Abstract
Plants are associated with various microorganisms throughout their life, including commensal, symbiotic and pathogenic microorganisms. Pathogens are genetically adapted to aggressively colonize and proliferate in host plants to cause disease. However, disease outbreaks occur only under permissive environmental conditions. The interplay between host, pathogen and environment is famously known as the “disease triangle”. Among the environmental factors, rainfall events, which often create a period of high atmospheric humidity, have repetitively been shown to promote disease outbreaks in plants, suggesting that water availability is crucial for pathogenesis. During pathogen infection, water-soaking spots on the infected leaves are frequently observed as an early symptom of disease. Recent studies showed that pathogenic bacteria dedicate specialized virulence proteins to create an aqueous living space inside the leaf apoplast under high humidity. Water availability in and likely associated other changes to the apoplastic environment can determine the success of potentially pathogenic microbes. These new findings reinforce the notion that the fight over water may be a major battleground between plants and pathogens. In this article, we will discuss the role of water availability on host-microbe interactions, with a focus on plant-bacterial interactions.
Keywords: high humidity, water-soaking, plant disease, plant immunity, stomata
INTRODUCTION
Water is essential for all living organisms. It functions as a solvent, a temperature buffer and a metabolite in living cells. Unlike animals, plants and microorganisms rely largely on their immediate surroundings for water. Land plants obtain water mainly from soil, whereas microbes that live in or on land plants gain water from the plant and/or water vapor from the atmosphere. Water availability is known to greatly impact plant diversity and microbial community structure (Lau and Lennon, 2012; Blazewicz et al., 2014; Jonas et al., 2015; Taketani et al., 2017).
Although plants are often surrounded by a magnitude of various microbes, most microorganisms cannot colonize plants. This is largely attributed to plants having evolved layers of active defense mechanisms that are effective in protecting themselves from most microbes. For example, plants have developed strong physical barriers such as a hydrophobic wall on mature roots, bark on stems and waxy cuticles on leaves to prevent microorganisms from entering plant tissues. At the cellular level, microbes can be detected by plasma-membrane-bound receptors (called pattern recognition receptors PRRs) on the cell surface. Each PRR recognizes a specific pathogen-associated molecular pattern (PAMP), which are broadly conserved in microbes, such as bacterial flagellin or elongation factor Tu (EF-Tu). Recognition of PAMPs triggers an ancient form of plant defense called pattern-triggered immunity (PTI), which halts the proliferation of most nonpathogenic microbes (reviewed in Segonzac and Zipfel, 2011; Macho and Zipfel, 2014; Li et al., 2016).
Over the course of plant-microbe co-evolution, some microorganisms have adapted to colonize and proliferate pathogenically in plants. As a major mechanism of pathogenesis, many pathogens translocate proteinaceous virulence proteins (called effectors) into host cells, targeting different components of pattern-triggered immunity and other forms of plant defense to disarm the plant (reviewed in Jones and Dangl; 2006, Grant et al., 2006; Rafiqi et al., 2012; Buttner and He, 2009). In addition, pathogenic bacteria utilize effectors to create a suitable living environment by redirecting sugar (Chen et al., 2010; Cohn et al., 2014; Cox et al., 2016) and water (Xin et al., 2016; Schwartz et al., 2017) into the extracellular space where many of them live inside the plant.
In addition to PRR-based surveillance system at the cell surface, plants have evolved an intracellular surveillance system. Specifically, intracellular immune receptors (also known as disease resistance (R) proteins) can detect microbial effector proteins inside the plant cell and trigger a second layer of plant defense termed effector-triggered immunity (ETI; reviewed in Chisholm et al., 2006; Jones et al., 2016). Effector-triggered immunity is generally a more robust form of plant defense compared to pattern-triggered immunity, as it is often accompanied by plant cell death known as the hypersensitive response (HR). The HR may help to restrict the proliferation of microbes from the infection sites (reviewed in Khan et al., 2016).
It has been long observed that the interaction between a virulent pathogen and a genetically susceptible host plant does not always lead to disease. For a pathogenic microbe to aggressively proliferate in a host plant, favorable environmental conditions are also required. The triangular interaction between pathogen, plant and environment is known as the “disease triangle” (Stevens 1960). Among the environmental factors that influence disease development, high atmospheric humidity has been repeatedly found to be associated with disease outbreaks (Miller et al., 1996; Pernezny et al., 2005; Schwartz 2011). In this article, we discuss the critical role of high humidity and water on host-microbe interactions. We will start with an overview of water transportation and homeostasis in land plants as a preamble to an in-depth discussion on the effect of water on microbes on the plant surface, microbial pathogenesis inside the plant and the effectiveness of plant defense.
A BRIEF OVERVIEW OF WATER TRANSPORTATION AND HOMEOSTASIS IN LAND PLANTS
Plants have evolved ways to uptake water and maintain water homeostasis to survive and grow. For land plants, soil provides a major water source. Soil pores between and within aggregates function as storage compartments for water. To maximize water uptake, land plants have developed an elaborated network of roots that spreads through the soil to gain access to water. Although all parts of the root system might be involved in absorbing water, root tips and root hairs account for bulk water uptake as those cell types are more permeable to water. Mature regions of the root, on the other hand, have developed specialized tissues named exodermis or hypodermis, which hinder efficient water permeability (Hose et al., 2001).
Upon absorption by the root hairs or epidermal cells, water traffics across the cortex, the endodermis and finally the pericycle before being unloaded into the vasculature for long distance transport in plants. From the root epidermis to the endodermis water moves through three pathways: apoplastic pathway, symplastic pathway and transmembrane pathway (Figure 1b). The apoplastic space includes plant cell walls and extracellular spaces in between the plasma membranes. In the apoplastic pathway, water is absorbed into root tissues through the cell wall of root hairs or epidermal cells and traffics apoplastically without going into cells. In the symplastic pathway, water is absorbed into root hairs or epidermal cells and traffics to the pericycle through the plasmodesmata, which are membrane-lined channels connecting between cells. On the other hand, if water enters and exits from one cell to the other directly through the plasma membrane, the route is known as the transmembrane pathway (Figure 1b; Steudle and Peterson 1998). The apoplastic and transmembrane movement of water are forced into the symplastic pathway at the endodermal cells because they are surrounded by Casparin strip (Figure 1b), which blocks water diffusion. Given the amount of water needed to move from the root to the shoot, water-transporting channel proteins, aquaporins, play an active role in facilitating movement of water across living cells (Kjellbom et al., 1999).
Figure 1. Movement of water from soil to the atmosphere through a plant.
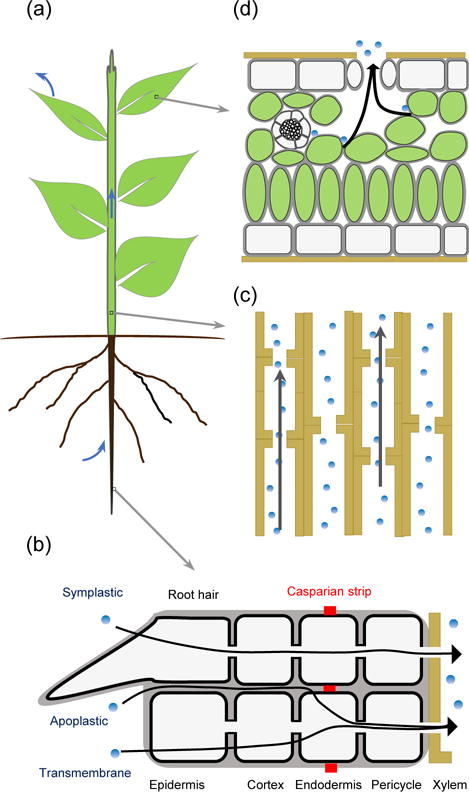
(a) A land plant uptakes water from soil by roots, distributes water through the xylem to other parts of the plant, and transpires water vapor into the atmosphere from the leaves. Root hairs and epidermal cells are mainly responsible for water uptake. Blue arrows indicate the water flow from soil to atmosphere via a plant. (b) Water enters root cells through three distinct pathways: apoplstic, symplastic and transmembrane pathways. All three pathways converge into a symplastic movement at the endodermis. (c) Water is unloaded into the xylem and subjected to long distant transport. (d) In the leaf, water leaves the vascular bundle and is distributed to mesophyll cells and epidermal cells. Water is then drawn into plant cell walls. The water vapor from cell walls move to the atmosphere through stomata during transpiration. Black arrows indicate the direction of the water flow in a plant.
After water traffics through the endodermis and the pericycle, it flows into the xylem by osmosis for long distance transport (Figure 1c). In higher plants, the xylem is mostly made of tracheids. During the xylem maturation process, cells undergo cell wall lignification and programmed cell death, creating cylinder-shaped pipes made of remaining cell walls with pits on the wall. The xylem is composed by vertically overlapped tracheids, whereas pits of the adjacent tracheids often align with each other, creating pit pairs. Pit pairs allow water to move through a low-resistance path between tracheids (Taiz and Zeiger, 2010). After water is transported to leaves, it exits the xylem and is distributed to cells in the leaf (Figure 1d).
Plants have developed a robust system to maintain water homeostasis under various environmental conditions. The upward water flow from root to shoot appeared to be governed by negative pressure (Wheeler and Stroock, 2008). It is generally believed that the base of the root possesses the positive pressure, whereas the leaf retains the negative pressure. The negative pressure in the leaf is created by the mesophyll cell walls, which are composed of hydrophilic materials (e.g., cellulose microfibrills and pectins; Taiz and Zeiger, 2010), and water transpiration through stomata (Landsberg and Waring 2017), which are microscopic pores in leaves involved in the uptake of carbon dioxide (CO2) necessary for photosynthesis. It is estimated that plants retain only around 5% of the water absorbed by the roots, and stomata are responsible for almost 97% plant water loss through the transpiration process (reviewed in Ruggiero et al., 2017). In addition, specialized water pores on the leaf edge, called hydathodes, have a minor role in plant water loss and water homeostasis in plants (Taiz and Zeiger, 2010).
Plant water content is subjected to environmental condition changes. The transpiration rate can be affected by environmental factors, including atmospheric humidity, temperature, light and wind velocity. When atmospheric humidity is low, plants reduce stomatal aperture to prevent excess water loss through transpiration. High atmospheric temperature, high light intensity and high wind velocity increase transpiration rate (Moreshet, 1970). Similarly, low atmospheric humidity, high temperature and strong wind would increase water evaporation from plant surfaces. Thus, water availability to microorganisms that live in or on the plant varies under different environmental conditions.
EFFECT OF WATER ON MICROBES PRIOR TO ENTERING PLANTS
Water is a prerequisite for microorganisms to grow and proliferate; however, most microorganisms do not possess mechanisms to actively uptake water. Instead, they rely on osmotically active substances in the cytoplasm to maintain a positive turgor. The osmotic gradient triggers water flux into the microorganisms in a hypotonic environment (Kempf and Bremer, 1998), where the exterior has higher water potential than the interior of the microorganisms.
Effect of water on rhizosphere microbes
Soil microorganisms are ubiquitous, but they thrive only where water is accessible. Low water content in soil has a profound effect on microbial inhabitants by affecting not only water availability, but also nutrient availability. Thus, water content in soil poses a major selective pressure in shaping soil microbial communities. Recent studies documented that soil relative humidity positively correlates with the richness of soil microbiota (Lau and Lennon, 2012; Blazewicz et al., 2014; Jonas et al., 2015; Taketani et al., 2017). Rhizosphere bacterial communities from semi-arid ecosystems show a drastic increase of bacterial abundance during the wet season, compared to the dry season. In addition, the dry season promoted a much higher population of desiccation-resistant bacteria (e.g., Actinobacteria), some of which can form spores to withstand desiccation stress. On the contrary, the wet season favored the growth of desiccation-sensitive bacteria (e.g., Proteobacteria; Taketani et al., 2017). Similar to rhizosphere bacterial populations, rhizosphere fungal populations are also affected by soil humidity (Lau and Lennon, 2012; Blazewicz et al., 2014).
Water flooding also affects rhizosphere microbes. In a controlled greenhouse experiment, a significant decrease in the overall microbial population was observed after flooding (Unger et al., 2009; Ferrando and Fernandez Scavino, 2015). Flooding might also change the movement and distribution of microbes in soils and damage plant tissues, potentially creating wounds for microbes to enter plants.
Effect of water on phyllosphere microbes
Compared to rhizosphere microorganisms, phyllosphere microbes dwelling on and within above-ground tissues are generally more vulnerable to water stress due to their closer proximity to the atmosphere. Phyllosphere microbes that live on the leaf surface (called epiphytic microbes) rely on trace amounts of water (also nutrients) available on that habitat. Since the vast area of the leaf surface is covered with waxy cuticles, which effectively block water transpiration, nutrient release and gas exchange, epiphytic microbes tend to live where trace amounts of water and nutrients are available. As discussed in the previous section, the majority of plant-associated water moves to the atmosphere through stomata; however, other aqueous pathways in the leaf cuticle have been shown to allow water transpiration (Schonherr, 2006). These minor aqueous pathways are found near the base of trichome and anticlinal cell walls, which are mostly located near the vascular tissues. Interestingly, these aqueous pathways are where epiphytic microbes tend to colonize (Monier and Lindow, 2003, Monier and Lindow, 2004), supporting the notion that microbes aggregate near water (probably also nutrient) sources on the leaf surface. In addition to survival, phyllosphere bacteria also require water for motility on the leaf surface. A positive correlation between leaf surface water abundance and bacterial motility (both swimming and twitching) has been reported (Beattie, 2011). Flagellar motility of Pseudomonas putida, a Gram-negative bacterium, requires liquid films thicker than 1.5 μm (Dechesne et al., 2010), suggesting that flagellum-dependent bacteria are able to move when the thickness of water films is greater than the size of a bacterium. Movement across the leaf surface increases the chance for bacteria to gain access into the leaf interior, where more water and nutrients are available.
Given that high atmospheric humidity might relieve epiphytic microbes from water stress on the leaf surface, it has been shown to shape the phyllosphere microbiome. In a field research, high atmospheric humidity shows a strong positive correlation with the abundance and richness of culturable fungi on the leaf surface (Talley et al., 2002). Under controlled laboratory conditions, high atmospheric humidity is required for the survival of newly infected Pseudomonas syringae bacteria on bean leaves (Monier and Lindow, 2005). Similarly, the population, spore germination and disease outbreak of filamentous pathogens are also influenced by atmospheric moisture and water availability on the leaf surface (Talley et al., 2002; Huber and Gillespie 1992). In particular, dormant fungal spores require water and/or elicitor cues (e.g., components of the cuticle; Serrano et al., 2014) to break dormancy. Upon germination, filamentous pathogens penetrate through the cuticle layer to gain access to stable sources of water and nutrients in the leaf apoplast (van der Does and Rep, 2017).
It has been well documented that heavy precipitation events increase water availability to phyllosphere microbes, which allows them to grow and multiply (Hirano and Upper, 2000). In addition, rainfall may liberate and disperse pathogens from the infected tissues to the surrounding tissues and plants. Wounds created on the leaf due to rainfalls also increase opportunity for the microbes to enter the plant. High atmospheric humidity increases cuticle permeability, which provides more water and nutrients to microbes; it promotes stomatal opening, which allows the microbes to enter the apoplast (Melotto et al., 2017).
Effect of humidity on stomatal defense
Although primarily serving as a portal for gas exchange and transpiration, stomata put plants at risk as foliar pathogens exploit stomata as entry sites to gain access into the apoplast (Melotto et al., 2006). To prevent pathogen invasion through stomata, the cells that make up stomata, guard cells, have evolved to recognize a variety of PAMPs, including flagellin, chitin, chitosan and oligogalacturonic acid (Arnaud and Hwang, 2015). Such recognition triggers downstream signaling events, ultimately resulting in narrowing of the stomatal aperture as a defense mechanism (Melotto et al., 2017).
Some pathogenic microbes have evolved specific virulence factors to actively manipulate the stomatal aperture. Many of these virulence factors, including the fungal toxin fusicoccin, the bacterial toxins coronatine and syringolin A, as well as an increasing list of proteinaceous effectors (e.g. AvrB, HopF2, HopM1, HopX1 and HopZ1), have been shown to promote stomatal opening to facilitate bacterial entry into the plant (reviewed in Melotto et al., 2017). Intriguingly, pathogenic microbes might also directly or indirectly induce stomatal closure at a later stage of infection presumably to maintain leaf apoplastic water potential for sustained multiplication. Consistent with this notion, pathogen-induced stomatal opening appears to be a transient phenomenon (Freeman and Beattie, 2009). It was reported that an effector of P. syringae, HopAM1, plays an important role to promote its virulence activity in water-stressed plants. HopAM1 was found to promote stomatal closure in an abscisic acid (ABA)-dependent manner (Goel et al., 2008). ABA is a plant hormone known to induce stomatal closure (Melotto et al., 2017). Together, the findings suggest that pathogenic bacteria deploy virulence factors to manipulate stomatal movements, allowing bacteria to enter the apoplast at the early infection stage. Once bacteria have entered the plant they induce stomatal closure, presumably to increase water availability for bacterial multiplication inside the leaf.
Although the pathogens actively manipulate stomatal movements using virulence factors, high atmospheric humidity also promotes stomatal opening. Therefore, some phyllosphere microbes that do not have active mechanisms of stomatal opening could potentially take advantage of high atmospheric humidity to gain entry into the plant. Recent studies have begun to shed light on the molecular mechanisms regulating humidity-dependent stomatal movement at the molecular level. High atmospheric humidity triggers the degradation of ABA by upregulating key enzymes involved in ABA catabolism, resulting in stomatal opening (Okamoto et al., 2009). Additionally, a recent study shows that bacteria-triggered stomatal closure is suppressed by high humidity due to early activation of jasmonate (JA) hormone signaling and suppression of salicylic acid (SA) defense signaling pathway within stomatal guard cells (Panchal et al., 2016). The effect of high humidity on JA and SA signaling pathways in the guard cells is observed as early as 15 minutes after the high humidity treatment (Panchal et al., 2016). JA and SA are plant hormones involved in plant defense (Campos et al., 2014; Yan and Dong, 2014). Together, these findings show that high humidity modulates positive and negative regulators of the stomatal aperture that could contribute to the abundance and movements of phyllosphere microbes.
EFFECT OF WATER ON MICROBES AFTER ENTERING PLANTS
Pathogenic bacteria create an aqueous living space in the leaf apoplast under high humidity
In addition to promoting survival and movement of microbes on the leaf surface and invasion of bacteria into the plant interior, a recent study reveals a crucial role of high atmospheric humidity in modulating bacterial population even after bacteria have entered the leaf apoplast (Xin et al., 2016). It has been long known that many bacterial pathogens induce water-soaked spots as an early disease symptom under the high atmospheric humidity condition (Johnson 1937). Water-soaking symptoms are also induced by other types of pathogens, including fungal and oomycete pathogens. For example, the fungal pathogen Magnaporthe oryzae, the causal agent of rice blast disease, induces water-soaked lesions during an early infection phase (Ahn et al., 2005). In 1937, it was reported that an artificially water-soaked apoplast allowed aggressive proliferation of Bacterium angulatum and B. tabacum in tomato, bean, apple, as well as other plants which are not normally susceptible to colonization by these microbes (Johnson 1937). A similar observation was made by Young in 1974. The pathogenic bacterium Pseudomonas phaseolicola as well as non-pathogenic bacteria P. lachrymans and P. syringae could multiply to similar levels in bean leaves when water is supplied to the apoplast during the infection (Yong 1974). These findings suggest that water in the apoplast can fundamentally change the host-microbe interactions (Ramos 2010).
A detailed study of water-soaking symptom development in the Arabidopsis-Pseudomonas syringae pv tomato (Pst) DC3000 pathosystem showed that water-soaking is a transient process that appears during the early infection (~24 hours; Figure 2a) and disappears before the appearance of late disease symptoms, including tissue chlorosis and necrotic lesions. In vivo imaging showed that water-soaked regions are where bacteria aggressively proliferate (Xin et al., 2016). Remarkably, unlike the virulent strain Pst DC3000, an avirulent strain, Pst DC3000 (avrRpt2), which activates effector-triggered immunity, fails to induce water-soaking symptom (Xin et al., 2016). This finding suggests that activation of the plant immune response can block the water-soaking process, possibly as an integral part of the plant defense mechanism against bacterial pathogenesis. In line with this intriguing finding, a previous study showed that virulent and avirulent bacteria experience different water stress levels in the leaf apoplast. Specifically, virulent bacteria Pst DC3000 experience suitable water potentials for pathogen multiplication in the leaf apoplast, whereas the avirulent bacteria Pst DC3000 (avrRpm1) experience a very high level of water stress in the resistant plant that would inhibit bacterial growth in vitro (Wright and Beattie, 2004). Together, these results suggest that activation of effector-triggered immunity may restrict water supply at the infection sites of avirulent bacteria.
Figure 2. Pathogenic bacteria create water-soaking spots on host plants during pathogenesis.
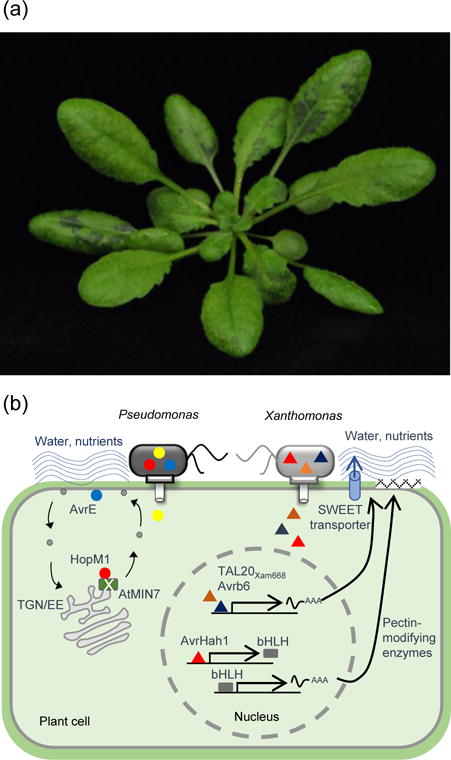
(a) An Arabidopsis plant is infected with a bacterial pathogen, Pst DC3000. The image was taken one day after infection. Dark areas on the leaves indicate water-soaking spots. (b) A model illustrates how pathogenic bacteria create an aqueous environment in the leaf apoplast to support their aggressive growth. Pst DC3000 utilizes two protein effectors, HopM1 and AvrE, to create an aqueous living space in the apoplast. Once inside the plant cell, HopM1 is targeted to the trans-Golgi network/early endosome (TGN/EE) and degrades a plant ARF-family guanine nucleotide exchange factor protein, AtMIN7, involved in vesicle trafficking. AvrE is localized to the plasma membrane. These two effectors likely affect the plant plasma membrane integrity, creating osmotic sinks to draw water (possibly nutrients) into the apoplast. X. gardneri, on the other hand, employs AvrHah1, which is a transcription activator-like (TAL) effector (TALE), to induce water-soaking symptoms in plants. AvrHah1 up-regulates expression of two basic helix-loop-helix (bHLH) transcription factors, which subsequently induce the expression of two genes that encode pectin-modifying enzymes. The actions of pectin-modifying enzymes might change the composition of plant cell walls, affecting the hygroscopicity of the cell walls. In addition, X. axonopodis pv. manihoti and X. citri subsp. malvacearum delivers TALEs TAL20Xam668 and Avrb6, respectively, to up-regulate expression of the sugar transporter genes SWEET in plants. By redirecting the distribution of sugar in their host plants, the pathogenic bacteria might facilitate their nutrition as well as increase osmotic potential in the apoplast, leading to an aqueous apoplast environment in the infected leaves.
How pathogens create an aqueous apoplast environment under high humidity is not clear, but specific pathogen virulence factors are required. In the case of bacteria, several effector proteins have been shown to be involved in developing water-soaking spots in host plants. The activities of effector proteins AvrE and HopM1 from Pst DC3000 as well as WtsE, an AvrE-family effector protein from Pantoea stewartii subsp. stewartii, causes an aqueous apoplast in Arabidopsis (Xin et al., 2016) and maize (Asselin et al., 2015; Ham et al., 2006), respectively. In addition, the Pst DC3000 avrE−hopM1− mutant, which lacks water-soaking inducing effectors, fails to cause an aqueous apoplast during infection (Xin et al., 2016). Interestingly, although AvrE and HopM1 share no amino acid sequence similarity, they are functionally redundant in Pst DC3000 pathogenesis (DebRoy et al., 2004). HopM1 targets and degrades a plant ARF-family guanine nucleotide exchange factor protein, AtMIN7, involved in vesicle trafficking (Nomura et al., 2006). Correspondingly, the Arabidopsis atmin7 mutation, which partially mimics the virulence action of HopM1, promotes spontaneous, albeit limited, water-soaked spots in certain Arabidopsis genotypes under high atmospheric humidity (Xin et al., 2016). These results suggest that AtMIN7 is normally involved in maintaining water homeostasis in the apoplast and that Pst DC3000 uses HopM1 to destroy the AtMIN7 protein as part of its mechanism to change water availability in the apoplast. How HopM1-mediated degradation of AtMIN7 and the molecular actions of AvrE and WtsE lead to an aqueous environment is not yet known. Given the functional role of AtMIN7 in regulating vesicle trafficking and maintaining the plasma membrane (PM) integrity and the PM localization of AvrE and WtsE (and its host target protein phosphatase 2A) in Arabidopsis (Xin et al., 2016; Jin et al., 2016), these bacterial effectors might manipulate the PM integrity and/or phosphorylation of host cells to possibly create osmotic sinks that draw water into the apoplast to benefit the bacteria (Figure 2b).
Xanthomonas gardneri, a bacterial pathogen, can also induce water-soaked disease symptom in tomato. AvrHah1, a transcription activator-like (TAL) effector (TALE) of X. gardneri was recently found to target two basic helix-loop-helix (bHLH) transcription factors in plants. Activation of these transcription factors causes expression of two pectin-modifying genes, a pectate lyase and a pectinesterase. Using designer TALEs (dTALEs), ectopic expression of the two bHLH transcriptions and the pectin lyase, but not the pectinesterase, ultimately lead to a water-soaked leaf apoplast upon bacterial infection. Strikingly, tobacco leaf infected with X. gardneri containing AvrHah1 can draw externally added water from the leaf surface into the apoplast (Schwartz et al., 2017). How the pectin lyase causes water-soaking remains to be elucidated? One possibility is that changes in plant cell wall property might affect the hygroscopicity of the cell wall, causing water accumulation in the leaf apoplast (Figure 2b).
In addition, two Xanthomonas species cause water-soaked disease symptom in their host plants via TALE-mediated induction of plant sugar transporters. X. axonopodis pv. manihotis, a pathogen that causes bacterial blight of cassava, delivers TAL20Xam668 to regulate expression of the sugar transporter gene MeSWEET10a in cassava (Cohn et al., 2014). On the other hand, X. citri subsp. malvacearum (Xcm), the causal agent of bacterial blight of cotton, secretes Avrb6 to induce the expression of the sugar transporter gene GhSWEET10 in cotton (Cox et al., 2017). These two studies indicate that the pathogenic bacteria could redirect the distribution of sugar in their host plants to not only facilitate their nutrition, but also to alter osmotic potential in the apoplast, resulting in an aqueous apoplast environment in the infected leaves. Together, the above-mentioned findings suggest that different pathogens have convergently evolved distinct mechanisms to establish an aqueous living space in the leaf apoplast, impacting different aspects of plant-bacterial interactions.
Pathogen-induced water soaking symptom may impact plant-microbe interactions beyond pathogenesis. Avrb6-induced water-soaking is associated with greatly increased release of Xcm bacteria from infected plant tissues (Yang et al., 1994). In addition, it was recently reported that water-soaking and necrosis in leaves infected by X. euvesicatoria and X. gardneri, can promote colonization of the human pathogen Salmonella enterica inside plant tissues (Potnis et al., 2015). This is interesting in light of the finding by Xin and colleagues (2016) that the levels and abundance of leaf endophytic bacterial communities are altered in plant genotypes that are prone to water-soaking under high humidity. Thus, apoplast water-soaking affects not only pathogenesis, but likely also affects the endogenous microbiome in the leaf apoplast. The observed promotion of human pathogen colonization by apoplast water-soaking illustrates the importance of elucidating the water-soaking mechanisms in understanding the dynamics of foodborne pathogens in plants.
High atmospheric humidity affects R gene-mediated hypersensitive response (HR)
High humidity has been shown to suppress the R gene-mediated hypersensitive response (HR), which involved rapid plant cell death at the site of pathogen infection. It was reported that high atmospheric humidity delays the HR in tomato plants expressing an R gene (Cf-4 or Cf-9) and a matching avirulence gene (Avr4 or Avr9) from a fungal pathogen Cladosporium fulvum (Wang et al., 2005). In addition, several Arabidopsis “autoimmune” mutants, in which immune responses are spontaneously activated without pathogen infections, show humidity-dependent phenotype. Arabidopsis mutants ssi1 and shl1 (Zhou et al., 2004; Noutoshi et al., 2005), caused by a gain-of-function mutation in R genes, cpn1/bon1 and cpr22 (Yoshioka et al., 2006; Mosher et al., 2010), exhibit retarded growth, chlorotic and enhanced disease resistance phenotypes under moderate humidity; however, the phenotypes are suppressed by high atmospheric humidity (>95%; Zhou et al., 2004; Noutoshi et al., 2005; Yoshioka et al., 2006; Mosher et al., 2010). In addition, the autoimmune mutants exhibit elevated defense hormone SA production when the plants are grown under moderate humidity; whereas the SA production is suppressed by high humidity (Zhou et al., 2004; Noutoshi et al., 2005; Mosher et al., 2010). However, as mentioned above, the avirulent bacterium Pst DC3000 (avrRpt2) still can mount effector-triggered immunity and block water-soaking symptom under high humidity condition, even though the macroscopic tissue collapse (i.e., HR) was effectively prevented (Xin et al., 2016). Together, the findings suggest that high humidity may negatively impacts some, but not all ETI-associated immune responses. Understanding how high atmospheric humidity negatively affects the function and activity of R genes and downstream signaling steps has significant practical implications in the deployment of R genes for disease control in the context of changing climate.
Effect of drought on plant resistance
The ongoing changes in climate conditions have significant implications to crop production around the globe. It is predicted that a warmer climate will increase the occurrence of prolonged drought and flooding (Wetherald and Manabe, 2002). Two recent studies showed that drought-stressed Arabidopsis and chickpea plants are more resistant to bacterial pathogens, Pst DC3000 and P. syringae pv. phaseolicola, respectively (Gupta et al., 2016; Sinha et al., 2016). Drought treatment or Pst DC3000 infection each is known to increase the accumulation of ABA in plants (reviewed in Helander et al., 2016; Ton et al., 2009). ABA promotes stomatal closure, which prevents bacteria from entering through stomata (Inoue and Kinoshita, 2017; Eisenach and de Angeli, 2017; Vialet-Chabrand et al., 2017). However, increased ABA levels are associated with enhanced susceptibility to Pst DC3000 in Arabidopsis at the post-invasive stage (de Torres-Zabala et al., 2007). Interestingly, when Arabidopsis plants are challenged with drought and pathogen simultaneously, ABA level remains unchanged, but the levels of defense hormones SA and JA are raised, providing an explanation why the combined stress leads to enhanced disease resistance in Arabidopsis (Gupta et al., 2017). In addition, drought-stressed chickpea exhibits higher resistance to a xylem-inhabiting bacterial pathogen, R. solanacearum (Sinha et al., 2016). In contrast, drought-treated rice becomes more susceptible to M. oryzae infection (Bidzinski et al., 2016). In this pathosystem, drought stress was found to suppress pattern-triggered immunity and effector-triggered immunity (Bidzinski et al., 2016). These contrasting findings suggest that severe water limitation (i.e., drought) might differentially affect plant defense and microbial pathogenesis in different pathosystems. Further study is needed to clarify the underlying causes using different pathosystems, which will shed light on the effect of drought stress on host-pathogen interactions.
Microbial-mediated drought tolerance in plants
Not only does drought alter plant responses to pathogens, drought-adapted microbial communities have been shown to be beneficial for overall plant fitness under drought stress regardless of their historical growth conditions (i.e., dry or wet; Lau and Lennon, 2012). In addition to the effect of the rhizosphere microbiome on plant responses to drought, inoculation of individual bacterial strains has also been shown to improve plant resistance to drought (reviewed in Ngumbi and Kloepper 2016). Plant-growth-promoting rhizobacterium (PGPR), Paenibacillus polymyxa, can protect Arabidopsis against drought stress and upregulate expression of drought-stress response genes (Timmusk and Wagner, 1999). Moreover, several bacterial strains (Bacillus sp., Pseudomonas sp., Acinetobacter sp., Sphingobacterium sp., Enterobacter sp., and Delftia sp.) isolated from drought-treated grapevine rootstocks can improve grapevine resistance to drought by increasing overall plant fitness (Salomon et al., 2014, Rolli et al., 2015). Among these bacteria, Bacillus licheniformis Rt4M10 and Pseudomonas fluorescens Rt6M10 can produce several plant hormones, including ABA, indole-3-acetic acid (IAA), and gibberellin (Salomon et al., 2014). ABA induces stomatal closure to reduce water consumption in plants, improving drought tolerance in plants (reviewed in Helander et al., 2016). Inoculation with the two ABA-producing bacterial strains significantly increases ABA level in the inoculated plants (Salomon et al., 2014), protecting the plants from drought stress. In addition to bacteria, an endophytic fungus, Piriformospora indica, can also improve drought tolerance and induces expression of a suite of drought stress-related genes in Arabidopsis (Sherameti et al., 2008). Together, the findings suggest that prolonged water deficit could drastically alter soil microbial community compositions and some of the drought-enriched microbial strains might be beneficial for their colonized plants in improving drought tolerance and water homeostasis.
Conclusion and outstanding questions
Clearly, water plays a fundamental role in modulating the biology of plants, microbes and their interactions. With changing climate conditions, it is increasingly relevant to understand how water availability and homeostasis in plants and their surrounding habitats impact diverse plant-microbe interactions. Current understanding of this topic is very limited with respect to both the scope and the depth. In the case of plant diseases, it is clear that the changing weather patterns will continue to impact the frequencies of disease outbreaks, as well as the emergence of new diseases and disappearance of some old diseases. There is renewed urgency to deepen our understanding of how changing environmental conditions, including temperature, humidity and microbiota, affect plant-pathogen interactions in diverse ecosystems, as such fundamental knowledge is needed to develop climate-resilient crop plants for future generations. Reductionist experiments in the laboratory have proven (and will continue) to be powerful in revealing some of the underlying mechanisms of plant-microbe interactions. However, the bulk of the current studies has been conducted under artificial environmental conditions that are quite far removed from what plants and microbes experience in nature. In crop fields, simultaneous exposure to biotic and abiotic stresses is likely the rule in a given plant-microbe interaction. As advances in new technologies have enabled the study of plant-microbe interactions at the molecular and cellular levels, it is important to devote substantially more efforts to investigate how plants interact with pathogenic, commensal and symbiotic microbes under conditions that closely simulate their native growth environment.
Significance Statement.
Environmental conditions are major determinants affecting disease outbreaks in field crops. Recent studies have begun to shed light on how pathogenic bacteria utilize virulence proteins to create an aqueous living space inside the leaf apoplast, under high humidity conditions, to aggressively colonize plant leaves.
Acknowledgments
We thank Brad Paasch, Danve Castroverde, YuTi Cheng, and Tanner Neher for critically reading and commenting on the manuscript. This work was supported by grants from the Gordon and Betty Moore Foundation (GBMF3037), the National Institute of General Medical Sciences (GM109928) and the Department of Agriculture – National Institute of Food and Agriculture (2015-67017-23360 and 2017-67017-26180) to S.Y.H., the National Institute of General Medical Sciences (K99GM115766) to K.A., and the National Science Foundation of China (31670273) to Y.J.J.
Footnotes
AUTHOR CONTRIBUTION
KA, YJJ and SYH wrote the paper.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- Ahn IP, Kim S, Kang S, Suh SC, Lee YH. Rice Defense Mechanisms Against Cochliobolus miyabeanus and Magnaporthe grisea Are Distinct. Phytopathol. 2005;95:1248–55. doi: 10.1094/PHYTO-95-1248. [DOI] [PubMed] [Google Scholar]
- Arnaud D, Hwang I. A sophisticated network of signaling pathways regulates stomatal defenses to bacterial pathogens. Mol Plant. 2015;8:566–81. doi: 10.1016/j.molp.2014.10.012. [DOI] [PubMed] [Google Scholar]
- Asselin JE, Lin J, Perez-Quintero AL, Gentzel I, Majerczak D, Opiyo SO, Zhao W, Paek SM, Kim MG, Coplin DL, Blakeslee JJ, Mackey D. Perturbation of maize phenylpropanoid metabolism by an AvrE family type III effector from Pantoea stewartii. Plant Physiol. 2015;167:1117–35. doi: 10.1104/pp.114.253120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie GA. Water relations in the interaction of foliar bacterial pathogens with plants. Annu Rev Phytopathol. 2011;49:533–55. doi: 10.1146/annurev-phyto-073009-114436. [DOI] [PubMed] [Google Scholar]
- Bidzinski P, Ballini E, Ducasse A, Michel C, Zuluaga P, Genga A, Chiozzotto R, Morel JB. Transcriptional Basis of Drought-Induced Susceptibility to the Rice Blast Fungus Magnaporthe oryzae. Front Plant Sci. 2016;7:1558. doi: 10.3389/fpls.2016.01558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazewicz SJ, Schwartz E, Firestone MK. Growth and death of bacteria and fungi underlie rainfall-induced carbon dioxide pulses from seasonally dried soil. Ecol. 2014;95:1162–72. doi: 10.1890/13-1031.1. [DOI] [PubMed] [Google Scholar]
- Buttner D, He SY. Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 2009;150:1656–64. doi: 10.1104/pp.109.139089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos ML, Kang JH, Howe GA. Jasmonate-triggered plant immunity. J Chem Ecol. 2014;40:657–675. doi: 10.1007/s10886-014-0468-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, Chermak D, Antony G, White FF, Somerville SC, Mudgett MB, Frommer WB. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–32. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–14. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Cohn M, Bart RS, Shybut M, Dahlbeck D, Gomez M, Morbitzer R, Hou BH, Frommer WB, Lahaye T, Staskawicz BJ. Xanthomonas axonopodis virulence is promoted by a transcription activator-like effector-mediated induction of a SWEET sugar transporter in cassava. Mol Plant Microbe Interact. 2014;27:1186–98. doi: 10.1094/MPMI-06-14-0161-R. [DOI] [PubMed] [Google Scholar]
- Cox KL, Meng F, Wilkins KE, Li F, Wang P, Booher NJ, Carpenter SCD, Chen L, Zheng H, Gao X, Zheng Y, Fei Z, Yu JZ, Isakeit T, Wheeler T, Frommer WB, He P, Bogdanove AJ, Shan L. TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton. Nature Commun. 2017;8:15588–15601. doi: 10.1038/ncomms15588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debroy S, Thilmony R, Kwack YB, Nomura K, He SY. A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci U S A. 2004;101:9927–32. doi: 10.1073/pnas.0401601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deTorres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez EP, Bogre L, Grant M. Psuedomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007;26:1434–1443. doi: 10.1038/sj.emboj.7601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechesne A, Wang G, Gulez G, Or D, Smets BF. Hydration-controlled bacterial motility and dispersal on surfaces. Proc Natl Acad Sci U S A. 2010;107:14369–72. doi: 10.1073/pnas.1008392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenach C, de Angeli A. Ion transport at the vacuole during stomatal movements. Plant Physiol. 2017;174:520–530. doi: 10.1104/pp.17.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando L, Fernandez Scavino A. Strong shift in the diazotrophic endophytic bacterial community inhabiting rice (Oryza sativa) plants after flooding. FEMS Microbiol Ecol. 2015;91:fiv104. doi: 10.1093/femsec/fiv104. [DOI] [PubMed] [Google Scholar]
- Goel AK, Lundberg D, Torres MA, Matthews R, Akimoto-Tomiyama C, Farmer L, Dangl JL, Grant SR. The Pseudomonas syringae type III effector HopAM1 enhances virulence on water-stressed plants. Mol Plant Microbe Interact. 2008;21:361–70. doi: 10.1094/MPMI-21-3-0361. [DOI] [PubMed] [Google Scholar]
- Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. Annu Rev Microbiol. 2006;60:425–49. doi: 10.1146/annurev.micro.60.080805.142251. [DOI] [PubMed] [Google Scholar]
- Gupta A, Dixit SK, Senthil-Kumar M. Drought Stress Predominantly Endures Arabidopsis thaliana to Pseudomonas syringae Infection. Front Plant Sci. 2016;7:808. doi: 10.3389/fpls.2016.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Hisano H, Hojo Y, Matsuura T, Ikeda Y, Mori IC, Senthil-Kumar Global profiling of phytohormone dynamics during combined drought and pathogen stress in Arabidopsis thaliana reveals ABA and JA as major regulators. Sci Rep. 2017;7:4017–4029. doi: 10.1038/s41598-017-03907-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham JH, Majerczak DR, Arroyo-Rodriguez AS, Mackey DM, Coplin DL. WtsE, an AvrE-family effector protein from Pantoea stewartii subsp. stewartii, causes disease-associated cell death in corn and requires a chaperone protein for stability. Mol Plant Microbe Interact. 2006;19:1092–102. doi: 10.1094/MPMI-19-1092. [DOI] [PubMed] [Google Scholar]
- Helander JD, Vaidya AS, Cutler SR. Chemical manipulation of plant water use. Bioorg Med Chem. 2016;24:493–500. doi: 10.1016/j.bmc.2015.11.010. [DOI] [PubMed] [Google Scholar]
- Hirano SS, Upper CD. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae-a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev. 2000;64:624–53. doi: 10.1128/mmbr.64.3.624-653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hose E, Clarkson DT, Steudle E, Schreiber L, Hartung W. The exodermis: a variable apoplastic barrier. J Exp Bot. 2001;52:2245–64. doi: 10.1093/jexbot/52.365.2245. [DOI] [PubMed] [Google Scholar]
- Huber L, Gillespie TJ. Modeling leaf wetness in relation to plant disease epidemiology. Annu Rev Phytopathol. 1992;30:553–77. [Google Scholar]
- Inoue S, Kinoshita T. Blue light regulation of stomatal opening and the plasma membrane H+-ATPase. Plant Physiol. 2017;174:531–538. doi: 10.1104/pp.17.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Ham JH, Hage R, Zho W, Soto-Hernandez J, Lee SY, Paek SM, Kim MG, Boone C, Coplin DL, Mackey D. Direct and Indirect Targeting of PP2A by Conserved Bacterial Type-III Effector Proteins. PLoS Pathog. 2016;12:e1005609. doi: 10.1371/journal.ppat.1005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas JL, Buhl DA, Symstad AJ. Impacts of weather on long-term patterns of plant richness and diversity vary with location and management. Ecol. 2015;96:2417–32. doi: 10.1890/14-1989.1. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–9. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Jones JD, Vance RE, Dangl JL. Intracellular innate immune surveillance devices in plants and animals. Science. 2016;354:aaf6395. doi: 10.1126/science.aaf6395. [DOI] [PubMed] [Google Scholar]
- Kempf B, Bremer E. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch Microbiol. 1998;170:319–30. doi: 10.1007/s002030050649. [DOI] [PubMed] [Google Scholar]
- Khan M, Subramaniam R, Desveaux D. Of guards, decoys, baits and traps: pathogen perception in plants by type III effector sensors. Curr Opin Microbiol. 2016;29:49–55. doi: 10.1016/j.mib.2015.10.006. [DOI] [PubMed] [Google Scholar]
- Kjellbom P, Larsson C, Johansson II, Karlsson M, Johanson U. Aquaporins and water homeostasis in plants. Trends Plant Sci. 1999;4:308–314. doi: 10.1016/s1360-1385(99)01438-7. [DOI] [PubMed] [Google Scholar]
- Landsberg J, Waring R. Water relations in tree physiology: where to from here? Tree Physiol. 2017;37:18–32. doi: 10.1093/treephys/tpw102. [DOI] [PubMed] [Google Scholar]
- Lau JA, Lennon JT. Rapid responses of soil microorganisms improve plant fitness in novel environments. Proc Natl Acad Sci U S A. 2012;109:14058–62. doi: 10.1073/pnas.1202319109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Meng X, Shan L, He P. Transcriptional Regulation of Pattern-Triggered Immunity in Plants. Cell Host Microbe. 2016;19:641–50. doi: 10.1016/j.chom.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Zipfel C. Plant PRRs and the Activation of Innate Immune Signaling. Mol Cell. 2014;54:263–272. doi: 10.1016/j.molcel.2014.03.028. [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–80. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Melotto M, Zhang L, Oblessuc PR, He SY. Stomatal Defense a Decade Later. Plant Physiol. 2017;174:561–571. doi: 10.1104/pp.16.01853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Rowe R, Riedel R. Bacterial spot, speck, and canker of tomatoes. Ohio State University Extension Fact Sheet. 1996 HYG-3120-96. [Google Scholar]
- Monier JM, Lindow SE. Pseudomonas syringae Responds to the Environment on Leaves by Cell Size Reduction. Phytopathol. 2003;93:1209–16. doi: 10.1094/PHYTO.2003.93.10.1209. [DOI] [PubMed] [Google Scholar]
- Monier JM, Lindow SE. Frequency, size, and localization of bacterial aggregates on bean leaf surfaces. Appl Environ Microbiol. 2004;70:346–55. doi: 10.1128/AEM.70.1.346-355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier JM, Lindow SE. Spatial organization of dual-species bacterial aggregates on leaf surfaces. Appl Environ Microbiol. 2005;71:5484–93. doi: 10.1128/AEM.71.9.5484-5493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreshet S. Effect of environmental factors on cuticular transpiration resistance. Plant Physiol. 1970;46:815–8. doi: 10.1104/pp.46.6.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher S, Moeder W, Nishimura N, Jikumaru Y, Joo SH, Urquhart W, Klessig DF, Kim SK, Nambara E, Yoshioka K. The lesion-mimic mutant cpr22 shows alterations in abscisic acid signaling and abscisic acid insensitivity in a salicylic acid-dependent manner. Plant Physiol. 2010;152:1901–13. doi: 10.1104/pp.109.152603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngumbi E, Kloepper J. Bacterial-mediated drought tolerance: Current and future prospects. Appl Soil Ecol. 2016;105:109–125. [Google Scholar]
- Nomura K, Debroy S, Lee YH, Pumplin N, Jones J, He SY. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–3. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- Noutoshi Y, Ito T, Seki M, Nakashita H, Yoshida S, Marco Y, Shirasu K, Shinozaki K. A single amino acid insertion in the WRKY domain of the Arabidopsis TIR-NBS-LRR-WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J. 2005;43:873–88. doi: 10.1111/j.1365-313X.2005.02500.x. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Tanaka Y, Abrams SR, Kamiya Y, Seki M, Nambara E. High humidity induces abscisic acid 8′-hydroxylase in stomata and vasculature to regulate local and systemic abscisic acid responses in Arabidopsis. Plant Physiol. 2009;149:825–34. doi: 10.1104/pp.108.130823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal S, Chitrakar R, Thompson BK, Obulareddy N, Roy D, Hambright WS, Melotto M. Regulation of Stomatal Defense by Air Relative Humidity. Plant Physiol. 2016;172:2021–2032. doi: 10.1104/pp.16.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernezny K, Zhang S. Bacterial speck of tomato. University of Florida IFAS Extension; 2005. p. 10. [Google Scholar]
- Potnis N, Colee J, Jones JB, Barak JD. Plant pathogen-induced water-soaking promotes Salmonella enterica growth on tomato leaves. Appl Environ Microbiol. 2015;81:8126–34. doi: 10.1128/AEM.01926-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiqi M, Ellis JG, Ludowici VA, Hardham AR, Dodds PN. Challenges and progress towards understanding the role of effectors in plant-fungal interactions. Curr Opin Plant Biol. 2012;15:477–82. doi: 10.1016/j.pbi.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Ramos ME. Diss UC-Berkeley. 2010. Bacterial growth in the plant apoplast is limited by nutrient availability. [Google Scholar]
- Rolli E, Marasco R, Vigani G, Ettoumi B, Mapelli F, Deangelis ML, Gandolfi C, Casati E, Previtali F, Gerbino R, Pierotti Cei F, Borin S, Sorlini C, Zocchi G, Daffonchio D. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ Microbiol. 2015;17:316–31. doi: 10.1111/1462-2920.12439. [DOI] [PubMed] [Google Scholar]
- Ruggiero A, Punzo P, Landi S, Costa A, Van Oosten MM, Grillo S. Improving plant water use efficiency through molecular genetics. Horticulture. 2017;3:31. doi: 10.3390/horticulturae3020031. [DOI] [Google Scholar]
- Salomon MV, Bottini R, De Souza Filho GA, Cohen AC, Moreno D, Gil M, Piccoli P. Bacteria isolated from roots and rhizosphere of Vitis vinifera retard water losses, induce abscisic acid accumulation and synthesis of defense-related terpenes in in vitro cultured grapevine. Physiol Plant. 2014;151:359–74. doi: 10.1111/ppl.12117. [DOI] [PubMed] [Google Scholar]
- Schonherr J. Characterization of aqueous pores in plant cuticles and permeation of ionic solutes. J Exp Bot. 2006;57:2471–91. doi: 10.1093/jxb/erj217. [DOI] [PubMed] [Google Scholar]
- Schwartz HF. Bacterial diseases of beans. Colorado State University Extension Fact Sheet No: 2.913 2011 [Google Scholar]
- Schwartz AR, Morbitzer R, Lahaye T, Staskawicz BJ. TALE-induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. Proc Natl Acad Sci U S A. 2017;114:E897–E903. doi: 10.1073/pnas.1620407114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C, Zipfel C. Activation of plant pattern-recognition receptors by bacteria. Curr Opin Microbiol. 2011;14:54–61. doi: 10.1016/j.mib.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Serrano M, Coluccia F, Torres M, L’haridon F, Metraux JP. The cuticle and plant defense to pathogens. Front Plant Sci. 2014;5:274. doi: 10.3389/fpls.2014.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherameti I, Tripathi S, Varma A, Oelmuller R. The root-colonizing endophyte Pirifomospora indica confers drought tolerance in Arabidopsis by stimulating the expression of drought stress-related genes in leaves. Mol Plant Microbe Interact. 2008;21:799–807. doi: 10.1094/MPMI-21-6-0799. [DOI] [PubMed] [Google Scholar]
- Sinha R, Gupta A, Senthil-Kumar M. Understanding the Impact of Drought on Foliar and Xylem Invading Bacterial Pathogen Stress in Chickpea. Front Plant Sci. 2016;7:902. doi: 10.3389/fpls.2016.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steudle E, Peterson CA. How does water get through roots. J Exp Bot. 1998;49:775–788. [Google Scholar]
- Stevens RB. Plant Pathology, an Advanced Treatise. 1960;3:357–429. [Google Scholar]
- Taketani RG, Lanconi MD, Kavamura VN, Durrer A, Andreote FD, Melo IS. Dry Season Constrains Bacterial Phylogenetic Diversity in a Semi-Arid Rhizosphere System. Microb Ecol. 2017;73:153–161. doi: 10.1007/s00248-016-0835-4. [DOI] [PubMed] [Google Scholar]
- Taiz L, Zeiger E. Plant Physiology. 5th. Sinauer Associates, Inc; Sunderland: 2010. Water balance of plants; pp. 85–105. [Google Scholar]
- Talley SM, Coley PD, Kursar TA. The effects of weather on fungal abundance and richness among 25 communities in the Intermountain West. BMC Ecol. 2002;2:7. doi: 10.1186/1472-6785-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk S, Wagner EG. The plant-growth-promoting rhizobacterium Paenibacillus polymyxa induces changes in Arabidopsis thaliana gene expression: a possible connection between biotic and abiotic stress responses. Mol Plant Microbe Interact. 1999;12:951–9. doi: 10.1094/MPMI.1999.12.11.951. [DOI] [PubMed] [Google Scholar]
- Ton J, Flors V, Mauch-Mani B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009;14:310–317. doi: 10.1016/j.tplants.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Van Der Does HC, Rep M. Adaptation to the Host Environment by Plant-Pathogenic Fungi. Annu Rev Phytopathol. 2017;55:427–450. doi: 10.1146/annurev-phyto-080516-035551. [DOI] [PubMed] [Google Scholar]
- Vialet-Chabrand S, Hills A, Wang Y, Griffiths H, Lew VL, Lawson T, Blatt MR, Rogers S. Global sensitivity analysis of OnGurad models identifies key hubs for transport interaction in stomatal dynamics. Plant Physiol. 2017;174:680–688. doi: 10.1104/pp.17.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Cai X, Zheng Z. High humidity represses Cf-4/Avr4- and Cf-9/Avr9-dependent hypersensitive cell death and defense gene expression. Planta. 2005;222:947–56. doi: 10.1007/s00425-005-0036-8. [DOI] [PubMed] [Google Scholar]
- Wetherald RT, Manabe S. Simulation of hydrologic changes associated with global warming. J Geophys Res. 2002;107(D19):4379. doi: 10.1029/2001JD001195. [DOI] [Google Scholar]
- Wheeler TD, Stroock AD. The transpiration of water at negative pressures in a synthetic tree. Nature. 2008;455:208–12. doi: 10.1038/nature07226. [DOI] [PubMed] [Google Scholar]
- Wright CA, Beattie GA. Pseudomonas syringae pv. tomato cells encounter inhibitory levels of water stress during the hypersensitive response of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101:3269–74. doi: 10.1073/pnas.0400461101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin XF, Nomura K, Aung K, Velasquez AC, Yao J, Boutrot F, Chang JH, Zipfel C, He SY. Bacteria establish an aqueous living space in plants crucial for virulence. Nature. 2016;539:524–529. doi: 10.1038/nature20166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Feyter RD, Gabriel DW. Host-specific symptoms and increased release of Xanthomonas citri and X. campestris pv. malvacearum from leaves are determined by the 102-bp tandem repeats of pthA and avrb6, respectively. Mol Plant Microbe Interact. 1994;3:345–355. [Google Scholar]
- Yang S, Dong X. Perception of the plant immune signal salicylic acid. Curr Opin Plant Biol. 2014;20:64–68. doi: 10.1016/j.pbi.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K, Moeder W, Kang HG, Kachroo P, Masmoudi K, Berkowitz G, Klessig DF. The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. Plant Cell. 2006;18:747–63. doi: 10.1105/tpc.105.038786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM. Effect of water on bacterial multiplication in plant tissue. New Zealand J Agri Res. 1974;17(1):115–119. doi: 10.1080/00288233.1974.10421089. [DOI] [Google Scholar]
- Zhou F, Menke FL, Yoshioka K, Moder W, Shirano Y, Klessig DF. High humidity suppresses ssi4-mediated cell death and disease resistance upstream of MAP kinase activation, H2O2 production and defense gene expression. Plant J. 2004;39:920–32. doi: 10.1111/j.1365-313X.2004.02180.x. [DOI] [PubMed] [Google Scholar]


