Abstract
Purpose
The cell cycle mediators Aurora A and B are targets of drugs currently in clinical development. As with other targeted therapies in breast cancer, response to therapy might be associated with target expression in tumors. We therefore assessed expression of Aurora A and B in breast tumors and studied associations with clinical/pathologic variables.
Experimental Design
Tissue microarrays containing primary specimens from 638 patients with 15-year follow-up were employed to assess expression of Aurora A and B using our automated quantitative analysis method; we used cytokeratin to define pixels as breast cancer (tumor mask) within the array spot and measured Aurora A and B expression within the mask using Cy5-conjugated antibodies.
Results
Aurora A and B expression was variable in primary breast tumors. High Aurora A expression was strongly associated with decreased survival (P = 0.0005). On multivariable analysis, it remained an independent prognostic marker. High Aurora A expression was associated with high nuclear grade and high HER-2/neu and progesterone receptor expression. Aurora B expression was not associated with survival.
Conclusions
Aurora A expression defines a population of patients with decreased survival, whereas Aurora B expression does not, suggesting that Aurora A might be the preferred drug target in breast cancer. Aurora A expression in early-stage breast cancer may identify a subset of patients requiring more aggressive or pathway-targeted treatment. Prospective studies are needed to confirm the prognostic role of Aurora A as well as the predictive role of Aurora A expression in patients treated with Aurora A inhibitors.
The Aurora kinase proteins are serine/threonine kinases that serve as regulators of mammalian cell division. They are important for cell cycle progression and are frequently overexpressed or mutated in human tumors, as reviewed (1, 2). They are localized to the centrosome during mitosis (3–5) and are involved in chromosome segregation and cytokinesis (6–8). Aurora A and B have been implicated in tumor formation and progression (1, 2, 7, 9) and are overexpressed in a variety of cell lines (10–13). Inhibition of Aurora kinases in xenograft models results in tumor regression (14), and overexpression of both Aurora A and B can induce centrosome amplification and aneuploidy (2, 11, 12).
Aurora kinases have diverse functions: Aurora A is predominantly involved in centrosome function and assembly of the mitotic spindle (15). It is critical for maintenance of genome instability (16) and phosphorylates tumor suppressors such as p53, thereby modulating their activities (1). Aurora B is a chromosome passenger protein, essential for chromosome segregation and required for histone H3 phosphorylation, chromosome orientation, spindle assembly checkpoint, and cytokinesis. Little is known about the function of Aurora C. It is also a chromosomal passenger protein involved in assembling the two meiotic spindles during spermatogenesis (7).
Inhibitors that target this family of kinases are currently in clinical development. These agents selectively target the enzymatic activity of Aurora kinases by occupying the catalytic ATP-binding site. Examples include ZM447439 (AstraZeneca; ref. 17), Hesoeradin (Boehringer; ref. 18), and VX-680 (Vertex; ref. 14).
High expression of Aurora A and B has been shown in small patient cohorts in several tumor types, including breast, lung, colon, prostate, pancreas, liver, skin, stomach, rectum, esophagus, endometrium, cervix, bladder, ovary, and thyroid cancers (for review, see refs. 1, 2, 10, 19). In these tumor types, expression levels were higher than in the corresponding normal tissues. Expression levels of Aurora C were not different in tumors compared with benign tissues (1). Aurora C is predominantly expressed in the testes (1); therefore, this work focuses on Aurora A and B.
Several studies have assessed the importance of Aurora A and B in breast cancer. Aurora A overexpression in mouse mammary epithelium has been shown to induce breast tumor formation (20). Polymorphisms in the Aurora A gene are associated with increased risk of primary breast cancer (21, 22), and these polymorphisms are synergistic in their effect on the risk of breast cancer in women with prolonged estrogen exposure (23). Aurora A is responsible for phosphorylation of BRCA1 and thus regulates the transition of cells from the G2 to M phase of cell cycle (24). Small studies have assessed expression of Aurora A in breast cancer specimens: Tanaka et al. (25) studied 33 cases of invasive ductal carcinoma and showed overexpression of Aurora A in 94% of cases. Miyoshi et al. showed elevated expression in 64% of breast cases by reverse transcription-PCR in 47 patients (26), and Hoque et al. showed decreased expression of Aurora A in invasive breast cancer when compared with adjacent in situ carcinoma in 37 cases (27). The largest cohort studied included 112 cases and showed no association between Aurora A expression and survival (28).
Aurora B is induced by estrogens in preclinical models, resulting in cell proliferation (29). Inhibition of Aurora B activity in breast cancer cell lines results in down-regulation of survivin and apoptosis (30). Inhibitors of Aurora B have activity in animal breast cancer models (31). We did not find any studies assessing expression patterns and prognostic value of Aurora B in breast cancer; however, increased risk of breast cancer has been shown in individuals harboring the Ser295Ser (885A>G) polymorphism in the Aurora B gene (32).
Given the importance of Aurora A and B in malignant progression and the current development of Aurora kinase inhibitors, our purpose was to assess expression patterns of these kinases in breast cancer and study the association with survival and other clinical variables. To the best of our knowledge, no large-scale quantitative studies have been conducted on Aurora A and B expression in clinical breast cancer specimens. To obtain more accurate, objective expression measures, we used our newly developed method of automated quantitative analysis (AQUA) of tissue microarrays. This method has been validated and has been proven to be more accurate than pathologist-based scoring of 3,3′-diaminobenzidine stain (33, 34). As is the case with some other targeted therapies, it is possible that response to Aurora kinase targeting drugs might be associated with expression levels of the target in tumors, and quantitative assays need to be developed to predict response.
Materials and Methods
Cell lines and Western blots
MDA-MB-231, MDA-MB-435, MDA-MB-436, MCF-7, T47D, SKBR3, BT-474, BT-20, and ZR-7510 human breast cancer cell lines were purchased from the American Type Culture Collection. Western blotting of protein extracts was done using standard methods. Aurora A and B were detected by overnight incubation with rabbit polyclonal anti-Aurora A IgG at 1:400 and with rabbit polyclonal anti-Aurora B IgG at 1:500. Both antibodies were purchased from Cell Signaling Technology. Protein loading was assessed using mouse anti-h-actin (Sigma-Aldrich) at 1:5,000.
Tissue microarray construction
Tissue microarrays were constructed as described previously (35). Node-negative (n = 319) and node-positive (n = 319) breast cancer cores, each measuring 0.6 mm in diameter, were spaced 0.8 mm apart. Specimens and clinical information were collected with approval of a Yale University Institutional Review Board. The cohort has been described and validated in previous publications (35). Estrogen receptor (ER) staining was positive in 52%, progesterone receptor staining (PR) was positive in 46%, and HER-2/neu staining was positive in 14%. Nuclear grade 3 (on a 1-3 scale) was seen in 28% of the specimens, and 59% were larger than 2 cm. The histologic subtypes included 72% invasive ductal carcinomas, 14% lobular carcinomas, and 14% mixed or other histology. The specimens were resected between 1962 and 1980, with a follow-up range between 4 months and 53 years and a mean follow-up time of 12.6 years. Age at diagnosis ranged from 24 to 88 years (mean, 58 years). Complete treatment history was not available for the entire cohort. Most patients were treated with local irradiation. None of the node-negative patients were given adjuvant systemic therapy. A minority of the node-positive patients (~15%) received chemotherapy, and ~27% of those treated after 1978 received tamoxifen. The time between tumor resection and tissue fixation was not available. A pathologist reviewed slides from all of the blocks to select representative areas of invasive tumor to be cored. The cores were placed on the tissue microarray using a Tissue Microarrayer (Beecher Instruments). The tissue microarrays were then cut to 5 Am sections and placed on glass slides using an adhesive tape-transfer system (Instrumedics) with UV cross-linking.
Immunohistochemistry
One set of two slides (each containing a core from different areas of the tumor for the same patient) was stained for each target marker (Aurora A and B). Staining was done for AQUA as described (35). Briefly, slides were deparaffinized in xylene and transferred through two changes of 100% ethanol. For antigen retrieval, the slides were boiled in a pressure cooker containing 6.5 mmol/L sodium citrate (pH 6.0). Endogenous peroxidase activity was blocked in a mixture of methanol and 2.5% hydrogen peroxide for 30 min at room temperature. To reduce nonspecific background staining, slides were incubated at room temperature for 30 min in 0.3% bovine serum albumin in 1× TBS. Slides were incubated at 4°C overnight in a humidity tray with the primary antibodies: anti-Aurora A antibody (at 1:40) or anti-Aurora B antibody (at 1:100) diluted in TBS containing 0.3% bovine serum albumin. Goat anti-rabbit horseradish peroxidase– decorated polymer backbone (Envision; DAKO) was used as a secondary reagent and Cy5-tyramide (Perkin-Elmer Life Science) was used to visualize the target. To create a tumor mask, slides were simultaneously incubated with a mouse anti-human cytokeratin antibody (DAKO) at 1:200 and were visualized with secondary Alexa 488 – conjugated goat anti-mouse antibodies (Molecular Probes). Coverslips were mounted with ProLong Gold Antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen).
Automated image acquisition and analysis
Images were analyzed using algorithms that have been extensively described (34). Multiple monochromatic, high-resolution (1,024 × 1,024 pixel, 0.5 μm) grayscale images were obtained for each histospot using the ×10 objective of an Olympus AX-51 epifluorescence microscope with an automated microscope stage and digital image acquisition driven by custom program and macro-based interfaces with IPLabs software (Scanalytics). Areas of tumor were distinguished from stroma by creating a mask with the cytokeratin signal tagged with Alexa 488. Coalescence of cytokeratin at the cell surface was used to identify the membrane/cytoplasm compartment within the tumor mask, whereas 4′,6-diamidino-2-phenylindole was used to identify the nuclear compartment within the tumor mask. The target markers (Aurora A and B) were visualized with Cy5 (red). Cy5 was used because its emission peak is outside the color spectrum of tissue autofluorescence. The target signal was scored on a scale of 0 to 255 (AQUA score) for the nuclear and cytoplasmic compartments. Two images (one in-focus and one out-of-focus) were taken of the compartment specific tags and the target marker. A percentage of the out-of-focus image was subtracted from the in-focus image for each pixel, representing the signal-to-noise ratio of the image. An algorithm described as Rapid Exponential Subtraction Algorithm was used to subtract the out-of-focus information in a uniform fashion for the entire microarray. Subsequently, the Pixel Locale Assignment for Compartmentalization of Expression algorithm was used to assign each pixel in the image to a specific subcellular compartment and the signal in each location is calculated. Pixels that cannot accurately be assigned to a compartment were discarded. The AQUA scores were calculated as the average signal intensity per unit of compartment area and expressed on a scale of 0 to 255.
Data analysis
JMP version 5.0 software was used (SAS Institute). The prognostic significance of variables was assessed using the Cox proportional hazards model with survival at 15 years as an endpoint. Associations between continuous AQUA scores of target expression and clinical and pathologic variables were assessed by unpaired t tests. Survival curves were generated using the Kaplan-Meier method, with significance evaluated using the Mantel-Cox log-rank test.
Results
Lysates from a panel of breast cancer cell lines were probed for Aurora A and B and revealed single 48- and 40-kDa bands, respectively. The intensity of the bands was more variable for Aurora B than for Aurora A as shown in Fig. 1.
Fig. 1.
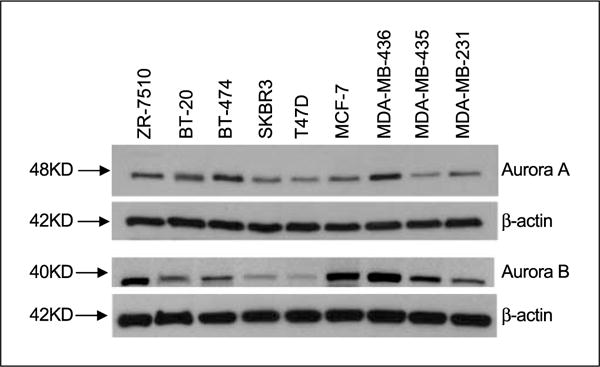
Western blots of a panel of breast cancer cell lines probed for expression of Aurora A and B.
To account for intratumor heterogeneity, two separate sets of slides, each containing a core from a different area of the tumor for each patient, were used to evaluate the expression of each marker. We did log regression analyses to assess the correlation between the two slides for Aurora A and B (Supplementary Fig. S1). The matching tumor spots on each array were highly correlated for the two markers (R = 0.67 for Aurora A and R = 0.63 for Aurora B).
Aurora A staining was stronger in the cytoplasm for most histospots as shown in Fig. 2. However, nuclear staining was also seen. The nuclear and cytoplasmic scores were strongly correlated (R = 0.87); therefore, the analysis is based on total Aurora A staining within the tumor mask. Aurora B staining was mainly nuclear as shown in Fig. 2.
Fig. 2.
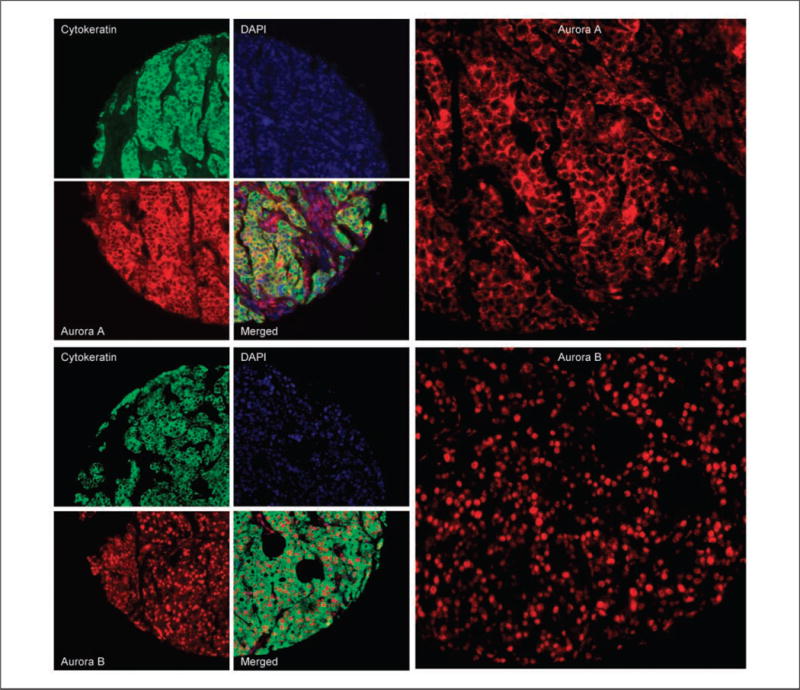
Immunoflourescent staining of Aurora A and B: predominantly cytoplasmic Aurora A (top) and nuclear Aurora B (bottom) staining in a breast cancer histospot using cytokeratin to the define tumor mask, 4′,6-diamidino-2-phenylindole to define the nuclear compartment, and Cy5 to define the target (Aurora A and B). Images on right panels represent higher magnifications of the histospots shown on the left.
AQUA scores ranged from 1.21 to 171.83 for Aurora A and from 4.81 to 158.45 for Aurora B, with a median score of 25.81 and 23.37, respectively. The staining patterns of Aurora A and B are shown in Fig. 2.
For each of the markers, the AQUA scores from the set of two slides were combined to give a single data set. Of the 638 patient tumor histospots on each slide stained for Aurora A, 530 were interpretable for both cores and 590 were interpretable for one core. Of the 638 patient tumor histospots on each slide stained for Aurora B, 568 were interpretable for both cores and 575 were interpretable for one core. Tumor spots were deemed uninterpretable if they had insufficient tumor cells, loss of tissue in the spot, or an abundance of necrotic tissue. For patients who had two interpretable histospots, a composite score was formed by taking the average of the two scores. For patients with only one interpretable core, the single score was used. The combined data set for Aurora A had scores for 533 patients with survival information. For Aurora B, we obtained scores for 517 patients with survival information.
To assess the association between Aurora A and B expression and other commonly used clinical and pathologic variables, we did ANOVA analyses as shown in Table 1. High Aurora A scores were associated with high nuclear grade, low PR expression, high HER-2/neu expression, and lymph node involvement. High Aurora B was associated with early age at diagnosis, high nuclear grade, low ER expression, and lymph node involvement. By Spearman’s ρ test, a weak association was found between Aurora A and B (ρ = 0.1125; P = 0.0084).
Table 1.
Association between Aurora A and B expression and commonly used clinical and pathologic variables
| Variable | Aurora A
|
Aurora B
|
||
|---|---|---|---|---|
| t test | P | t test | P | |
| Tumor size >2 cm | 0.863 | 0.388 | 0.943 | 0.346 |
| Age at diagnosis <50 y | 1.358 | 0.175 | 4.521 | <0.0001 |
| Nuclear grade (2-3 vs 1) | 2.542 | 0.011 | 1.980 | 0.0483 |
| ER (0 vs 1-3) | 1.017 | 0.309 | 2.776 | 0.0057 |
| PR (0 vs 1-3) | 2.630 | 0.008 | 0.617 | 0.5378 |
| HER-2/neu (3 vs 0-2) | 2.861 | 0.004 | 0.447 | 0.655 |
| Nodal positivity | 2.06 | 0.039 | 2.87 | 0.004 |
Using the Cox univariate survival analysis of raw AQUA scores, we found that high total (nuclear and cytoplasmic) Aurora A expression was associated with decreased breast cancer–specific survival in the entire cohort (P = 0.0005) and the node-negative subset (P = 0.0035). There was no association between Aurora B and breast cancer–specific survival (P = 0.98). Supplementary Table S1 shows the associations between continuous scores for Aurora A and B and survival in the entire cohort and in the node-negative and node-positive subsets.
Continuous AQUA scores were then divided arbitrarily into quartiles, reflecting the use of routine statistical divisions in the absence of underlying justification for division of expression levels. Kaplan-Meier survival curves were generated for the Aurora A and B scores as shown in Fig. 3. Log-rank analysis revealed a statistically significant association with survival for Aurora A in the entire cohort (P = 0.0003) and in the node-negative subset (P = 0.0012) and an association of borderline significance in the node-positive subset (P = 0.041).
Fig. 3.
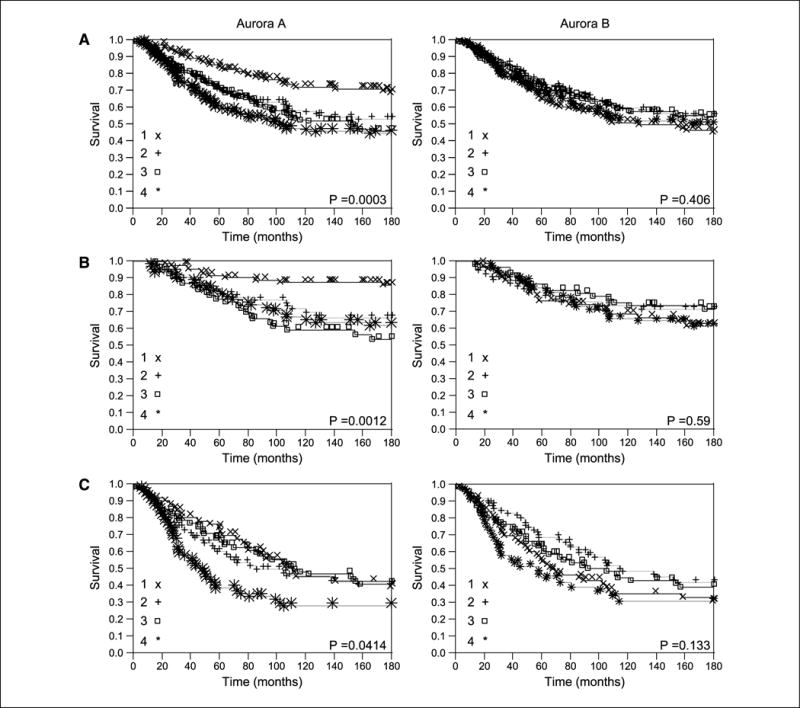
Kaplan-Meier survival curves for Aurora A and B AQUA scores divided by quartiles for the entire cohort of patients (A), node-negative patients (B), and node-positive patients (C).1, first quartile; 2, second quartile; 3, third quartile; 4, fourth quartile.
Using the Cox proportional hazards model, we did multivariate analysis to assess the independent predictive value of Aurora A expression. High Aurora A expression remained a strong independent prognostic marker as shown in Table 2. To further validate that this marker has added prognostic value to standard markers (nodal status, tumor size, nuclear grade, ER, PR, HER-2/neu, and age), we introduced a novel cross-validation graphical method for detecting differences between the predicted survival probability distribution of validation sets and the actual survival probability as displayed in Fig. 4. We randomly split the data set 10,000 times into training (80% of cases) and validation (20% of cases) sets. To simplify the graphical presentation, we reduced each validation set to have an equal number of survivors and nonsurvivors at 15 years. For each training set, we determined the regression coefficients of the multivariate Cox model and derived the survival probability for each of the cases in the corresponding validation set. Censored patients with survival times shorter than 15 years were excluded from this analysis. The curves in Fig. 4A show the fraction of cases that are still alive at 15 years (actual survival probability) in each of the 100 bins along the predicted survival probability axis (X axis). In an optimal survival model, the fraction of actual survivors should be equal to the predicted survival probability in the validation sets (dashed curve). To derive the other curves shown in Fig. 4A, we used three multivariate Cox models: (a) a model with four binary covariates, nodal status, age (<50 or ≥50), nuclear grade (1 versus 2-3), and tumor size (<2 or ≥2 cm); (b) a model with the above four covariates and ER, PR, and HER-2/neu; and (c) a model containing the continuous Aurora A scores and all the covariates in model b. Overall, the curve corresponding to model c is closer to the optimal survival model (dashed curve) than model b; in turn, model b is closer to the optimal curve than model a. To assess the quality of each of these models relative to the optimal model, we used two measures: (a) correlation between the curve associated with the model of interest and the optimal curve (the closer to 1, the better) and (b) the root mean square of the residuals between the curve of interest and the optimal curve (the closer to 0, the better). The correlations between the curves of models a to c and the optimal curve were 0.9041, 0.9410, and 0.9604, respectively. The root mean square scores of these three models were 0.0110, 0.0097, and 0.0079, respectively. To support these observations from a statistical standpoint, we repeated this analysis 100 times using a bootstrap procedure. For each bootstrap run, we evaluated the fraction of cases that are still alive at 15 years in each of the bins along the predicted survival probability axis. We then presented the results for models b and c using box plots for the first 20 bins in the low survival probability range (Fig. 4B). We did not include model a, as it was clearly inferior to models b and c. Figure 4B provides a convincing statistical display of the superiority of model c over model b in the predicted survival probability range of <0.2, as the actual survival probability (black squares) is contained within each of the interquartile ranges of model c (black boxes) but is underestimated by model b (gray boxes). We tested the significance of these observations as follows: (a) we computed a vector of the absolute distances between the predicted survival probabilities and the actual survival at the given bin of interest for model b and a similar vector for model c and (b) we applied the Wilcoxon rank-sum test for equal medians to these two vectors. In the poor prognostic range, model c was superior. For example, at the bin associated with the [0.11-0.12] predicted survival, the median of the absolute distances from the (actual survival) midpoint 0.115 in model b is 0.0844, whereas the corresponding median in model c is 0.0461. The P value for this observation is <0.00001, and similar P values were seen for other bins in the predicted survival probability range of <0.2.
Table 2.
Multivariable Cox proportional hazards analysis of Aurora A and other commonly used clinical and pathologic variables
| Variable | 95% Confidence interval | P |
|---|---|---|
| Tumor size >2 cm | 0.542-0.770 | <0.00001 |
| Age at diagnosis <50 y | 0.773-1.089 | 0.341 |
| Nuclear grade (1 vs 2-3) | 0.809-1.205 | 0.965 |
| ER (0 vs 1-3) | 1.044-1.444 | 0.0127 |
| PR (0 vs 1-3) | 0.915-1.243 | 0.410 |
| HER-2/neu (0-2 vs 3) | 0.666-1.050 | 0.116 |
| Nodal positivity | 0.531-0.731 | <0.00001 |
| Aurora A | 1.007-1.021 | 0.0001 |
Fig. 4.
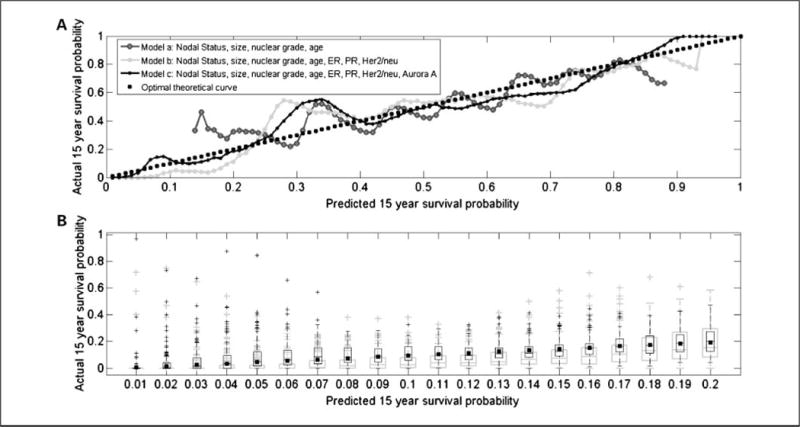
A, actual survival as a function of predicted survival probabilities evaluated in a cross-validation approach. The curves shown correspond to three multivariate Cox models: (a) a model with four binary covariates, nodal status, age (>50 or ≥50), nuclear grade (1 versus 2-3), and tumor size (<2 or ≥2 cm); (b) a model with the above four covariates and ER, PR, and HER-2/neu; and (c) a model containing the continuous Aurora A scores and all the covariates in model b. The quality of each model is detected visually in a manner similar to Q-Q plots and is estimated by similarity (correlation) or dissimilarity (root mean square of residuals) between the curve and the optimal dashed (y = x) line. The curve involving Aurora A has the highest correlation and lowest root mean square of residuals with respect to the optimal dashed (y = x) line. B, box plots of actual survival as a function of predicted survival probabilities evaluated using a bootstrap procedure applied to the cross-validation approach of A. Gray and black box plots, bootstrap results for models b and c, respectively. The bins shown on the X axis are associated with the low survival probability range of [0.0-0.2]. These bins correspond to the first 20 bins of A. The figure shows the superiority of model c over model b in this range, as the actual survival probability (black squares) is contained within each of the interquartile ranges of model c (black boxes) but underestimated by model b (gray boxes).
Discussion
In this work, we assessed expression of Aurora A and B in a quantitative fashion on a large cohort of primary breast cancer specimens and evaluated the association with breast cancer– specific survival. Our method of analysis gives objective and continuous measures of expression rather than routinely used pathologist-based divisions of staining into nominal scores of 0, 1, 2, and 3 or “positive/negative.” Our results were reproducible when using a second set of arrays with different cores from tumors of the same patients.
High Aurora A expression was strongly associated with decreased survival in the entire cohort (P = 0.0005) and the node-negative subset (P = 0.0035). The association with survival in the node-positive subset was not significant by Cox univariate analysis. High Aurora A expression retained its independence as a prognostic factor on multivariable analysis. There was no significant association between Aurora B expression and survival in our cohort.
A published study by Royce et al. found no association between survival and Aurora A staining in a cohort of 112 patients, of which 31% had node-negative disease (28). They did find an association of borderline significance (P = 0.05) between nuclear grade and Aurora A expression. Our results differ from those, and this might be attributable to the larger sample size in our study, the larger proportion of node-negative cases, or the ability of AQUA to detect subtle differences in expression not discernable by the naked eye when performing immunohistochemical analyses by 3,3′-diaminobenzidine staining.
In this work, we introduced a novel cross-validation graphical method for detecting differences between the predicted survival probability distribution of validation sets and the actual survival. The method provides further evidence that the addition of Aurora A scores leads to improvement in predicting survival probability. Moreover, this graphical method has additional advantages for interpreting the quality of the model, which could be especially useful for making treatment decisions for patients with very high and very low predicted survival probabilities. For instance, in Fig. 4A, in regions where the curve is above (or below) the optimal curve, the model is overpessimistic (or overoptimistic), because the actual survival is higher (or lower) than the predicted survival probability. In the model that involves Aurora A (model c), the instances with predicted survival probabilities >0.87 have even higher actual survival. Therefore, the certainty that they may not need chemotherapy is even greater than the model prediction. In addition, in the poor survival probability range of <0.2, the model that includes Aurora A is clearly superior to the model that does not (Fig. 4B), suggesting that these patients might benefit from an aggressive therapeutic approach.
Aurora A and B are ~70% homologous, yet their subcellular distribution and function differ. In our study, Aurora A staining was cytoplasmic and nuclear as has been reported in the literature (27, 28, 36). Aurora B staining was predominantly nuclear, again consistent with reports in the literature (37, 38). The lack of association between expression levels of these two Aurora family members in human breast tumors likely reflects their divergent functions as reviewed by Fu et al. (2).
Staging of primary breast cancer is important for determining prognosis and selection of patients for adjuvant chemotherapy, which reduces the risk of relapse and death. Molecular markers in the primary specimen that are highly associated with survival, such as Aurora A, could supplement standard staging information and enable us to identify node-negative breast cancer patients at high risk for disease recurrence, thus avoiding the toxicity associated with chemotherapy for the vast majority of patients who are cured by local therapy alone (39, 40). Similarly, such markers could be used to avoid aggressive therapy in the smaller subset of node-positive patients who are cured without additional systemic therapy (41).
In addition to a potentially important role as a prognostic marker, our findings have important implications for the therapeutic application of Aurora kinase inhibitors in breast cancer. At present, it is unclear whether there is an association between expression levels of Aurora A and B and response to Aurora kinase targeting therapy. Many (but not all) targeted therapies are more effective in tumors that overexpress the target, particularly when activity of the target is associated with disease aggression. The Aurora kinase inhibitors that have entered the clinic have variable specificity for the different family members. For example, PHA-739358 (Merck) is in phase I trials for solid tumors and inhibits Aurora A, AZD-1152 (AstraZeneca) inhibits Aurora B, and MK-0457 (Merck) is in phase II studies for colorectal cancer and inhibits Aurora A and B.4,5 In breast cancer, the association between high Aurora A expression and disease aggression suggests that Aurora A targeting agents might be more effective than Aurora B targeting therapies, and this will need to be validated in preclinical models and in breast cancer patients.
Careful and rational strategies are needed to target the Aurora kinases in breast cancer. For example, the fact that Aurora A expression was associated with decreased survival within the node-negative cohort suggests that the biological events, which are simultaneously permissive for metastatic behavior and also lead to high Aurora A expression, occur early in the course of the disease. High Aurora A expressers might respond differently to adjuvant chemotherapy than low expressers as was recently shown in an ovarian cancer study in which high Aurora A expressers had worse survival when receiving non-taxane-based adjuvant chemotherapy (36). Combined with our data, this suggests that the addition of agents that target Aurora A to adjuvant chemotherapy for poor prognosis node-negative or node-positive breast cancer with high Aurora A expression might decrease the likelihood of disease recurrence. If these agents are well tolerated, this approach will improve the therapeutic ratio for patients. Therefore, appropriate selection of patients, characterization of individual tumor biology, and perhaps intervention with additional rationally targeted agents might be necessary to achieve the best results.
In summary, we have shown a strong association between high Aurora A expression and decreased survival in primary breast cancer both among the entire patient cohort and among the node-negative patients. No association was seen between Aurora B expression and survival, suggesting that targeting Aurora A might be more beneficial in breast cancer. Prospective studies are needed to confirm the prognostic role of Aurora A. Further work is needed to establish the association between Aurora A expression and response to therapy that targets this molecule. All future clinical trials using Aurora A inhibitors for breast cancer should stratify patients based on Aurora A expression.
Supplementary Material
Acknowledgments
Grant support: NIH grant K0-8 ES11571 and the Breast Cancer Alliance (R.L. Camp), NIH grants R33 CA 106709 and R33 CA 110511 (D.L. Rimm), Susan G. Komen Foundation (H.M. Kluger, Y. Kluger) and the Breast Cancer Alliance (H.M. Kluger).
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
R.L. Camp and David L. Rimm are founders stockholders and consultants of HistoRx, a private corporation to which Yale University has given exclusive rights to produce and distribute software and technologies embedded in AQUA. Yale University retains patent rights for the AQUA technology. The other authors declare that they have no competing interests.
References
- 1.Keen N, Taylor S. Aurora-kinase inhibitors as anticancer agents. Nat Rev Cancer. 2004;4:927–36. doi: 10.1038/nrc1502. [DOI] [PubMed] [Google Scholar]
- 2.Fu J, Bian M, Jiang Q, Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res. 2007;5:1–10. doi: 10.1158/1541-7786.MCR-06-0208. [DOI] [PubMed] [Google Scholar]
- 3.Kimura M, Kotani S, Hattori T, et al. Cell cycle-dependent expression and spindle pole localization of a novel human protein kinase, Aik, related to Aurora of Drosophila and yeast Ipl1. J Biol Chem. 1997;272:13766–71. doi: 10.1074/jbc.272.21.13766. [DOI] [PubMed] [Google Scholar]
- 4.Terada Y, Tatsuka M, Suzuki F, Yasuda Y, Fujita S, Otsu M. AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J. 1998;17:667–76. doi: 10.1093/emboj/17.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura M, Matsuda Y, Yoshioka T, Sumi N, Okano Y. Identification and characterization of STK12/Aik2: a human gene related to Aurora of Drosophila and yeast IPL1. Cytogenet Cell Genet. 1998;82:147–52. doi: 10.1159/000015089. [DOI] [PubMed] [Google Scholar]
- 6.Adams RR, Carmena M, Earnshaw WC. Chromosomal passengers and the (Aurora) ABCs of mitosis. Trends Cell Biol. 2001;11:49–54. doi: 10.1016/s0962-8924(00)01880-8. [DOI] [PubMed] [Google Scholar]
- 7.Carmena M, Earnshaw WC. The cellular geography of Aurora kinases. Nat Rev Mol Cell Biol. 2003;4:842–54. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 8.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 9.Bischoff JR, Anderson L, Zhu Y, et al. A homologue of Drosophila Aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–65. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katayama H, Ota T, Jisaki F, et al. Mitotic kinase expression and colorectal cancer progression. J Natl Cancer Inst. 1999;91:1160–2. doi: 10.1093/jnci/91.13.1160. [DOI] [PubMed] [Google Scholar]
- 11.Tatsuka M, Katayama H, Ota T, et al. Multinuclearity and increased ploidy caused by overexpression of the Aurora- and Ipl1-like midbody-associated protein mitotic kinase in human cancer cells. Cancer Res. 1998;58:4811–6. [PubMed] [Google Scholar]
- 12.Zhou H, Kuang J, Zhong L, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–93. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 13.Sasai K, Katayama H, Stenoien DL, et al. Aurora-C kinase is a novel chromosomal passenger protein that can complement Aurora-B kinase function in mitotic cells. Cell Motil Cytoskeleton. 2004;59:249–63. doi: 10.1002/cm.20039. [DOI] [PubMed] [Google Scholar]
- 14.Harrington EA, Bebbington D, Moore J, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–7. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 15.Marumoto T, Zhang D, Saya H. Aurora-A—a guardian of poles. Nat Rev Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- 16.Warner SL, Bearss DJ, Han H, Von Hoff DD. Targeting Aurora-2 kinase in cancer. Mol Cancer Ther. 2003;2:589–95. [PubMed] [Google Scholar]
- 17.Ditchfield C, Johnson VL, Tighe A, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–80. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauf S, Cole RW, La Terra S, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–94. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dewar H, Tanaka K, Nasmyth K, Tanaka TU. Tension between two kinetochores suffices for their bi-orientationonthe mitotic spindle. Nature. 2004;428:93–7. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Zhou YX, Qiao W, et al. Overexpression of Aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene. 2006;25:7148–58. doi: 10.1038/sj.onc.1209707. [DOI] [PubMed] [Google Scholar]
- 21.Cox DG, Hankinson SE, Hunter DJ. Polymorphisms of the AURKA (STK15/Aurora kinase) gene and breast cancer risk (United States) Cancer Causes Control. 2006;17:81–3. doi: 10.1007/s10552-005-0429-9. [DOI] [PubMed] [Google Scholar]
- 22.Fletcher O, Johnson N, Palles C, et al. Inconsistent association between the STK15 F31I genetic polymorphism and breast cancer risk. J Natl Cancer Inst. 2006;98:1014–8. doi: 10.1093/jnci/djj268. [DOI] [PubMed] [Google Scholar]
- 23.Dai Q, Cai QY, Shu XO, et al. Synergistic effects of STK15 gene polymorphisms and endogenous estrogen exposure in the risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:2065–70. [PubMed] [Google Scholar]
- 24.Ouchi M, Fujiuchi N, Sasai K, et al. BRCA1 phosphorylation by Aurora-A in the regulation of G2 to M transition. J Biol Chem. 2004;279:19643–8. doi: 10.1074/jbc.M311780200. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka T, Kimura M, Matsunaga K, Fukada D, Mori H, Okano Y. Centrosomal kinase AIK1 is overexpressed in invasive ductal carcinoma of the breast. Cancer Res. 1999;59:2041–4. [PubMed] [Google Scholar]
- 26.Miyoshi Y, Iwao K, Egawa C, Noguchi S. Association of centrosomal kinase STK15/BTAK mRNA expression with chromosomal instability in human breast cancers. Int J Cancer. 2001;92:370–3. doi: 10.1002/ijc.1200. [DOI] [PubMed] [Google Scholar]
- 27.Hoque A, Carter J, Xia W, et al. Loss of Aurora A/STK15/BTAK overexpression correlates with transition of in situ to invasive ductal carcinoma of the breast. Cancer Epidemiol Biomarkers Prev. 2003;12:1518–22. [PubMed] [Google Scholar]
- 28.Royce ME, Xia W, Sahin AA, et al. STK15/Aurora-A expression in primary breast tumors is correlated with nuclear grade but not with prognosis. Cancer. 2004;100:12–9. doi: 10.1002/cncr.11879. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz-Cortes ZT, Kimmins S, Monaco L, Burns KH, Sassone-Corsi P, Murphy BD. Estrogen mediates phosphorylation of histone H3 in ovarian follicle and mammary epithelial tumor cells via the mitotic kinase, Aurora B. Mol Endocrinol. 2005;19:2991–3000. doi: 10.1210/me.2004-0441. [DOI] [PubMed] [Google Scholar]
- 30.Kamagata C, Tsuji N, Moriai M, Kobayashi D, Watanabe N. 15-Deoxy-Δ(12,14)-prostaglandin J2 inhibits G2-M phase progression in human breast cancer cells via the down-regulation of cyclin B1 and survivin expression. Breast Cancer Res Treat. 2007;102:263–73. doi: 10.1007/s10549-006-9336-3. [DOI] [PubMed] [Google Scholar]
- 31.Soncini C, Carpinelli P, Gianellini L, et al. PHA-680632, a novel Aurora kinase inhibitor with potent antitumoral activity. Clin Cancer Res. 2006;12:4080–9. doi: 10.1158/1078-0432.CCR-05-1964. [DOI] [PubMed] [Google Scholar]
- 32.Tchatchou S, Wirtenberger M, Hemminki K, et al. Aurora kinases A and B and familial breast cancer risk. Cancer Lett. 2007;247:266–72. doi: 10.1016/j.canlet.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Camp RL, Dolled-Filhart M, King BL, Rimm DL. Quantitative analysis of breast cancer tissue microarrays shows that both high and normal levels of HER2 expression are associated with poor outcome. Cancer Res. 2003;63:1445–8. [PubMed] [Google Scholar]
- 34.Camp RL, Chung GG, Rimm DL. Automated sub-cellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–7. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 35.Dolled-Filhart M, McCabe A, Giltnane J, Cregger M, Camp RL, Rimm DL. Quantitative in situ analysis of h-catenin expression in breast cancer shows decreased expression is associated with poor outcome. Cancer Res. 2006;66:5487–94. doi: 10.1158/0008-5472.CAN-06-0100. [DOI] [PubMed] [Google Scholar]
- 36.Lassmann S, Shen Y, Jutting U, et al. Predictive value of Aurora-A/STK15 expression for late stage epithelial ovarian cancer patients treated by adjuvant chemotherapy. Clin Cancer Res. 2007;13:4083–91. doi: 10.1158/1078-0432.CCR-06-2775. [DOI] [PubMed] [Google Scholar]
- 37.Zeng WF, Navaratne K, Prayson RA, Weil RJ. Aurora B expression correlates with aggressive behaviour in glioblastoma multiforme. J Clin Pathol. 2007;60:218–21. doi: 10.1136/jcp.2006.036806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurai M, Shiozawa T, Shih HC, et al. Expression of Aurora kinases A and B in normal, hyperplastic, and malignant human endometrium: Aurora B as a predictor for poor prognosis in endometrial carcinoma. Hum Pathol. 2005;36:1281–8. doi: 10.1016/j.humpath.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 39.Fisher B, Dignam J, Wolmark N, et al. Tamoxifen and chemotherapy for lymph node-negative, estrogen receptor-positive breast cancer. J Natl Cancer Inst. 1997;89:1673–82. doi: 10.1093/jnci/89.22.1673. [DOI] [PubMed] [Google Scholar]
- 40.Fisher B, Anderson S, Tan-Chiu E, et al. Tamoxifen and chemotherapy for axillary node-negative, estrogen receptor-negative breast cancer: findings from National Surgical Adjuvant Breast and Bowel Project B-23. J Clin Oncol. 2001;19:931–42. doi: 10.1200/JCO.2001.19.4.931. [DOI] [PubMed] [Google Scholar]
- 41.Fisher B, Redmond C, Legault-Poisson S, et al. Postoperative chemotherapy and tamoxifen compared with tamoxifen alone in the treatment of positive-node breast cancer patients aged 50 years and older with tumors responsive to tamoxifen: results from the National Surgical Adjuvant Breast and Bowel Project B-16. J Clin Oncol. 1990;8:1005–18. doi: 10.1200/JCO.1990.8.6.1005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


