in patients with stages I-III NSCLC. The 7 patients with METamp tumors had a significantly shorter median time to distant metastasis (11.6 months) than did patients with METneg (43.8 months; P =.004) or METcng (37.0 months) tumors (Table 3 and Figure 2), indicating that METamp is a risk factor for earlier distant metastases in patients with NSCLC.
Figure 2. Kaplan-Meier curves comparing time to distant metastasis using the optimized reporting criteria.
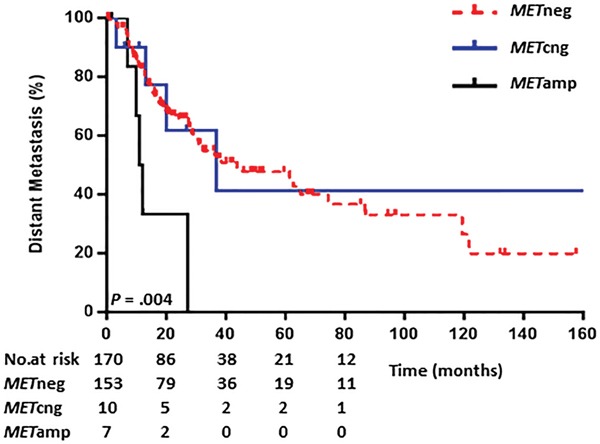
Note: Distant metastasis included bone, brain, liver, adrenal, contralateral lobe, tumor with pleural nodules, or malignant pleural effusion.
METamp is an independent predictor of worse OS in patients with stages I-III NSCLC
Multivariate analyses revealed among patients with stages I-III tumors that patients with adenocarcinoma had longer OS than did patients with other tumor types (hazard ratio [HR], 0.85; 95% confidence interval [CI], 0.30 - 0.90; P =.019). Patients with stage III tumors showed a higher risk for distant metastases (HR, 2.25; 95% CI, 1.24 - 4.09; P =.008) and for shorter OS (HR, 3.33; 95% CI, 1.73–6.42; P <.001) than did patients with stage I disease. Patients with METamp tumors had significantly shorter OS than did patients with METneg tumors (HR, 3.26; 95% CI, 1.15 - 9.23; P =.026). In addition, METamp was found to be highly associated with distant metastases (HR, 4.86; 95% CI, 1.85 - 12.75; P =.001) (Table 3 and Figure 2). However, in patients with stage IV disease, METamp showed no significant impact on outcomes in either the univariate or the multivariate analyses (Table 4).
MET overexpression assessed by IHC is not associated with clinical outcomes
Of 202 patients with MET-IHC data available, the median OS was 24.5 months in patients with MET overexpression versus 36.5 months observed in patients with a normal MET expression (P =.31). We further assessed the potential clinical impact of MET overexpression in patients with different disease stages. In 92 patients with stage I-III disease, the median OS was 51.6 months in those with tumors with normal MET expression (n = 41) versus 54.9 months in patients with tumors with MET overexpression (n = 51) (P =.97). In 110 patients with stage IV disease, the median OS was 16.3 months in those with tumors that showed normal MET expression (n = 37) versus 20.4 months in those patients with tumors with MET overexpression (n = 73) (P =.67) (Supplementary Figure 1). Our data indicate that MET overexpression alone does not impact clinical outcome.
DISCUSSION
In this study, we evaluated data on MET-CN status in 384 NSCLC patients with various disease stages and identified associations between MET-CN and clinical outcome. To resolve the conflicting MET-FISH reporting criteria in the literature, we stratified our patient cohort into 5 groups based on MET-CN and MET/CEP7 ratio and identified associations between these groups and outcomes. We focused initially on patients with stage I-III tumors because METamp has been reported to be associated with stage IV tumors [6]. On the basis of overall survival, we optimized our MET-FISH reporting criteria by integrating MET-CN and MET/CEP7 ratio and reclassifying the patients into 3 groups: METamp, METcng, and METneg. These new, optimized criteria merged groups 4 and 5 (MET-CN ≥ 5 and MET/CEP7 ratio ≥ 2.0) into one group, METamp, because they had similar OS (Supplementary Table 1). These results were consistent with previous reports [6]. Moreover, patients with METcng (MET/CEP7 ratio < 2.0 and MET-CN ≥ 4 to < 5) had markedly better OS than did patients in the other groups, suggesting that patients with METcng constitute a distinct group of NSCLC patients. We also found that patients with METamp had a higher risk of distant metastasis than did patients in the other groups.
According to the MET-FISH reporting criteria we suggest, the overall frequency of METamp in this study cohort was about 8%. We noted that METamp was significantly more common in patients with stage IV disease than in patients without distant metastases. We confirmed that METamp was an independent risk factor for poor OS in patients with early-stage (I-III) NSCLC (28.1 months vs. 134.35 months in METcng and 51.6 months in METneg, P =.015), consistent with results reported previously [6]. Importantly, all patients with METamp and stage I-III disease in this study cohort developed distant metastases within 1 year of diagnosis, indicating that METamp status predicts early distant metastases in patients with stages I-III NSCLC. Although 34 patients in the stage I-III disease group were positive for EGFR mutation, when this subset was compared with those who had no EGFR mutations, there was no significant impact observed on OS or progression to distant metastasis.
Only limited clinical data have been published on MET-CN status and its clinical implications in patients with stage IV NSCLC [3, 6, 16]. In this study, 214 patients with stage IV disease were investigated. We found that METamp was highly associated with stage IV NSCLC, but did not further negatively impact clinical outcomes when compared with the METneg category for patients with the same disease stage. However, we found that patients with stage IV disease and METcng had much longer OS durations than did those with either METneg or METamp (66.2 months vs. 17.5 months in METneg vs. 22.8 months in METamp, P =.053).
The METcng category identified in this cohort likely represents a distinct group of NSCLC patients, similar to that identified by Cappuzzo et al. [6], although that study classified patients with MET-CN ≥ 4 to < 5 as METneg and for that reason did not provide full clinical data for all patients. We speculate that the METcng category may represent patients with multiple polysomies e.g. near-tetrasomies as indicated by the copy numbers. Most of the 17 METcng patients in this cohort, who presumably had near-tetrasomy of chromosome 7, also showed evidence of having near-tetrasomies of chromosomes 2 (by ALK-FISH), 6 (by ROS1-FISH), or 10 (by RET-FISH) (data not shown). METcng coinciding with CN gains of ALK, ROS1, and RET likely resulted from genome-wide polysomies, particularly near-tetraploidy. Similar findings have been reported in a study of 47 patients with EGFR-negative lung cancer, in which 13 patients with polysomy 7 showed improved progression-free survival [25]. To further confirm the FISH findings, we performed OncoScan microarray (Affymetrix) on a subset of patients based on the availability of FFPE tumor blocks including 5 METcng samples. SNP array data showed an overall good concordance with FISH, and the normalized genomic profiles in cases with METcng showed significantly lower genomic copy number complexity comparing the METneg and METamp groups. Although our microarray sample number was small, such METcng cases with a low frequency of somatic copy number alterations (SCNAs) [1, 2] could be clinically less aggressive and likely associated with a better outcome, however, the underline mechanisms remain to be explored and a genome wide assessment of SCNAs in a large study is needed to confirm our findings.
On the basis of these observations, we argue against treating patients who have METcng with MET inhibitors. Several clinical trials have failed to show any positive effect of MET inhibitors on patient survival. We speculate that these studies may have inaccurately identified patients with METcng as having METamp. For example, in one phase II study of 37 MET-positive patients [23], MET positivity was defined as MET-CN ≥ 4 in over 40% of cells; only 3 of the 37 patients with a MET/CEP7 ratio > 2.0 would have been interpreted as having METamp using the criteria we suggest in this study. Similarly, in a phase III study, only 4 of 54 patients had a MET/CEP7 ratio > 2.0 [22]. We speculate that the patients enrolled in these two trials may have had METcng, not “real” METamp, and that this may explain the poorer response among these patients to MET inhibitors. Our results highlight the clinical importance of accurate MET-CN assessment using standardized and optimized reporting criteria in determining eligibility for clinical trials of MET inhibitors.
Cases with a MET-CN at the borderline of METneg and METcng (3.8-3.9) or of METcng and METamp (4.8-4.9) can be challenging for risk stratification, as some of these results could be explained by tumor heterogeneity which are not uncommon in many cancers. However, in such cases, the overall estimated copy numbers i.e. by FISH for the assessment of oncogene amplifications are often underestimated [26]. Using the definition of METamp in this study, the results are in line with what have been reported. In addition, using our suggested reporting criteria, those cases with tumor cells with clustering MET-FISH signals in > 10% cells would be considered to be positive and this can be considered as alternative method to address tumor heterogeneity to at least some degree. To further address tumor heterogeneity, we recommend reflex testing by repeating FISH or reflex testing using alternative assays, such as chromogenic in situ hybridization [27], microarray-based technology [28, 29], comprehensive molecular characterizations or sequencing based single cell analysis, etc. to accurately determine relevant copy numbers in cells with tumor heterogeneity as previously reported [1, 2, 30]. We are aware of that FISH based testing i.e. the MET-FISH cannot assess genome-wide SCNAs that are often associated with treatment resistance or disease progression in NSCLS, and an integrated genomic approach to accurately assess SCNAs could be utilized in the clinical settings in near future [1, 2].
Over half of the patients in the current cohort were also assessed for MET expression status by IHC. Our results show that MET overexpression were not associated with disease stages, although patients with METamp did have the higher number of cases with MET overexpression compared with the METneg and METcng subgroups (Supplementary Table 5). Unlike the ideal concordance observed HER2 amplification and HER2 overexpression reported previously [31] the overall concordance between MET-CN and MET expression observed in this study was low (~ 45%) which was consistent with what has been reported by others [32]. Similar discordance also has been observed in gastric-intestinal cancer studies by tissue microarray (unpublished data). In addition, the clinical impact of MET overexpression has been controversial, with some studies showing negative impact [33] and others showing the opposite results [32]. Our data did not show independent impact by MET overexpression alone assessed by IHC. We propose that standardized IHC reporting criteria is also needed [34] and that more studies should be conducted to determine if MET protein overexpression assessed by IHC alone is more informative than the MET-CN by FISH [33].
In conclusion, we suggest that the optimized MET-FISH reporting criteria that are useful for stratifying risk in patients with all stages and histological types of NSCLC. Our results indicated that METamp is strongly associated with stage IV tumors and that MET-FISH testing can identify patients who are eligible for treatment with MET inhibitors [35]. Furthermore, METamp is an independent predictor of poorer prognosis in patients with stages I-III NSCLC. METamp is also a reliable predictor of distant metastases in patients with early stages NSCLC and, such patients may be candidates for more intensive treatments, such as combination therapy using MET inhibitors after surgery. Finally, we found that METcng could represent an independent prognostic group of patients with NSCLC; however, more data are needed to confirm this observation.
PATIENTS AND METHODS
Patients
We retrospectively reviewed the records of all patients with NSCLC who had been tested for MET using FISH between February 1, 2010 and December 31, 2015 at MD Anderson Cancer Center. All patients diagnosed with NSCLC for whose detailed clinicopathologic data are available were included. Patients with ALK receptor tyrosine kinase (ALK), ROS proto-oncogene 1 receptor tyrosine kinase (ROS1), or ret proto-oncogene (RET) gene rearrangements were excluded from the study. Clinical disease stage for all patients was determined using the National Comprehensive Cancer Network staging system for NSCLC [36]. This study was approved by the Institutional Review Board.
FISH analysis
MET-copy-number (MET-CN) status was assessed using a dual-color FISH probe set (CymoGenDX, Irvine, CA) targeting MET and CEP7on formalin-fixed, paraffin-embedded tissue sections from tumor specimens following established standard laboratory procedures. MET-CN and the number of CEP7 per nucleus were scored in 60 cells and the mean MET/CEP7 ratio was calculated for each specimen. A subset of FFPE tissue specimens from at least 20 normal individuals was included to establish normal cutoff values following laboratory standard procedures (e.g. chromosome 7 aneuploidy such as monosomy or trisomy/tetrasomy/polysomy 7). The initial reporting criteria used at our institution during the study period, which we designate here as “historical” MET-FISH reporting criteria, classified cases into two categories, METamp and MET-negative (METneg), with a cutoff of MET/CEP7 ratio of 2.0. A sample was considered to have METamp if the mean MET/CEP7 ratio was ≥ 2.0 or if the MET/CEP7 ratio was < 2.0 but the MET-CN was ≥ 20 copies/cell or MET signal clusters were seen in more than10% of tumor cells [24].
Immunohistochemistry (IHC) analysis
MET immunohistochemical staining (MET-IHC) was evaluated using a Benchmark Ultra Autostainer (Ventana, Tucson, AZ) with anti-total c-MET (SP44) rabbit monoclonal primary antibody (Ventana, Tucson, AZ), following the manufacturer’s instructions. Staining was scored by determining the percentage of cells showing weak (1+), moderate (2+), or strong (3+) membranous staining. MET overexpression was considered as positive if ≥ 50% of tumor cells showing cellular membrane staining at an intensity of “2+” or “3+” [37]. Scoring was performed independently by two individuals and any discrepant cases were re-evaluated for the final interpretation.
Statistical analysis
Patient and tumor characteristics, including demographics, tumor type, and MET-CN, were summarized using frequencies, percentages, and distributions. Categorical variables were compared using chi-square and Fisher exact tests, and continuous variables were compared using the Student t test. OS was calculated from the date of first diagnosis to the date of last follow-up or death of the patient. The log-rank test and Kaplan-Meier curves were employed to compare OS between subgroups. GraphPad Prism software version 6 (GraphPad Software, La Jolla, CA) was used for the survival analyses. Multivariable analyses, including Cox proportional hazards regression analysis, were performed using SPSS software version 9.3 (SPSS, Inc., Chicago, IL). All tests were 2-sided when appropriate, and differences were considered significant at P <.05.
SUPPLEMENTARY MATERIALS FIGURE AND TABLES
Acknowledgments
The authors wish to thank the technologists in the Clinical Cytogenetics and Molecular Diagnostic Laboratories at MD Anderson Cancer Center for their contributions. Authors also wish to thank the Department of Scientific Publications for their support.
Abbreviations
- NSCLC
Non-small cell lung cancer
- MET
mesenchymal-epithelial transition factor
- MET-CN
MET copy number
- METneg
MET negative
- METcng
MET copy number gain
- METamp
MET amplification
- FISH
fluorescence in situ hybridization
- IHC
immunohistochemistry
- OS
overall survival
- HR
hazard ratio
- CI
confidence interval
Footnotes
Author contributions
XL designed the study. LF, ZT, and CL collected data. LF, HC, HY, LJM, and XL analyzed the data and wrote the paper. SH, LJM, SH, MR, HC, NK, and PL reviewed the pathology data. DH reviewed the clinical data. All authors approved the paper.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
FUNDING
This work was supported in part by the Chapman Foundation and by the National Institutes of Health through Cancer Center Support Grant P30 CA016672 to MD Anderson Cancer Center. The following Cancer Center Support Grant core resources were used: Bioinformatics Shared Resource, Clinical Trials Support Resource. This work was also supported by the Natural Science Foundation of China (No.81603566) and Natural Science Foundation of Jiangsu Province (No. BK20141034).
REFERENCES
- 1.Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355:eaaf8399. doi: 10.1126/science.aaf8399. https://doi.org/10.1126/science.aaf8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TB, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, Salm M, Horswell S, Escudero M, et al. TRACERx Consortium Tracking the Evolution of Non- Small-Cell Lung Cancer. N Engl J Med. 2017;376:2109–21. doi: 10.1056/NEJMoa1616288. https://doi.org/10.1056/NEJMoa1616288 [DOI] [PubMed] [Google Scholar]
- 3.Jin Y, Sun PL, Kim H, Seo AN, Jheon S, Lee CT, Chung JH. MET gene copy number gain is an independent poor prognostic marker in Korean stage I lung adenocarcinomas. Ann Surg Oncol. 2014;21:621–8. doi: 10.1245/s10434-013-3355-1. https://doi.org/10.1245/s10434-013-3355-1 [DOI] [PubMed] [Google Scholar]
- 4.Tanaka A, Sueoka-Aragane N, Nakamura T, Takeda Y, Mitsuoka M, Yamasaki F, Hayashi S, Sueoka E, Kimura S. Co-existence of positive MET FISH status with EGFR mutations signifies poor prognosis in lung adenocarcinoma patients. Lung Cancer. 2012;75:89–94. doi: 10.1016/j.lungcan.2011.06.004. https://doi.org/10.1016/j.lungcan.2011.06.004 [DOI] [PubMed] [Google Scholar]
- 5.Kowalczuk O, Kozlowski M, Niklinska W, Kisluk J, Niklinska BJ, Niklinski J. Increased MET Gene Copy Number but Not mRNA Level Predicts Postoperative Recurrence in Patients with Non-Small Cell Lung Cancer. Transl Oncol. 2014;7:605–12. doi: 10.1016/j.tranon.2014.08.002. https://doi.org/10.1016/j.tranon.2014.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cappuzzo F, Marchetti A, Skokan M, Rossi E, Gajapathy S, Felicioni L, Del Grammastro M, Sciarrotta MG, Buttitta F, Incarbone M, Toschi L, Finocchiaro G, Destro A, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol. 2009;27:1667–74. doi: 10.1200/JCO.2008.19.1635. https://doi.org/10.1200/jco.2008.19.1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma PC, Maulik G, Christensen J, Salgia R. c-Met: structure, functions and potential for therapeutic inhibition. Cancer Metastasis Rev. 2003;22:309–25. doi: 10.1023/a:1023768811842. [DOI] [PubMed] [Google Scholar]
- 8.Gherardi E, Youles ME, Miguel RN, Blundell TL, Iamele L, Gough J, Bandyopadhyay A, Hartmann G, Butler PJ. Functional map and domain structure of MET, the product of the c-met protooncogene and receptor for hepatocyte growth factor/scatter factor. Proc Natl Acad Sci U S A. 2003;100:12039–44. doi: 10.1073/pnas.2034936100. https://doi.org/10.1073/pnas.2034936100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ou SH, Kwak EL, Siwak-Tapp C, Dy J, Bergethon K, Clark JW, Camidge DR, Solomon BJ, Maki RG, Bang YJ, Kim DW, Christensen J, Tan W, et al. Activity of crizotinib (PF02341066), a dual mesenchymal-epithelial transition (MET) and anaplastic lymphoma kinase (ALK) inhibitor, in a non-small cell lung cancer patient with de novo MET amplification. J Thorac Oncol. 2011;6:942–6. doi: 10.1097/JTO.0b013e31821528d3. https://doi.org/10.1097/JTO.0b013e31821528d3 [DOI] [PubMed] [Google Scholar]
- 10.Xia N, An J, Jiang QQ, Li M, Tan J, Hu CP. Analysis of EGFR, EML4-ALK, KRAS, and c-MET mutations in Chinese lung adenocarcinoma patients. Exp Lung Res. 2013;39:328–35. doi: 10.3109/01902148.2013.819535. https://doi.org/10.3109/01902148.2013.819535 [DOI] [PubMed] [Google Scholar]
- 11.Cappuzzo F, Janne PA, Skokan M, Finocchiaro G, Rossi E, Ligorio C, Zucali PA, Terracciano L, Toschi L, Roncalli M, Destro A, Incarbone M, Alloisio M, et al. MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann Oncol. 2009;20:298–304. doi: 10.1093/annonc/mdn635. https://doi.org/10.1093/annonc/mdn635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toschi L, Cappuzzo F. Clinical implications of MET gene copy number in lung cancer. Future Oncol. 2010;6:239–47. doi: 10.2217/fon.09.164. https://doi.org/10.2217/fon.09.164 [DOI] [PubMed] [Google Scholar]
- 13.Park S, Langley E, Sun JM, Lockton S, Ahn JS, Jain A, Park K, Singh S, Kim P, Ahn MJ. Low EGFR/MET ratio is associated with resistance to EGFR inhibitors in non-small cell lung cancer. Oncotarget. 2015;6:30929–38. doi: 10.18632/oncotarget.5131. https://doi.org/10.18632/oncotarget.5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dziadziuszko R, Wynes MW, Singh S, Asuncion BR, Ranger-Moore J, Konopa K, Rzyman W, Szostakiewicz B, Jassem J, Hirsch FR. Correlation between MET gene copy number by silver in situ hybridization and protein expression by immunohistochemistry in non-small cell lung cancer. J Thorac Oncol. 2012;7:340–7. doi: 10.1097/JTO.0b013e318240ca0d. https://doi.org/10.1097/JTO.0b013e318240ca0d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park S, Choi YL, Sung CO, An J, Seo J, Ahn MJ, Ahn JS, Park K, Shin YK, Erkin OC, Song K, Kim J, Shim YM, et al. High MET copy number and MET overexpression: poor outcome in non-small cell lung cancer patients. Histol Histopathol. 2012;27:197–207. doi: 10.14670/HH-27.197. [DOI] [PubMed] [Google Scholar]
- 16.Okuda K, Sasaki H, Yukiue H, Yano M, Fujii Y. Met gene copy number predicts the prognosis for completely resected non-small cell lung cancer. Cancer Sci. 2008;99:2280–5. doi: 10.1111/j.1349-7006.2008.00916.x. https://doi.org/10.1111/j.1349-7006.2008.00916.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noonan SA, Berry L, Lu X, Gao D, Baron AE, Chesnut P, Sheren J, Aisner DL, Merrick D, Doebele RC, Varella-Garcia M, Camidge DR. Identifying the Appropriate FISH Criteria for Defining MET Copy Number-Driven Lung Adenocarcinoma through Oncogene Overlap Analysis. J Thorac Oncol. 2016;11:1293–304. doi: 10.1016/j.jtho.2016.04.033. https://doi.org/10.1016/j.jtho.2016.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutterbach B, Zeng Q, Davis LJ, Hatch H, Hang G, Kohl NE, Gibbs JB, Pan BS. Lung cancer cell lines harboring MET gene amplification are dependent on Met for growth and survival. Cancer Res. 2007;67:2081–8. doi: 10.1158/0008-5472.CAN-06-3495. https://doi.org/10.1158/0008-5472.can-06-3495 [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Yang Y, Ye YC, Shi QF, Chai K, Tashiro S, Onodera S, Ikejima T. Activation of ERK-p53 and ERK-mediated phosphorylation of Bcl-2 are involved in autophagic cell death induced by the c-Met inhibitor SU11274 in human lung cancer A549 cells. J Pharmacol Sci. 2012;118:423–32. doi: 10.1254/jphs.11181fp. [DOI] [PubMed] [Google Scholar]
- 20.Tanizaki J, Okamoto I, Okamoto K, Takezawa K, Kuwata K, Yamaguchi H, Nakagawa K. MET tyrosine kinase inhibitor crizotinib (PF-02341066) shows differential antitumor effects in non-small cell lung cancer according to MET alterations. J Thorac Oncol. 2011;6:1624–31. doi: 10.1097/JTO.0b013e31822591e9. https://doi.org/10.1097/JTO.0b013e31822591e9 [DOI] [PubMed] [Google Scholar]
- 21.Spigel DR, Ervin TJ, Ramlau RA, Daniel DB, Goldschmidt JH, Jr, Blumenschein GR, Jr, Krzakowski MJ, Robinet G, Godbert B, Barlesi F, Govindan R, Patel T, Orlov SV, et al. Randomized phase II trial of Onartuzumab in combination with erlotinib in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2013;31:4105–14. doi: 10.1200/JCO.2012.47.4189. https://doi.org/10.1200/jco.2012.47.4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scagliotti G, von Pawel J, Novello S, Ramlau R, Favaretto A, Barlesi F, Akerley W, Orlov S, Santoro A, Spigel D, Hirsh V, Shepherd FA, Sequist LV, et al. Phase III Multinational, Randomized, Double-Blind, Placebo-Controlled Study of Tivantinib (ARQ 197) Plus Erlotinib Versus Erlotinib Alone in Previously Treated Patients With Locally Advanced or Metastatic Nonsquamous Non-Small-Cell Lung Cancer. J Clin Oncol. 2015;33:2667–74. doi: 10.1200/JCO.2014.60.7317. https://doi.org/10.1200/jco.2014.60.7317 [DOI] [PubMed] [Google Scholar]
- 23.Sequist LV, von Pawel J, Garmey EG, Akerley WL, Brugger W, Ferrari D, Chen Y, Costa DB, Gerber DE, Orlov S, Ramlau R, Arthur S, Gorbachevsky I, et al. Randomized phase II study of erlotinib plus tivantinib versus erlotinib plus placebo in previously treated non-small-cell lung cancer. J Clin Oncol. 2011;29:3307–15. doi: 10.1200/JCO.2010.34.0570. https://doi.org/10.1200/JCO.2010.34.0570 [DOI] [PubMed] [Google Scholar]
- 24.de Melo Gagliato D, Jardim DL, Falchook G, Tang C, Zinner R, Wheler JJ, Janku F, Subbiah V, Piha-Paul SA, Fu S, Hess K, Roy-Chowdhuri S, Moulder S, et al. Analysis of MET genetic aberrations in patients with breast cancer at MD Anderson Phase I unit. Clin Breast Cancer. 2014;14:468–74. doi: 10.1016/j.clbc.2014.06.001. https://doi.org/10.1016/j.clbc.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toffalorio F, de Marinis F, Conforti F, Spitaleri G, Catania C, Noberasco C, Lazzari C, Vecchio F, Stufano V, Barberis M, De Pas T. Erlotinib efficacy in NSCLC patients with high polysomy of chromosome 7 and EGFR/KRas wild-type tumors. J Thorac Oncol. 2015;10:392–6. doi: 10.1097/JTO.0000000000000355. https://doi.org/10.1097/jto.0000000000000355 [DOI] [PubMed] [Google Scholar]
- 26.Radziuviene G, Rasmusson A, Augulis R, Lesciute-Krilaviciene D, Laurinaviciene A, Clim E, Laurinavicius A. Automated Image Analysis of HER2 Fluorescence In Situ Hybridization to Refine Definitions of Genetic Heterogeneity in Breast Cancer Tissue. Biomed Res Int. 2017;2017:2321916. doi: 10.1155/2017/2321916. https://doi.org/10.1155/2017/2321916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metzger ML, Behrens HM, Boger C, Haag J, Kruger S, Rocken C. MET in gastric cancer—discarding a 10% cutoff rule. Histopathology. 2016;68:241–53. doi: 10.1111/his.12745. https://doi.org/10.1111/his.12745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Singh RR, Lu X, Huo L, Yao H, Aldape K, Abraham R, Virani S, Mehrotra M, Mishra BM, Bousamra A, Albarracin C, Wu Y, et al. Genome-wide copy number aberrations and HER2 and FGFR1 alterations in primary breast cancer by molecular inversion probe microarray. Oncotarget. 2017;8:10845–57. doi: 10.18632/oncotarget.14802. https://doi.org/10.18632/oncotarget.14802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luthra R, Chen H, Roy-Chowdhuri S, Singh RR. Next-Generation Sequencing in Clinical Molecular Diagnostics of Cancer: Advantages and Challenges. Cancers (Basel) 2015;7:2023–36. doi: 10.3390/cancers7040874. https://doi.org/10.3390/cancers7040874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yates LR. Intratumoral heterogeneity and subclonal diversification of early breast cancer. Breast. 2017;34:S36–S42. doi: 10.1016/j.breast.2017.06.025. https://doi.org/10.1016/j.breast.2017.06.025 [DOI] [PubMed] [Google Scholar]
- 31.Chao WR, Lee MY, Ruan A, Sheng HP, Hsu JD, Han CP, Koo CL. Assessment of HER2 Status Using Immunohistochemistry (IHC) and Fluorescence In Situ Hybridization (FISH) Techniques in Mucinous Epithelial Ovarian Cancer: A Comprehensive Comparison between ToGA Biopsy Method and ToGA Surgical Specimen Method. PLoS One. 2015;10:e0142135. doi: 10.1371/journal.pone.0142135. https://doi.org/10.1371/journal.pone.0142135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tran TN, Selinger CI, Kohonen-Corish MR, McCaughan B, Kennedy C, O’Toole SA, Cooper WA. Alterations of MET Gene Copy Number and Protein Expression in Primary Non-Small-Cell Lung Cancer and Corresponding Nodal Metastases. Clin Lung Cancer. 2016;17:30-8 e1. doi: 10.1016/j.cllc.2015.08.002. https://doi.org/10.1016/j.cllc.2015.08.002 [DOI] [PubMed] [Google Scholar]
- 33.Song Z, Wang X, Zheng Y, Su H, Zhang Y. MET Gene Amplification and Overexpression in Chinese Non-Small-Cell Lung Cancer Patients Without EGFR Mutations. Clin Lung Cancer. 2017;18:213-9 e2. doi: 10.1016/j.cllc.2016.09.011. https://doi.org/10.1016/j.cllc.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 34.Guo B, Cen H, Tan X, Liu W, Ke Q. Prognostic value of MET gene copy number and protein expression in patients with surgically resected non-small cell lung cancer: a meta-analysis of published literatures. PLoS One. 2014;9:e99399. doi: 10.1371/journal.pone.0099399. https://doi.org/10.1371/journal.pone.0099399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drilon A, Cappuzzo F, Ou SI, Camidge DR. Targeting MET in Lung Cancer: Will Expectations Finally Be MET? J Thorac Oncol. 2017;12:15–26. doi: 10.1016/j.jtho.2016.10.014. https://doi.org/10.1016/j.jtho.2016.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ettinger DS, Wood DE, Akerley W, Bazhenova LA, Borghaei H, Camidge DR, Cheney RT, Chirieac LR, D’Amico TA, Demmy TL, Dilling TJ, Govindan R, Grannis FW, Jr, et al. Non-small cell lung cancer, version 1.2015. J Natl Compr Canc Netw. 2014;12:1738–61. doi: 10.6004/jnccn.2014.0176. [DOI] [PubMed] [Google Scholar]
- 37.Koeppen H, Yu W, Zha J, Pandita A, Penuel E, Rangell L, Raja R, Mohan S, Patel R, Desai R, Fu L, Do A, Parab V, et al. Biomarker analyses from a placebo-controlled phase II study evaluating erlotinib+/−onartuzumab in advanced non-small cell lung cancer: MET expression levels are predictive of patient benefit. Clin Cancer Res. 2014;20:4488–98. doi: 10.1158/1078-0432.CCR-13-1836. https://doi.org/10.1158/1078-0432.ccr-13-1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


