Abstract
FOXQ1 is a member of the forkhead-box transcription factor family that has important functions in development, cancer, aging, and many cellular processes. The role of FOXQ1 in cancer biology has raised intense interest, yet much remains poorly understood. We investigated the possible function of the two zebrafish orthologs (foxq1a and foxq1b) of human FOXQ1 in innate immune cell development and function. We employed CRISPR-Cas9 targeted mutagenesis to create null mutations of foxq1a and foxq1b in zebrafish. Using a combination of molecular, cellular, and embryological approaches, we characterized single and double foxq1a bcz11 and foxq1b bcz18 mutants. This study provides the first genetic mutant analyses of zebrafish foxq1a and foxq1b. Interestingly, we found that foxq1a, but not foxq1b, was transcriptionally regulated during a bacterial response, while the expression of foxq1a was detected in sorted macrophages and upregulated in foxq1a-deficient mutants. However, the transcriptional response to E. coli challenge of foxq1a and foxq1b mutants was not significantly different from that of their wildtype control siblings. Our data shows that foxq1a may have a role in modulating bacterial response, while both foxq1a and foxq1b are not required for the development of macrophages, neutrophils, and microglia. Considering the implicated role of FOXQ1 in a vast number of cancers and biological processes, the foxq1a and foxq1b null mutants from this study provide useful genetic models to further investigate FOXQ1 functions.
Introduction
FOXQ1 is a member of the forkhead-box (FOX) transcription factor gene family, which contains a winged helix DNA binding domain and has important functions in development, cancer, aging, cell cycle regulation, cell migration, and other diverse cellular processes [1–5]. Previous studies in mammals show that the functions of FOXQ1 include promoting epithelial differentiation [6–11], inhibiting smooth muscle differentiation [12], mediating hair development [13,14], controlling gastric acid production and secretion in stomach mucous cells [15,16], regulating glucose metabolism [17], as well as possible immunological functions [18,19]. Additionally, upregulation of FOXQ1 is found in a large host of cancer types, possibly to regulate cell proliferation and invasion, including breast [7,20,21], colorectal [22–25], pancreatic [26,27], gastric [28–31], bladder [32], liver [33–36], lung [37,38], ovarian [39,40], and glioma [41]. With these findings, FOXQ1 has generated much interest into its functions and mechanisms.
In light of the interactions of innate immune cells, particularly macrophages, in tumor development, progression, and metastasis [42–48], and possible roles of FOXQ1 in regulating these processes [49,50], we generated gene knockouts of the two zebrafish orthologs (foxq1a and foxq1b) of human FOXQ1 using CRIPSR-Cas9 targeted mutagenesis. Using single and double foxq1a and foxq1b mutants, we investigated whether FOXQ1 is critical for the development and function of macrophages and other innate immune cells. Establishing a function for FOXQ1 in innate immune regulation can provide new understanding of its role in various cancers.
This study provides the first genetic mutant analyses of zebrafish foxq1a and foxq1b. We found RNA expression of both copies of foxq1 in the developing craniofacial structures as previously reported [51,52], in addition to foxq1a expression in sorted macrophages as well as in the yolk region. Neither single nor double foxq1a and foxq1b mutants had overt gross morphological defects or notable deficiency in the development of macrophages, neutrophils, and microglia. Furthermore, foxq1a and foxq1b mutants exhibited a typical transcriptional response to bacterial challenge, but foxq1a is interestingly downregulated during this response in wildtype. Considering the implicated role of FOXQ1 in a vast number of cancers and biological processes, the foxq1a and foxq1b mutants generated in this study provide new genetic models to further dissect the functions of FOXQ1.
Results
Generation of foxq1a bcz11 and foxq1b bcz18 null alleles using CRISPR-Cas9
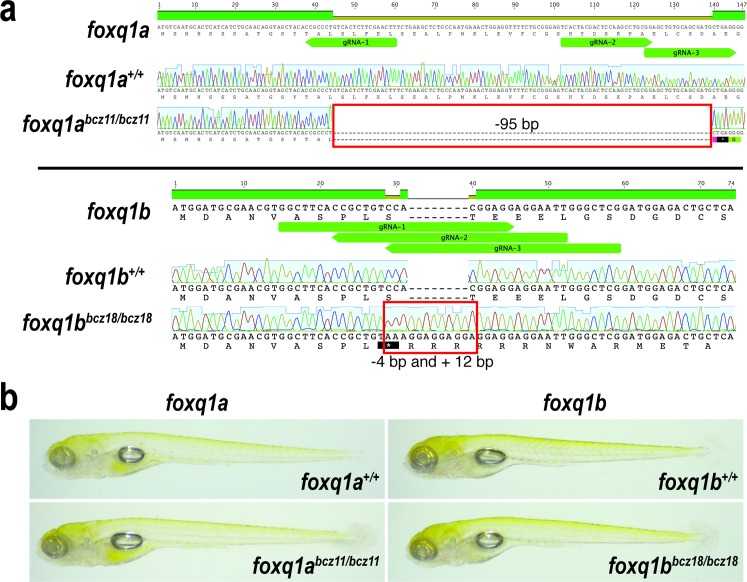
We employed the highly effective CRISPR-Cas9 mediated targeted mutagenesis in zebrafish as previously described [53,54] to create loss-of-function mutations at the beginning of foxq1a and foxq1b genes. We isolated two mutant alleles of interest: foxq1abcz11 and foxq1bbcz18 (Fig 1A). These are frameshift mutations that lead to a premature stop codon at the beginning of each respective gene: bcz11 has a large 95 base pair (bp) deletion in foxq1a that is immediately followed by a premature stop codon, and bcz18 has a 4 bp deletion and a 12 bp insertion in foxq1b that causes a premature stop codon within the start of the indel (Fig 1A). RT-PCR and sequencing analysis confirmed expression of the expected nonsense transcripts from the bcz11 and bcz18 mutations, and no alternative splicing was found (S1 Fig). The data indicate that bcz11 and bcz18 represent null alleles of their respective genes by producing nonsense mRNAs that would prevent Foxq1a and Foxq1b protein expression, respectively. For each gene target, three guide RNAs (gRNAs) were used in combination to increase the efficiency of the mutagenesis and the probability of generating a large indel mutation (Fig 1A). At 5 days post-fertilization (dpf) when zebrafish have well established organ systems and are free-swimming larvae, we examined the gross morphology of the homozygous mutants (Fig 1B). We found that both foxq1abcz11 and foxq1bbcz18 homozygous mutants were indistinguishable from wildtype siblings (Fig 1B), displaying normal-sized inflated swim bladders, and grossly normal head and body structures that were no different from wildtype. However, we cannot exclude the possibility that craniofacial and specific organ phenotypes may be present in the mutants without further detailed analysis, in regions that normally express foxq1a and foxq1b [51,52]. The viability of bcz11 and bcz18 mutants, at least up to larval stages, appeared to be normal, as we consistently found ~25% of the progenies to be mutants from a heterozygous incross.
Fig 1. Sequence analysis of foxq1abcz11 and foxq1bbcz18 mutations generated by CRISPR-Cas9 targeting.
a Top, DNA and amino acid sequences of the 5’ end of foxq1a coding region. DNA sequencing chromatograms show the expected foxq1a sequence in the wildtype sibling, and a large 95 base pair deletion (red box) in the homozygous bcz11 mutant, which causes a premature stop (black box with an asterisk) at the beginning of the gene. Bottom, DNA and amino acid sequences of the 5’ end of foxq1b coding region. DNA chromatograms show wildtype sequence in foxq1b wildtype sibling, while the homozygous bcz18 mutant carries a 4 bp deletion and 12 bp insertion (red box) causing a nonsense mutation at the beginning of the gene. gRNA, target sites of the guide RNAs used. b Whole mount live imaging of 5 dpf larvae show normal gross morphological development of foxq1abcz11 and foxq1bbcz18 mutants as compared to wildtype siblings (at least 10 animals were analyzed per genotype group).
Characterization of foxq1a and foxq1b gene expressions
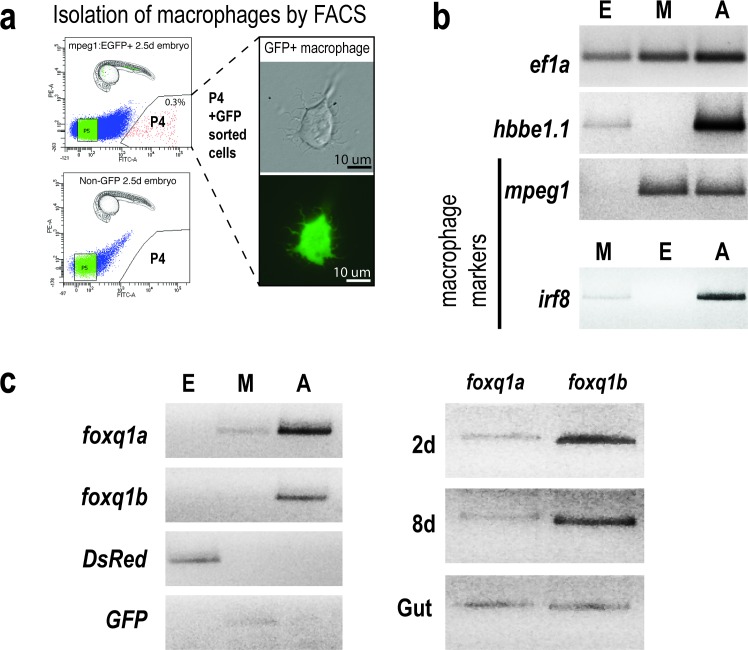
The expression patterns of foxq1a and foxq1b in zebrafish remain largely unanalyzed. Only two studies to date have examined their expressions in specific tissues of interest; one showed foxq1b expression in the developing jaws at 2 dpf [51] and the other reported that foxq1a may be expressed in the cranial suture in juvenile zebrafish (6 weeks post-fertilization) [52]. To better define the possible functions of these genes in zebrafish, we expanded the characterization of their expressions to various stages of development from 1 to 8 dpf, and tested whether they were expressed in macrophages (Fig 2). Using fluorescence-activated cell sorting (FACS), we isolated macrophages (M) based on GFP expression from 2.5 dpf zebrafish embryos carrying the macrophage-specific transgene mpeg1:EGFP (Fig 2A). To assess their expression in other cell types as additional references, we sorted erythrocytes (E) based on DsRed expression from erythrocyte-specific gata1:DsRed transgenic embryos as well as the remaining non-fluorescent cells (A) (Fig 2B and 2C). We verified the identity of the sorted cell populations by RT-PCR analysis using known markers and fluorescent reporter genes for the E, M, and A cells (Fig 2B and 2C). The data shows that while both genes are expressed from 2 to 8 dpf (Fig 2C), only foxq1a is expressed by macrophages and neither gene is expressed by erythrocytes (Fig 2C). Using RNA in situ hybridization, we found expression of foxq1a in the cranial region at 1 dpf that persists in the jaw at 2.5 dpf when it is also found in the body tissue along the yolk, and on the yolk (S2 Fig). By contrast, foxq1b was primarily found to be expressed in the ventral jaw region at 2.5 dpf (S2 Fig). While foxq1a RNA expression was found in macrophages isolated from wildtype embryos (Fig 2C), its level may be too low to be detectable by whole mount RNA in situ hybridization as we were unable to visualize its expression in macrophages by in situ (S2 Fig). Because FOXQ1 is known to be highly expressed in the mammalian gastrointestinal tract [15,16,55,56], we examined whether foxq1a and foxq1b are expressed in the adult zebrafish gut, and indeed found them both to be expressed (Fig 2C).
Fig 2. Gene expression analysis of foxq1a and foxq1b.
a Isolation of macrophages based on GFP expression by FACS in mpeg1:EGFP transgenic zebrafish embryos at 2.5 dpf. Non-fluorescent embryos were used as a negative control for gating. Top left, P4 shows the cell fraction sorted as GFP+ macrophages, also shown in the right panels in brightfield and green channel. b RT-PCR analysis of gene markers validated the different cell populations sorted by FACS, as denoted by E, erythrocytes; M, macrophages; A, all remaining non-fluorescent cells. The following genes were used: translation elongation factor 1 (ef1a) as a reference marker, hemoglobin beta embryonic-1.1 (hbbe1.1) as an erythrocyte marker, and macrophage expressed gene 1 (mpeg1) and interferon regulatory factor 8 (irf8) as well-established macrophage markers in zebrafish. As expected, the ‘A’ cells expressed all genes, while erythrocytes ‘E’ cells expressed hbbe1.1, ef1a, and DsRed genes and macrophage ‘M’ cells expressed macrophage markers, GFP, and ef1a. c Using the sorted cell populations, we found expression of foxq1a in macrophages and neither gene in erythrocytes. Both genes are expressed at 2 to 8 dpf of development as well as in the adult gut tissue.
Single and double foxq1abcz11 and foxq1bbcz18 mutants show normal macrophage, neutrophil, and microglia development
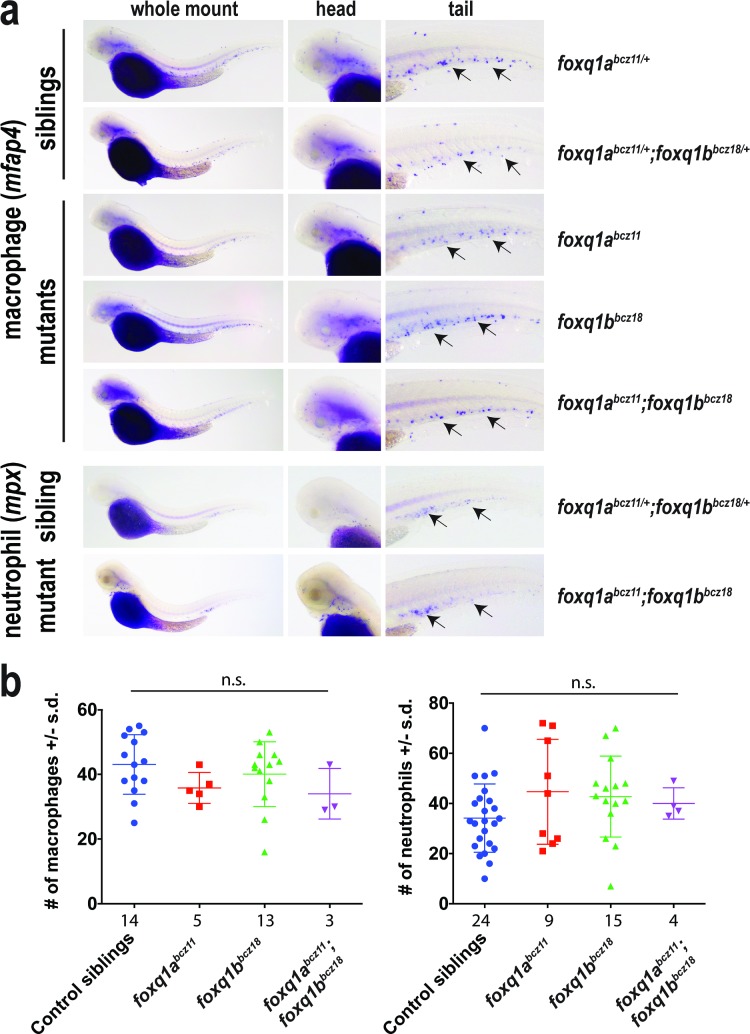
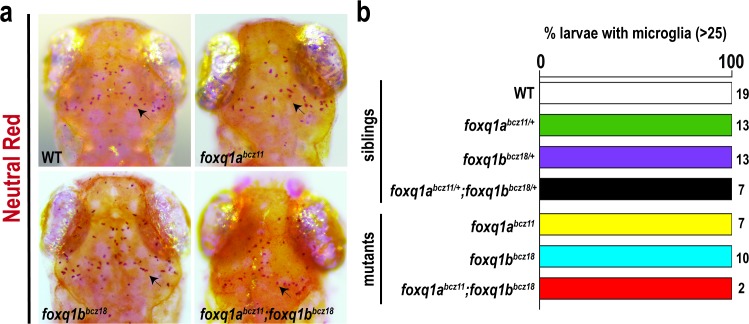
In light of the finding that foxq1a is expressed in macrophages and the implicated mammalian FOXQ1 function in tumor biology that may involve innate immune cells [19], we asked whether the development of macrophages and neutrophils may be altered in the zebrafish foxq1 mutants. By whole mount RNA in situ hybridization using a macrophage marker (mfap4) [57,58] and neutrophil marker (mpx) [59,60], we found no apparent difference in formation or distribution of these innate immune cells during development at 2.5 dpf in foxq1abcz11 mutants and foxq1bbcz18 mutants (Fig 3A). In case of redundant or compensatory functions of foxq1a and foxq1b, we examined the double foxq1abcz11;foxq1bbcz18 homozygous mutants, but also found no marked differences compared to wildtype siblings (Fig 3A). Quantification of macrophage and neutrophil numbers in the tail region further substantiates our finding that the mutants are not significantly different from siblings in innate immune cell development (Fig 3B). Since it is possible to have defects in macrophages at later time points, we evaluated the development of microglia, an important derivative of embryonic macrophages. We used a vital neutral red stain assay that has been shown to specifically label microglia in the larval zebrafish brain [61–63] as cells with dark red aggregates (Fig 4A). We found that all foxq1abcz11, foxq1bbcz18 and foxq1abcz11;foxq1bbcz18 mutants exhibited the typical pattern (Fig 4A) and numbers (Fig 4B) of microglia in the larval midbrain as those in wildtype and heterozygous control siblings and previously described for wildtype [61–65]. Taken together, the data indicates that foxq1a and foxq1b are not required for the proper development of macrophages, neutrophils, and microglia.
Fig 3. Single and double foxq1abcz11 and foxq1bbcz18 mutants have normal macrophage and neutrophil development.
a Whole mount in situ hybridization of 2.5 dpf single and double foxq1abcz11 and foxq1bbcz18 mutants and their siblings using RNA probes for mfap4 (macrophage marker) and mpx (neutrophil marker). Arrows, macrophages or neutrophils in the caudal hematopoietic tissue in the embryo tail. b Scatter plots showing number of macrophages and neutrophils in the tail region for each embryo quantified in the different genotype categories. n, number of embryos analyzed beneath each scatter plot. Plots report average ± standard deviation (s.d.). Multiple unpaired t-tests comparing between control siblings and each of the mutants groups were conducted with correction for multiple comparisons using the Sidak-Bonferroni method and without assuming equal variance to determine statistical significance. n.s., no statistical significance as defined by p > 0.05 was found in all the comparisons.
Fig 4. Neutral red analysis shows normal microglia development in single and double foxq1a and foxq1b mutants at 4 dpf.
a Neutral red staining in 4 dpf wildtype (WT) sibling, foxq1abcz11, foxq1bbcz18, and foxq1abcz11;foxq1bbcz18 mutants shows a stereotypical pattern of microglia in the midbrain (arrow). b Bar graph shows all larvae analyzed in all genotype groups had normal numbers of microglia (>25). Number of larvae analyzed (n) shown to the right of the corresponding bar.
foxq1a and foxq1b null mutants exhibit typical transcriptional response to Escherichia coli brain challenge
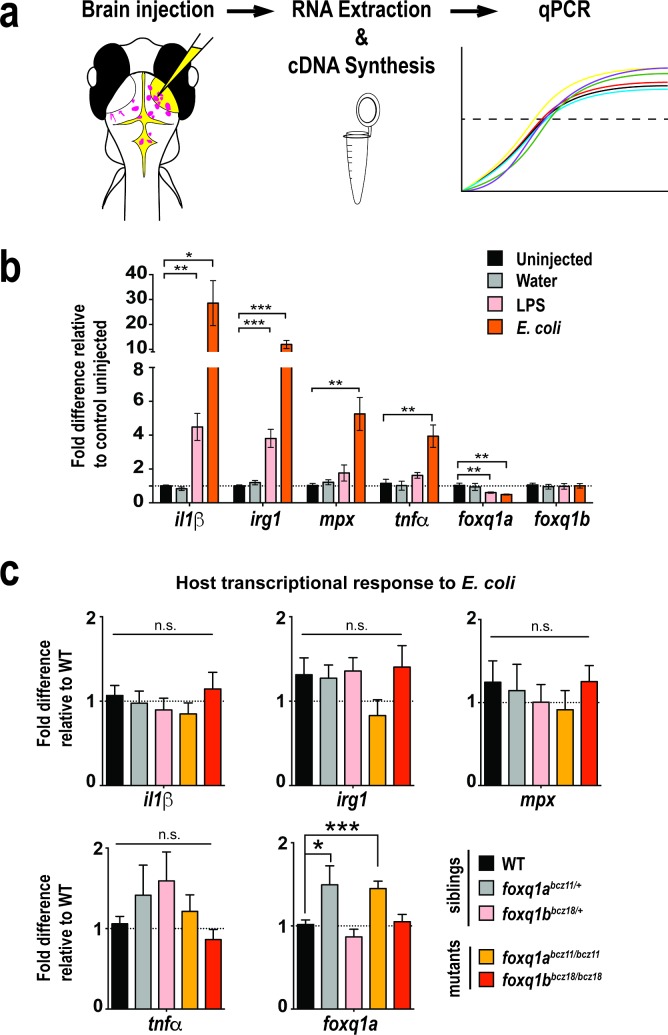
While foxq1a and foxq1b are not essential for innate immune cell development, they may affect the function of these cells, such as their ability to mediate an immune response. We developed a brain challenge assay (Fig 5A) to trigger innate immune activation by introducing live E. coli bacteria into the brain tectum of zebrafish larvae. We used this assay to address whether foxq1a, foxq1b, or both may regulate systemic response to E. coli challenge. We validated the effectiveness of our assay to trigger immune activation by showing that after brain challenge with bacteria or LPS at 6 hours post injection (hpi), we indeed found significant elevation of several innate immune activation genes systemically: interleukin 1 beta (il1β) [61,62,66], immune responsive gene (irg1) [67], neutrophil gene (mpx) [68], and tumor necrosis factor alpha (tnfα) [69] but not in water injected or uninjected controls (Fig 5B). We also tested whether foxq1a and foxq1b themselves could be transcriptionally regulated during a bacterial response to implicate a possible role in this process. Interestingly, we found that foxq1a, but not foxq1b, was significantly downregulated by about a factor of 2 in wildtype larvae after LPS or bacteria injection (Fig 5B).
Fig 5. foxq1abcz11 and foxq1bbcz18 mutants exhibit wildtype transcriptional response to E. coli challenge in the larval brain.
a Graphical representation of the brain challenge assay. E. coli was injected into right brain tectum, and at 6 hours post injection (hpi), injected larvae were individually processed for RNA extraction, genotyping, cDNA synthesis, and qPCR analysis. b Bar chart shows significant upregulation of immune activation genes il1β, irg1, mpx, and tnfα after bacteria challenge at 6 hpi in wildtype larvae compared with uninjected controls. Water injected wildtype larvae exhibit no induction of activation genes, whereas LPS injected wildtype animals have a significant upregulation of il1β and irg1. After microbial activation by E. coli or LPS, a significant downregulation of foxq1a was found by about a factor of 2. No significant change was found for foxq1b after immune challenge. Data from wildtype injections validate the efficacy of the assay to activate the innate immune system. Dotted line marks no fold difference at 1. At least 6 or more independent biological samples were measured per category. c Bar plot shows transcriptional changes of target genes after E. coli injection in the brain at 6 hpi for control siblings and foxq1a and foxq1b mutants. n = 9–14 independent biological samples were measured per genotype. All error bars show standard error of means. Statistical significance in b and c was determined by multiple unpaired t-tests comparing between uninjected or sibling controls and the experimental groups with correction for multiple comparisons using the Sidak-Bonferroni method and without assuming equal variance. n.s., no statistical significance as defined by p > 0.05. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To analyze the immune response of foxq1 mutants, we employed the brain challenge assay on 4 dpf larval progenies of foxq1abcz11/+ and foxq1bbcz18/+ single and double heterozygous incrosses (Fig 5C). We found that the expression levels of immune activation genes (il1β, irg1,mpx, and tnfα) in foxq1 mutants at 6 hpi were not significantly different from WT siblings (Fig 5C). However, we cannot exclude the possibility that double foxq1a and foxq1b mutants may have deficient or aberrant transcriptional response to E. coli. Since foxq1a transcriptional level was downregulated in wildtype response to E. coli (Fig 5B), we also assessed whether this occurred normally in foxq1 mutants as compared to WT siblings. qPCR data shows that foxq1b mutants had similar foxq1a expression as WT controls after bacterial exposure, but both heterozygous and homozygous foxq1a mutants had a modest but significant increase of ~50% in foxq1a RNA level (Fig 5C). The foxq1a upregulation was also found in foxq1a mutants at steady state, suggesting that the increase during the bacterial response may be due to an already higher baseline level rather than necessarily the bacterial exposure (S3 Fig). The results indicate that foxq1a and foxq1b may not be required for mediating an appropriate transcriptional response to E. coli challenge, but foxq1a may affect other aspects of this bacterial response as it is transcriptionally regulated.
Discussion
Our study used zebrafish genetic mutants to investigate the possibility that FOXQ1 may have a role in innate immune cell development and function. Using our newly generated null mutations of the zebrafish homologs of human FOXQ1, foxq1a and foxq1b, we reveal that these genes are not required for the normal development and distribution of macrophages, neutrophils, and microglia. Furthermore, our data suggests that these two genes are not necessary for a stereotypical transcriptional response to E. coli exposure, albeit deficiency in both copies of foxq1 was not assessed. It is possible, however, that foxq1a and foxq1b may have important roles in other innate immune functions such as during an infection or injury that were not examined in this study, which include cell migration and recruitment, phagocytosis, and host response to pathogens, viruses, fungi, and environmental toxins. Since foxq1a expression was found in macrophages and downregulated during a bacterial response, foxq1a may have a role in innate immunity that remains to be fully explored in zebrafish. Furthermore, we show that foxq1a is upregulated in the foxq1a mutants at steady state and during a bacterial response. This suggests that foxq1a may be forming a negative feedback loop in negatively regulating its own transcription.
The foxq1abcz11 and foxq1bbcz18 mutants will provide useful genetic models to address previously unanswered questions regarding function of zebrafish foxq1 in craniofacial development, possible gastrointestinal processes, and later adult organogenesis [51,52]. Consistent with the possibility that FOXQ1 has multiple roles in development, FOXQ1/Foxq1 is highly expressed in mouse stomach and kidney, and human gastric mucosa, bladder, salivary glands, trachea, intestines, and liver [2,16,70]. In addition, murine Foxq1 deletion mutants have deficiencies in gastric acid secretion and hair differentiation, and show partial penetrance for embryonic lethality [16,70]. The zebrafish foxq1 mutants can also be invaluable for uncovering mechanisms mediating the role of FOXQ1 in cancer biology that remains poorly understood.
Materials and methods
Zebrafish lines and embryos
Embryos from wildtype (TL and AB), transgenic lines: mpeg1:EGFP [58,71] and gata1:DsRed [72], and heterozygous lines: foxq1abcz11/+, foxq1bbcz18/+, and foxq1abcz11/+ foxq1bbcz18/+ were raised at 28.5°C, and staged as described [73]. Homozygous mutants were derived from either single heterozygous incrosses or from double heterozygous incrosses. The latter yielded double foxq1abcz11 foxq1bbcz18 mutants at a ratio of ~ 1/16. Embryos were treated with 0.003% 1-phentyl-2-thiourea (PTU) to inhibit pigmentation in sterilized zebrafish system water. This study was carried out in accordance with the animal protocol (16–160) approved by the Institute Animal Care and Use Committee at UNC Chapel Hill.
Neutral red analysis
Microglia were scored in live larvae by neutral red vital dye staining assay as previously described [61,62]. In brief, 3 dpf larvae were stained with neutral red by immersion in system water supplemented with 2.5 μg/ml neutral red and 0.003% PTU at 28.5°C for 1 hour, followed by 1–2 water changes, and then analyzed 2–3 hours later using a stereomicroscope.
Whole mount RNA in situ hybridization
In situ hybridization was performed using standard methods as described in [61,62,74]. Antisense riboprobes were transcribed from: mfap4 plasmid previously cloned [61], mpx (full-length, Open Biosystems clone 6960294) [61], foxq1a plasmid (pCES150), which was made from a 877 bp fragment cloned from a wildtype zebrafish embryo cDNA library using primers F476: GCACTCATCATCTGCAACAGGTA and R1353: TGATATCCCGCGGTTGCAGG. Anti-sense foxq1a RNA probe was made from pCES150 digested with HindIII and transcribed by T7 RNA polymerase. To make the negative control RNA probe, foxq1a sense strand sequence was transcribed by T3 RNA polymerase using a PCR template made from the foxq1a plasmid pCES150. foxq1b anti-sense RNA probe was made from a PCR-based template encompassing 754 bp of coding sequence using forward primer: GTGTTGCGAGATCCCTCGCGTC and reverse primer: GTAAACACTGTGCAGTGGCGCGTC, where the reverse primer also included the T7 RNA polymerase recognition site.
RNA extraction, RT-PCR and quantitative PCR (qPCR)
Total RNA was extracted from individual larvae using the RNAqueous-Micro Isolation Kit (Ambion). cDNA was made using oligo dT primer and SuperScript IV reverse transcriptase (Invitrogen). qPCR was performed on the Applied Biosystems QuantStudio Flex 6 Real-Time PCR System using probe-based assays or SYBR Green. The delta–delta ct method was used to determine the relative levels of mRNA expression between experimental samples and controls. ef1a was used as the reference gene for determination of relative expression of all target genes. Sequences of the qPCR probes and primer sets used in qPCR and RT-PCR analyses are listed in Table 1.
Table 1. Probes and primer sequences for qPCR and RT-PCR.
| ef1a | probe | CTGGAGACAGCAAGAACGACCCAC | |
| F primer | ACATCCGTCGTGGTAATGTG | ||
| R primer | TGATGACCTGAGCGTTGAAG | ||
| il1b | probe | TCCGTCAAATGTCCCGGTTGGTTTA | |
| F primer | ACCGGCAGCTCCATAAAC | ||
| R primer | GGTGTCTTTCCTGTCCATCTC | ||
| irg1 | probe | TTTAACACCGTGCTTCAGTGCAGC | |
| F primer | ACTGGGAGTGGGTTTGATTG | ||
| R primer | GCATACTGAGGTGGAAGAGATG | ||
| mpx | probe | TATAGCATGGTCACGCCCTCTTTGC | |
| F primer | TGCCTTCACATCCCACATAG | ||
| R primer | GGAGCAGACAATCCACAGAA | ||
| tnfa | F primer | GCGCTTTTCTGAATCCTACG | |
| R primer | TGCCCAGTCTGTCTCCTTCT | ||
| foxq1a | probe | TAAGGGATCCTTCGAGACCGTGGG | |
| F primer | GTGCGCCATAATCTGTCTCTAA | ||
| R primer | CGGGTTCAGCATCCAGTAAT | ||
| foxq1b | probe | ATGGGATTTCCGCCACAGAGCA | |
| F primer | TCCTCACTTTCTGCCAGTTG | ||
| R primer | ACATTCGACACCTCAGAACTG | ||
| DsRed | F primer | TCCGAGGACGTCATCAAGGAGTTC | sorted cells |
| R primer | GGCGGGGTGCTTCACGTACAC | sorted cells | |
| foxq1a | foxq1a_F531 | GCAGAAGGAGAAAGCGCATTAG | sorted cells |
| foxq1a_R773 | AGGATCGAAGCATGACATACGG | sorted cells | |
| foxq1b | foxq1b_F540 | CAGCGAGTACACTTTTGCAGAC | sorted cells |
| foxq1b_R839 | AATCTGCTCTGTGGCGGATAG | sorted cells | |
| GFP | F primer | TATATCATGGCCGACAAGCA | sorted cells |
| R primer | CTGGGTGGCTCAGGTAGTGG | sorted cells | |
| hbbe1.1 | hbbe1.1_F139 | GCTCTGGCAAGGTGTCTCATC | sorted cells |
| hbbe1.1_R465 | AGCGATGAATTTCTGGAAAGCG | sorted cells | |
| irf8 | irf8-218-F | CAGCGACATGGAAGACCAGATTG | sorted cells |
| irf8-647-R | GTGGTCACCATGTTGTCCACCATC | sorted cells | |
| mpeg1 | mpeg1_F1 | ACAACACCACCTTGTTACACTCT | sorted cells |
| mpeg1_R1 | ACAACTGCTGGATTTGGTCAATG | sorted cells | |
| foxq1a | foxq1a_F476 | TGATATCCCGCGGTTGCAGG | 2dpf/8dpf/gut |
| foxq1a_R1353 | GCACTCATCATCTGCAACAGGTA | 2dpf/8dpf/gut | |
| foxq1b | foxq1b_F159 | ATGGATGTGAACGTGGCTTCA | 2dpf/8dpf/gut |
| foxq1b_R961 | GGGTGAAATCCAGCCTCTTGTA | 2dpf/8dpf/gut |
CRISPR-Cas9 targeted mutagenesis of foxq1a and foxq1b
The target genes were foxq1a (NCBI accession: NM_001243344.1; Gene ID: 100537750) and foxq1b (NCBI accession: NM_212907.1; Gene ID: 405843). Co-injection of Cas9 mRNA and guide RNAs (gRNAs) was conducted in wildtype 1-cell stage zebrafish embryos. Cas9 mRNA was transcribed from XbaI linearalized pT3TS-nCas9n plasmid (Addgene #46757) [53] using mMessage mMachine T3 Kit (Ambion) according to the manufacturer’s instructions. CRISPR targets for gRNA designs were identified using CHOPCHOP (http://chopchop.cbu.uib.no) [54]. The following gene-specific oligonucleotides using T7 promoter were used to make gRNAs as previously described [54]; gene-specific target sequences are underlined: foxq1a gRNA-1 5’-TAATACGACTCACTATAGGGTTCGAAGAGTGACAGGGGTTTTAGAGCTAGAAATAGCAAG-3’, foxq1a gRNA-2 5’-TAATACGACTCACTATAGGACTACGACTCCAAGCCTGGTTTTAGAGCTAGAAATAGCAAG-3’, foxq1a gRNA-3 5’- TAATACGACTCACTATAGGAGTTGTGCAGCGATGCTGGTTTTAGAGCTAGAAATAGCAAG-3’, foxq1b gRNA-1 5’-TAATACGACTCACTATAGGCTTCACCGCTGTCCACGGGTTTTAGAGCTAGAAATAGCAAG-3’, foxq1b gRNA-2 5’-TAATACGACTCACTATAGGTTCCTCCTCCGTGGACAGGTTTTAGAGCTAGAAATAGCAAG-3’, foxq1b gRNA-3 5’-TAATACGACTCACTATAGGAGCCCAATTCCTCCTCCGGTTTTAGAGCTAGAAATAGCAAG-3’. In vitro transcription of gRNAs from assembled oligonucleotides was conducted using the HiScribe T7 Quick High Yield RNA Synthesis Kit (NEB). Injected clutches of embryos validated to contain CRISPR mediated mutagenesis by a T7 endonuclease assay were raised as F0 fish. Incross and outcross of F0 adults produced F1 progenies, which were genotyped and sequenced at adult stages to identify F1 founders carrying mutations of interest. These founders were isolated and outcrossed to raise new mutant lines, which yielded the foxq1abcz11/+ and foxq1bbcz18/+ heterozygous fish used in this study.
Cell dissociation and fluorescence activated cell sorting (FACS)
Dissociation of pools of 25–50 2.5 dpf transgenic zebrafish embryos either carrying the macrophage reporter (mpeg1:EGFP) or the erythrocyte reporter (gata1:DsRed) was conducted using a protocol adapted from [75]. Fluorescently tagged cells were sorted from non-fluorescent cells using the BD FACSAria cell sorter equipped with three lasers. Non-fluorescent control embryos were used for setting gates in FACS for GFP and DsRed positive cells. BD FACSDiva V8.0.1 software was used for cell sorting and FACS plots. Total RNA was extracted from the different populations of cells (GFP positive, GFP negative, DsRed positive, DsRed negative) using the RNAqueous-Micro Isolation Kit (Ambion), and made into cDNA for RT-PCR analyses as described above.
Genotyping bcz11 and bcz18 alleles
To genotype foxq1abcz11, the following primers were used: foxq1a F 5’-CGGTGTTTTTGTGACTTGATTT-3’ and foxq1a R 5’-AAGAGTATGGAGGTTTGGGTCTC-3’ in a PCR assay that yielded differentially sized products to distinguish the mutant allele which has a 95 bp deletion compared with the wildtype allele. The expected sizes for mutant band is 240 bp and wildtype band is 335 bp. To genotype foxq1bbcz18, a PCR and restriction enzyme combined assay was used. Primers foxq1b F 5’-AAGCAACTCATCTGACCTGACA-3’ and foxq1b R 5’-GGGTCTACGCGTATATGGTTTC-3’ were used to yield PCR products of 217 bp, followed by a digest with BsaJI, which only recognizes the wildtype sequence.
Immune challenge
Bacteria were prepared from 3 mL overnight cultures, each derived from a single Escherichia coli colony, which were centrifuged and re-suspended in 500 μL of PBS. The 500 μL bacteria mix (~1.6 x 106 cfu/μL) was supplemented with 1 μL of 5 mg/ml AlexaFluor 568 or AlexaFluor 488 conjugated dextran (10 kDa) from Invitrogen. Other reagents used for brain injections were: ultra-pure water supplemented with 0.05 mg/ml fluorescently labeled dextran (10 kDa) for the control water samples, and lipopolysaccharides (LPS) derived from 0111:B4 E. coli (L3024 Sigma) at 4.5 mg/ml was supplemented with 0.05 mg/ml fluorescently labeled dextran (10 kDa). For all injections, 1 nL of the reagent mix was injected into the right brain tectum of 4 dpf larvae, and the fluorescent dextran was used to validate the injection site. Larvae that were successfully injected were collected 6 hours post injection (hpi) and immediately processed for total RNA lysis. RNA extraction and subsequent steps for qPCR analyses are described above. All collected larvae derived from heterozygous intercrosses were genotyped after RNA extraction.
Supporting information
Primers used to amplify the approximate full-length coding region of each gene were: start-F 5’-GCACTCATCATCTGCAACAGGTA and end-R 5’-TGATATCCCGCGGTTGCAGG for foxq1a (~900 bp); and start-F 5’-AAGCAACTCATCTGACCTGACA and end-R 5’-GTAAACACTGTGCAGTGGCGCGTC for foxq1b (~1150 bp). (a) Inverted DNA gel image of the RT-PCR analysis of foxq1a mRNA shows the expected ~900 bp transcript in WT siblings and a single mutant transcript with a 95 bp deletion in homozygous bcz11 mutants, indicating no alternative splicing in the mutants. Gel image shows 2 independent samples per genotype. (b) Sanger sequencing analysis of the foxq1a transcript from four bcz11 mutants shows the expected 95 bp deletion that caused a frameshift leading to an early stop codon at the beginning of the gene. (c) RT-PCR analysis of the foxq1b transcript in WT and bcz18 mutants shows the expected product size of ~1150 bp and no apparent splice variants. Gel image shows 2 independent samples per genotype. (d) Sanger sequencing analysis of the foxq1b transcript from four bcz18 mutants confirms the indel mutation that leads to a frameshift and early stop codon. aa, amino acid; sib, sibling; Mut, mutant.
(TIF)
a-d foxq1a gene expression. a Expression in ventral head region is found in 1.5 dpf embryo (arrow). b Prominent expression at 2.5 dpf in the craniofacial region (arrows, higher magnification in c), on the yolk (arrows, higher magnification in d), and along the body abutting the yolk. Yolk expression is not found in all embryos analyzed whereas the craniofacial expression is. e-f As a negative control, whole mount in situ hybridization using the sense RNA probe for foxq1a does not show any expression at all stages analyzed from 1–3 dpf. g foxq1b expression is prominent in the craniofacial jaw region. Left, lateral view (arrow). Right, ventral view (arrows).
(TIF)
A modest upregulation of foxq1a RNA expression at ~ 44% on average is found in foxq1a mutants at 4 dpf at steady state in the absence of any immune challenge. n = 6 independent biological samples were measured per genotype. All error bars show standard error of means.
(TIF)
Acknowledgments
We are grateful to Ana Meireles for insightful discussions and sharing of reagents for implementing the CRISPR-Cas9 system, Patrick Autissier for conducting the FACS sorting in the Boston College Biology Core facility, and Keri Sullivan for technical support. This work was supported by start-up funds to CES from Boston College and UNC-Chapel Hill, and NIH NIGMS grant 1R35GM124719 to CES.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by start-up funds to CES from Boston College and UNC-Chapel Hill, and NIH NIGMS grant 1R35GM124719 to CES. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14: 142–146. [PubMed] [Google Scholar]
- 2.Bieller A, Pasche B, Frank S, Glaser B, Kunz J, Witt K, et al. Isolation and characterization of the human forkhead gene FOXQ1. http://dx.doi.org/101089/10445490152717596. 2001;20: 555–561. doi: 10.1089/104454901317094963 [DOI] [PubMed] [Google Scholar]
- 3.Katoh M, Katoh M. Human FOX gene family (Review). Int J Oncol. Spandidos Publications; 2004;25: 1495–1500. doi: 10.3892/ijo.25.5.1495 [PubMed] [Google Scholar]
- 4.Wotton KR, Mazet F, Shimeld SM. Expression of FoxC, FoxF, FoxL1, and FoxQ1 genes in the dogfish Scyliorhinus canicula defines ancient and derived roles for Fox genes in vertebrate development. Dev Dyn. 2008;237: 1590–1603. doi: 10.1002/dvdy.21553 [DOI] [PubMed] [Google Scholar]
- 5.Jackson BC, Carpenter C, Nebert DW, Vasiliou V. Update of human and mouse forkhead box FOX gene families. Human Genomics 2010 4:5. BioMed Central; 2010;4: 345 doi: 10.1186/1479-7364-4-5-345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, Meng F, Liu G, Bin Zhang, Zhu J, Wu F, et al. Forkhead Transcription Factor Foxq1 Promotes Epithelial–Mesenchymal Transition and Breast Cancer Metastasis. Cancer Res. American Association for Cancer Research; 2011;71: 1292–1301. doi: 10.1158/0008-5472.CAN-10-2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Meng F, Liu G, Zhang B, Zhu J, Wu F. Cross-species expression profiling identifies Forkhead-Box Q1 as a promoter of epithelial-mesenchymal transition and breast cancer metastasis. 2011.
- 8.Fan D-M, Feng X-S, Qi P-W, Chen Y-W. Forkhead factor FOXQ1 promotes TGF-β1 expression and induces epithelial-mesenchymal transition. Mol Cell Biochem. 2014;397: 179–186. doi: 10.1007/s11010-014-2185-1 [DOI] [PubMed] [Google Scholar]
- 9.Feuerborn A, Srivastava PK, Küffer S. The Forkhead factor FoxQ1 influences epithelial differentiation. Journal of cellular …. 2011. doi: 10.1002/jcp.22385/full [DOI] [PubMed] [Google Scholar]
- 10.Feuerborn A. The Forkhead factor FoxQ1 influences epithelial plasticity and modulates TGF-beta1 signalling. 2010.
- 11.Qiao Y, Jiang X, Lee ST, Karuturi RKM, Hooi SC, Yu Q. FOXQ1 Regulates Epithelial-Mesenchymal Transition in Human Cancers. Cancer Res. American Association for Cancer Research; 2011;71: 3076–3086. doi: 10.1158/0008-5472.CAN-10-2787 [DOI] [PubMed] [Google Scholar]
- 12.Hoggatt AM, Kriegel AM, Smith AF, Herring BP. Hepatocyte nuclear factor-3 homologue 1 (HFH-1) represses transcription of smooth muscle-specific genes. J Biol Chem. 2000;275: 31162–31170. doi: 10.1074/jbc.M005595200 [DOI] [PubMed] [Google Scholar]
- 13.Hong HK, Noveroske JK, Headon DJ, Liu T, Sy MS, Justice MJ, et al. The winged helix/forkhead transcription factor Foxq1 regulates differentiation of hair in satin mice. Genesis. 2001;29: 163–171. doi: 10.1002/gene.1020 [DOI] [PubMed] [Google Scholar]
- 14.Potter CS, Peterson RL, Barth JL, Pruett ND. Evidence that the satin hair mutant gene Foxq1 is among multiple and functionally diverse regulatory targets for Hoxc13 during hair follicle differentiation. Journal of Biological …. 2006. [DOI] [PubMed] [Google Scholar]
- 15.Verzi MP, Khan AH, Ito S, Shivdasani RA. Transcription Factor Foxq1 Controls Mucin Gene Expression and Granule Content in Mouse Stomach Surface Mucous Cells. Gastroenterology. 2008;135: 591–600. doi: 10.1053/j.gastro.2008.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goering W, Adham IM, Pasche B, Manner J, Ochs M, Engel W, et al. Impairment of gastric acid secretion and increase of embryonic lethality in Foxq1-deficient mice. Cytogenet Genome Res. 2008;121: 88–95. doi: 10.1159/000125833 [DOI] [PubMed] [Google Scholar]
- 17.Cui Y, Qiao A, Jiao T, Zhang H, Xue Y, Zou Y, et al. The hepatic FOXQ1 transcription factor regulates glucose metabolism in mice. Diabetologia. 2016;59: 2229–2239. doi: 10.1007/s00125-016-4043-z [DOI] [PubMed] [Google Scholar]
- 18.Jonsson H, Peng SL. Forkhead transcription factors in immunology. Cell Mol Life Sci. 2005;62: 397–409. doi: 10.1007/s00018-004-4365-8 [DOI] [PubMed] [Google Scholar]
- 19.Wang P, Lv C, Zhang T, Liu J, Yang J, Guan F, et al. FOXQ1 regulates senescence-associated inflammation via activation of SIRT1 expression. Cell Death Dis. 2017;8: e2946 doi: 10.1038/cddis.2017.340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S-H, Kaschula CH, Priedigkeit N, Lee AV, Singh SV. Forkhead Box Q1 Is a Novel Target of Breast Cancer Stem Cell Inhibition by Diallyl Trisulfide. J Biol Chem. 2016;291: 13495–13508. doi: 10.1074/jbc.M116.715219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sehrawat A, Kim SH, Vogt A, Singh SV. Suppression of FOXQ1 in benzyl isothiocyanate-mediated inhibition of epithelial–mesenchymal transition in human breast cancer cells. Carcinogenesis. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abba M, Patil N, Rasheed K, Nelson LD, Mudduluru G. A direct regulation of Twist1 by FOXQ1 promotes colorectal cancer metastasis. 2013. [DOI] [PubMed]
- 23.Weng W, Okugawa Y, Toden S, Toiyama Y, Kusunoki M, Goel A. FOXM1 and FOXQ1 Are Promising Prognostic Biomarkers and Novel Targets of Tumor-Suppressive miR-342 in Human Colorectal Cancer. Clin Cancer Res. 2016;22: 4947–4957. doi: 10.1158/1078-0432.CCR-16-0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng X, Luo Z, Kang Q, Deng D, Wang Q, Peng H, et al. FOXQ1 mediates the crosstalk between TGF-β and Wnt signaling pathways in the progression of colorectal cancer. Cancer Biol Ther. 2015;16: 1099–1109. doi: 10.1080/15384047.2015.1047568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu JY, Wu XY, Wu GN, Liu F-K, Yao X-Q. FOXQ1 promotes cancer metastasis by PI3K/AKT signaling regulation in colorectal carcinoma. Am J Transl Res. 2017;9: 2207–2218. [PMC free article] [PubMed] [Google Scholar]
- 26.Bao B, Azmi AS, Aboukameel A, Ahmad A. Pancreatic cancer stem-like cells display aggressive behavior mediated via activation of FoxQ1. Journal of Biological …. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhan H-X, Xu J-W, Wang L, Wu D, Zhang G-Y, Hu S-Y. FoxQ1 is a Novel Molecular Target for Pancreatic Cancer and is Associated with Poor Prognosis. Curr Mol Med. 2015;15: 469–477. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Liu Y, Zhang J, Cui X, Li G, Wang J, et al. FOXQ1 promotes gastric cancer metastasis through upregulation of Snail. Oncol Rep. 2016;35: 3607–3613. doi: 10.3892/or.2016.4736 [DOI] [PubMed] [Google Scholar]
- 29.Xiang X-J, Deng J, Liu Y-W, Wan L-Y, Feng M, Chen J, et al. MiR-1271 Inhibits Cell Proliferation, Invasion and EMT in Gastric Cancer by Targeting FOXQ1. Cell Physiol Biochem. 2015;36: 1382–1394. doi: 10.1159/000430304 [DOI] [PubMed] [Google Scholar]
- 30.Guo J, Yan Y, Yan Y, Guo Q, Zhang M, Zhang J, et al. Tumor-associated macrophages induce the expression of FOXQ1 to promote epithelial-mesenchymal transition and metastasis in gastric cancer cells. Oncol Rep. 2017. doi: 10.3892/or.2017.5877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soleimani F, Hajjari M, Mohammad Soltani B, Behmanesh M. Up-Regulation of FOXC2 and FOXQ1 Is Associated with The Progression of Gastric-Type Adenocarcinoma. Cell J. 2017;19: 66–71. doi: 10.22074/cellj.2017.4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ZHU Z, Zhu Z, ZHU Z, PANG Z, XING Y, WAN F, et al. Short hairpin RNA targeting FOXQ1 inhibits invasion and metastasis via the reversal of epithelialmesenchymal transition in bladder cancer. Int J Oncol. 2013;42: 1271–1278. doi: 10.3892/ijo.2013.1807 [DOI] [PubMed] [Google Scholar]
- 33.Xia L, Wu K, Fan D. 581 Sox12, a Direct Target of FoxQ1, Promotes Hepatocellular Carcinoma Metastasis Through up-Regulating TWIST1 and FGFBP1. Gastroenterology; 2015. [DOI] [PubMed] [Google Scholar]
- 34.Huang W, Chen Z, Shang X, Tian D, Wang D, Wu K, et al. Sox12, a direct target of FoxQ1, promotes hepatocellular carcinoma metastasis through up-regulating Twist1 and FGFBP1. Hepatology. 2015;61: 1920–1933. doi: 10.1002/hep.27756 [DOI] [PubMed] [Google Scholar]
- 35.Wang W, He S, Ji J, Huang J, Zhang S. The prognostic significance of FOXQ1 oncogene overexpression in human hepatocellular carcinoma. Pathology-Research and…. 2013. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Yang Y, Yang T, Yuan S, Wang R, Pan Z, et al. Double-negative feedback loop between microRNA-422a and forkhead box (FOX)G1/Q1/E1 regulates hepatocellular carcinoma tumor growth and metastasis. Hepatology. 6 ed. 2015;61: 561–573. doi: 10.1002/hep.27491 [DOI] [PubMed] [Google Scholar]
- 37.Xiao B, Liu H, Gu Z, Ji C. Expression of microRNA-133 inhibits epithelial-mesenchymal transition in lung cancer cells by directly targeting FOXQ1. Arch Bronconeumol. 2016;52: 505–511. doi: 10.1016/j.arbres.2015.10.016 [DOI] [PubMed] [Google Scholar]
- 38.Feng J, Zhang X, Zhu H, Wang X, Ni S, Huang J. FoxQ1 Overexpression Influences Poor Prognosis in Non-Small Cell Lung Cancer, Associates with the Phenomenon of EMT. Kalinichenko VV, editor. PLoS ONE. 2012;7: e39937 doi: 10.1371/journal.pone.0039937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herlinger AL, Gao M, Wu RC, Wang TL, Rangel L. Abstract A80: NAC1 attenuates BCL6 negative autoregulation and functions as a BCL6 coactivator of FOXQ1 transcription in ovarian cancer (OVCA). 2016. [DOI] [PMC free article] [PubMed]
- 40.Gao M, Shih I-M, Wang T-L. The role of forkhead box Q1 transcription factor in ovarian epithelial carcinomas. Int J Mol Sci. 2012;13: 13881–13893. doi: 10.3390/ijms131113881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun H-T, Cheng S-X, Tu Y, Li X-H, Zhang S. FoxQ1 Promotes Glioma Cells Proliferation and Migration by Regulating NRXN3 Expression. Jiang T, editor. PLoS ONE. 2013;8: e55693 doi: 10.1371/journal.pone.0055693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown JM, Recht L, Strober S. The Promise of Targeting Macrophages in Cancer Therapy. Clin Cancer Res. 2017;23: 3241–3250. doi: 10.1158/1078-0432.CCR-16-3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia L, Chen Z, Liu J, Shang X, Nie Y. Foxq1-ccl2 signaling-induced macrophage recruitment promotes hepatocellular carcinoma metastasis. Journal of …; 2013. [Google Scholar]
- 44.Ostuni R, Kratochvill F, Murray PJ, Natoli G. Macrophages and cancer: from mechanisms to therapeutic implications. Trends Immunol. 2015;36: 229–239. doi: 10.1016/j.it.2015.02.004 [DOI] [PubMed] [Google Scholar]
- 45.Petty AJ, Yang Y. Tumor-associated macrophages: implications in cancer immunotherapy. Immunotherapy. 2017;9: 289–302. doi: 10.2217/imt-2016-0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis CE, Harney AS, Pollard JW. The Multifaceted Role of Perivascular Macrophages in Tumors. Cancer Cell. 2016;30: 365 doi: 10.1016/j.ccell.2016.07.009 [DOI] [PubMed] [Google Scholar]
- 47.Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14: 399–416. doi: 10.1038/nrclinonc.2016.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27: 462–472. doi: 10.1016/j.ccell.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaneda H, Arao T, Tanaka K, Tamura D, Aomatsu K. Overexpression of FOXQ1 is important factor in tumorigenicity and tumor growth. 2010.
- 50.Li Y, Zhang Y, Yao Z, Li S, Yin Z, Xu M. Forkhead box Q1: A key player in the pathogenesis of tumors (Review). Int J Oncol. 2016;49: 51–58. doi: 10.3892/ijo.2016.3517 [DOI] [PubMed] [Google Scholar]
- 51.Planchart A, Mattingly CJ. 2,3,7,8-Tetrachlorodibenzo-p-dioxin upregulates FoxQ1b in zebrafish jaw primordium. Chem Res Toxicol. 2010;23: 480–487. doi: 10.1021/tx9003165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Topczewska JM, Shoela RA, Tomaszewski JP, Mirmira RB, Gosain AK. The Morphogenesis of Cranial Sutures in Zebrafish. PLoS ONE. 2016;11: e0165775 doi: 10.1371/journal.pone.0165775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jao L-E, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA. 2013;110: 13904–13909. doi: 10.1073/pnas.1308335110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gagnon JA, Valen E, Thyme SB, Huang P, Ahkmetova L, Pauli A, et al. Efficient Mutagenesis by Cas9 Protein-Mediated Oligonucleotide Insertion and Large-Scale Assessment of Single-Guide RNAs. Riley B, editor. PLoS ONE. 2014;9: e98186 doi: 10.1371/journal.pone.0098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willet SG, Mills JC. Stomach Organ and Cell Lineage Differentiation: from Embryogenesis to Adult Homeostasis. Cell Mol Gastroenterol Hepatol. 2016;2: 546–559. doi: 10.1016/j.jcmgh.2016.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim TH, Shivdasani RA. Stomach development, stem cells and disease. Development. 2016;143: 554–565. doi: 10.1242/dev.124891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walton EM, Cronan MR, Beerman RW, Tobin DM. The macrophage-specific promoter mfap4 allows live, long-term analysis of macrophage behavior during mycobacterial infection in zebrafish. Gong Z, editor. PLoS ONE. 2015;10: e0138949 doi: 10.1371/journal.pone.0138949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zakrzewska A, Cui C, Stockhammer OW, Benard EL, Spaink HP, Meijer AH. Macrophage-specific gene functions in Spi1-directed innate immunity. Blood. 2010;116: e1–11. doi: 10.1182/blood-2010-01-262873 [DOI] [PubMed] [Google Scholar]
- 59.Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood. 2001;98: 3087–3096. doi: 10.1182/blood.V98.10.3087 [DOI] [PubMed] [Google Scholar]
- 60.Mathias JR, Dodd ME, Walters KB, Yoo SK, Ranheim EA, Huttenlocher A. Characterization of zebrafish larval inflammatory macrophages. Developmental & Comparative Immunology. 2009;33: 1212–1217. doi: 10.1016/j.dci.2009.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shiau CE, Monk KR, Joo W, Talbot WS. An anti-inflammatory NOD-like receptor is required for microglia development. Cell Rep. 2013;5: 1342–1352. doi: 10.1016/j.celrep.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shiau CE, Kaufman Z, Meireles AM, Talbot WS. Differential Requirement for irf8 in Formation of Embryonic and Adult Macrophages in Zebrafish. Wen Z, editor. PLoS ONE. 2015;10: e0117513 doi: 10.1371/journal.pone.0117513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herbomel P, Thisse B, Thisse C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev Biol. 2001;238: 274–288. doi: 10.1006/dbio.2001.0393 [DOI] [PubMed] [Google Scholar]
- 64.Meireles AM, Shiau CE, Guenther CA, Sidik H, Kingsley DM, Talbot WS. The phosphate exporter xpr1b is required for differentiation of tissue-resident macrophages. Cell Rep. 2014;8: 1659–1667. doi: 10.1016/j.celrep.2014.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen K, Sidik H, Talbot WS. The Rag-Ragulator Complex Regulates Lysosome Function and Phagocytic Flux in Microglia. Cell Rep. Elsevier; 2016;14: 547–559. doi: 10.1016/j.celrep.2015.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ogryzko NV, Hoggett EE, Solaymani-Kohal S, Tazzyman S, Chico TJA, Renshaw SA, et al. Zebrafish tissue injury causes upregulation of interleukin-1 and caspase-dependent amplification of the inflammatory response. DMM Disease Models and Mechanisms. 2014;7: 259–264. doi: 10.1242/dmm.013029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hall CJ, Boyle RH, Astin JW, Flores MV, Oehlers SH, Sanderson LE, et al. Immunoresponsive gene 1 augments bactericidal activity of macrophage-lineage cells by regulating β-oxidation-dependent mitochondrial ros production. Cell Metab. 2013;18: 265–278. doi: 10.1016/j.cmet.2013.06.018 [DOI] [PubMed] [Google Scholar]
- 68.Yan B, Han P, Pan L, Lu W, Xiong J, Zhang M, et al. IL-1β and reactive oxygen species differentially regulate neutrophil directional migration and basal random motility in a zebrafish injury-induced inflammation model. Journal of Immunology. 2014;192: 5998–6008. doi: 10.4049/jimmunol.1301645 [DOI] [PubMed] [Google Scholar]
- 69.Zelova H, Hosek J. TNF-alpha signalling and inflammation: interactions between old acquaintances. Inflamm Res. 2013;62: 641–651. doi: 10.1007/s00011-013-0633-0 [DOI] [PubMed] [Google Scholar]
- 70.Frank S, Zoll B. Mouse NHF-3/fork head homolog-1-like gene: Structure, chromosomal location, and expression in adult and embryonic kidney. DNA and Cell Biology. 1998;17: 679–688. doi: 10.1089/dna.1998.17.679 [DOI] [PubMed] [Google Scholar]
- 71.Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood. 2011;117: e49–56. doi: 10.1182/blood-2010-10-314120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol. 2003;4: 1238–1246. doi: 10.1038/ni1007 [DOI] [PubMed] [Google Scholar]
- 73.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203: 253–310. doi: 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- 74.Pogoda H-M, Sternheim N, Lyons DA, Diamond B, Hawkins TA, Woods IG, et al. A genetic screen identifies genes essential for development of myelinated axons in zebrafish. Dev Biol. 2006;298: 118–131. doi: 10.1016/j.ydbio.2006.06.021 [DOI] [PubMed] [Google Scholar]
- 75.Manoli M, Driever W. Fluorescence-activated cell sorting (FACS) of fluorescently tagged cells from zebrafish larvae for RNA isolation. Cold Spring Harb Protoc. 2012;2012: pdb.prot069633. doi: 10.1101/pdb.prot069633 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used to amplify the approximate full-length coding region of each gene were: start-F 5’-GCACTCATCATCTGCAACAGGTA and end-R 5’-TGATATCCCGCGGTTGCAGG for foxq1a (~900 bp); and start-F 5’-AAGCAACTCATCTGACCTGACA and end-R 5’-GTAAACACTGTGCAGTGGCGCGTC for foxq1b (~1150 bp). (a) Inverted DNA gel image of the RT-PCR analysis of foxq1a mRNA shows the expected ~900 bp transcript in WT siblings and a single mutant transcript with a 95 bp deletion in homozygous bcz11 mutants, indicating no alternative splicing in the mutants. Gel image shows 2 independent samples per genotype. (b) Sanger sequencing analysis of the foxq1a transcript from four bcz11 mutants shows the expected 95 bp deletion that caused a frameshift leading to an early stop codon at the beginning of the gene. (c) RT-PCR analysis of the foxq1b transcript in WT and bcz18 mutants shows the expected product size of ~1150 bp and no apparent splice variants. Gel image shows 2 independent samples per genotype. (d) Sanger sequencing analysis of the foxq1b transcript from four bcz18 mutants confirms the indel mutation that leads to a frameshift and early stop codon. aa, amino acid; sib, sibling; Mut, mutant.
(TIF)
a-d foxq1a gene expression. a Expression in ventral head region is found in 1.5 dpf embryo (arrow). b Prominent expression at 2.5 dpf in the craniofacial region (arrows, higher magnification in c), on the yolk (arrows, higher magnification in d), and along the body abutting the yolk. Yolk expression is not found in all embryos analyzed whereas the craniofacial expression is. e-f As a negative control, whole mount in situ hybridization using the sense RNA probe for foxq1a does not show any expression at all stages analyzed from 1–3 dpf. g foxq1b expression is prominent in the craniofacial jaw region. Left, lateral view (arrow). Right, ventral view (arrows).
(TIF)
A modest upregulation of foxq1a RNA expression at ~ 44% on average is found in foxq1a mutants at 4 dpf at steady state in the absence of any immune challenge. n = 6 independent biological samples were measured per genotype. All error bars show standard error of means.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.