Abstract
Nonalcoholic fatty liver disease (NAFLD) is becoming the major chronic liver disease in many countries. Its pathogenesis is multifactorial but twin and familial studies indicate significant heritability, which is not fully explained by currently-known genetic susceptibility loci. Notably, mutations in genes encoding nuclear lamina proteins, including lamins, cause lipodystrophy syndromes that include NAFLD. We hypothesized that variants in lamina-associated proteins predispose to NAFLD and used a candidate gene-sequencing approach to test for variants in 10 nuclear lamina-related genes in a cohort of 37 twin and sibling pairs: 21 individuals with, and 53 without, NAFLD. Twelve heterozygous sequence variants were identified in four lamina-related genes (ZMPSTE24/TMPO/SREBF1/SREBF2). The majority of NAFLD patients (>90%) had at least one variant, compared to <40% of controls (P<0.0001). When only insertions/deletions and changes in conserved residues were considered, the difference between the groups was similarly striking (>80% versus <25%; P<0.0001). Presence of a lamina variant segregated with NAFLD independent of the PNPLA3 I148M polymorphism. Several variants were found in TMPO, which encodes the lamina-associated polypeptide-2 (LAP2) that has not previously been associated with liver disease. One of these, a frameshift insertion that generates truncated LAP2, abrogated lamin-LAP2 binding, caused LAP2 mislocalization, altered endogenous lamin distribution, increased lipid droplet accumulation after oleic acid treatment in transfected cells, and led to cytoplasmic association with the ubiquitin-binding protein p62/SQSTM1.
Conclusion
Novel variants in nuclear lamina-related genes were identified in a cohort of twins and siblings with NAFLD. One novel variant, which results in a truncated LAP2 protein and a dramatic phenotype in cell culture, represents the first association of TMPO/LAP2 variants with NAFLD and underscores the potential importance of the nuclear lamina in NAFLD.
Keywords: steatosis, lipodystrophy, laminopathy, nucleus, genetics
Introduction
Nonalcoholic fatty liver disease (NAFLD) is now the most common form of liver disease in many countries and includes a spectrum from simple steatosis to steatohepatitis which can lead to fibrosis and ultimately cirrhosis (1–4). It is estimated that NAFLD-related cirrhosis may soon be the most common indication for liver transplant listing in the United States (5). Effective therapies for NAFLD are limited (1,6), in part due to an incomplete understanding of its pathogenesis. Twin and familial aggregation studies suggest up to 50% heritability which is not accounted for by the susceptibility loci identified to date (7–11). One possible explanation for this discrepancy is that there are genetic variants that predispose to development of NAFLD but have not yet been identified.
More than 70 human diseases have been linked to mutations in genes that encode intermediate filament proteins (IF), with no effective targeted therapy (12–14). While there are >60 cytoplasmic IF, the nuclear IF include the A- and B-type lamins which are encoded by three genes, LMNA/LMNB1/LMNB2. Lamins form the major components of the nuclear lamina, which is intimately associated with the inner nuclear membrane and helps maintain the nuclear structural integrity while providing a link between the cytoskeleton and the genome (15).
At least 15 disorders are caused by mutations in the genes encoding lamins (15–17). These diseases affect a number of organ systems, and in some cases the same mutation results in different phenotypes (18,19), suggesting that genetic modifiers and/or tissue-specific regulation of gene expression impact disease phenotypes. A subset of patients with germline mutations in LMNA develop partial lipodystrophy syndromes characterized by aberrant body fat distribution, hyperlipidemia, insulin resistance, and hepatic steatosis (20,21). Notably, in a French cohort of 87 patients with metabolic syndrome, including many with NAFLD, three patients harbored heterozygous mutations in LMNA or ZMPSTE24 (of five tested genes that encode A-type lamins or enzymes involved in lamin maturation), and >10% of the tested patients had abnormal leukocyte nuclear morphology (22,23).
Given the nearly universal liver disease in patients with lamin-related lipodystrophy and the abnormal nuclear morphology in some patients with metabolic syndrome and NAFLD (22), we hypothesized that some patients with NAFLD, but without lipodystrophy, might have variants in genes encoding lamins or lamina-related proteins that predispose to their development of NAFLD. We examined a cohort of twin and sibling pairs with NAFLD and identified several heterozygous variants in lamina-related genes, including a novel truncation variant in TMPO, which encodes lamina-associated polypeptide 2 (LAP2). This truncation manifests a dramatic phenotype in cell culture and is the first association of TMPO/LAP2 variants with NAFLD.
Materials and Methods
Study design and participants
Study participants were recruited through the NAFLD Research Center (University of California, San Diego), as part of a prospective study of twin pairs as previously reported (7,9,11) (see Supplementary Methods). The majority (>80%) of the cohort consisted of twin pairs, with the remainder consisting of sibling-sibling pairs; while non-twin pairs were excluded from the NAFLD heritability analysis described previously (7), they were included in this study to increase the probability of detecting rare variants. Hepatic steatosis was quantified noninvasively by magnetic resonance imaging-determined proton-density fat-fraction (MRI-PDFF), with NAFLD defined as MRI-PDFF ≥5% without apparent secondary cause of hepatic steatosis (e.g., alcohol or steatogenic medication use, other liver disease causes). Liver fibrosis was quantified by MR elastography-determined stiffness (MRE-stiffness) (7). All participants provided written informed consent, and the research protocol was approved by the Institutional Review Board. No samples were obtained from executed prisoners or other institutionalized persons.
Next-generation DNA sequencing for variant identification
Genomic DNA was isolated using the Qiagen (Redwood City, CA) DNeasy Blood and Tissue Kit. Next-generation sequencing was performed with a custom-designed TruSeq amplicon panel using a MiSeq instrument (Illumina, San Diego, CA). Variants were annotated based on RefSeq v.69 gene models and matched with 1000 Genomes Phase-3 population frequency data and dbNSFP v2.0 functional annotations (24,25). Variants were confirmed by Sanger DNA sequencing using primers designed to amplify 400–600 base-pair regions of genomic DNA flanking variants of interest. The same primers were used for PCR amplification and sequencing (Table S1).
PNPLA3 variant genotyping
A subset of twin and sibling pairs were previously genotyped for the PNPLA3 I148M variant (7). The remainder were genotyped using a Thermo Fisher (Waltham, MA) SNP Genotyping assay (catalog number 4351379).
Plasmids and transfection
Plasmids containing human TMPO (LAP2α) and LMNA open reading frames were purchased from Origene (Rockville, MD). The open reading frames were subcloned into pCMV6 vectors with amino-terminal green fluorescent protein (GFP) or myc/DDK tag (Origene) using SgfI and MluI restriction enzymes, with a stop codon engineered immediately 3′ to the MluI site. Mutants of LAP2α were generated using the QuikChange II site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). The truncated variant of LAP2 (1-99) was generated via PCR amplification of base-pairs 1–297 of the TMPO open reading frame using primers engineered with flanking SgfI and MluI restriction sites, then subcloned into pCMV6 with GFP or myc/DDK tag similar to full-length LAP2α. Lipofectamine-2000 (Life Technologies, Carlsbad, CA) was used for transfection of Huh7 cells (American Type Culture Collection, Manassas, VA) (plated at ~60–70% confluency 1d prior to transfection).
Immunofluorescence
Two days after transfection, cells were washed, then fixed with a 1:1 acetone and methanol mixture (−20°C, 10min). Following fixation, washing then blocking, primary antibodies (Table S2) were added (overnight, 4°C). After washing, fluorescently-tagged secondary antibody (Alexa Fluor® 488 or 594; Thermo Fisher) was added (1h, 22°C), followed by DNA staining using 4′,6-diamidino-2-phenylindole (DAPI) (Life Technologies). Stained cells were visualized with a Zeiss AXIO Imager.M2 microscope, and images were acquired with a 40X objective.
Fatty acid treatment
Oleic acid (Sigma-Aldrich, St. Louis, MO) stock solution (50mM) was prepared in isopropanol (26). Huh7 cells were plated on lysine-coated glass chamber slides and transfected 1d after plating. After 2d, the culture medium was changed to DMEM containing 1% fatty acid-free BSA (Sigma-Aldrich) and oleic acid (500 μM) or vehicle. Cells were fixed (4% paraformaldehyde, 15min, 22°C) and permeabilized (0.1% Triton X-100, 5min). Blocking, antibody incubation, and DAPI mounting were performed, and 10μM BODIPY 493/503 (Thermo Fisher) was added at the primary antibody step and incubated overnight (4°C). Lipid staining was quantified by counting and measuring intracellular lipid droplets using the Zeiss Zen 2.3 lite software package (Fig. S1).
Semi-native and denaturing gels
Cells were solubilized in lysis buffer (1% Triton X-100, 50mM Tris pH 8.0, 150mM NaCl, protease inhibitor cocktail (Sigma-Aldrich)). After centrifugation (12,000xg, 10min, 4°C), the supernatant protein concentration was determined (bicinchoninic acid assay, Thermo Fisher). The supernatant was then diluted in Novex 2X Tris-glycine sample buffer with (denaturing gels) or without (semi-native gels) SDS (Invitrogen, Carlsbad, CA) with or without β-mercaptoethanol (2%). In parallel, the Triton-insoluble pellet was solubilized in SDS-containing sample buffer (95°C, 2min). Proteins were resolved using 4–20% Novex Tris-glycine native gels (Invitrogen) with running buffer containing 0.1% SDS, and visualized by silver (Thermo Fisher) or Coomassie staining.
Immunoprecipitation and immunoblotting
GFP or GFP-tagged lamin A was co-expressed in cultured cells with myc-tagged LAP2α. Two days after transfection, cells were solubilized in Triton lysis buffer (4°C, 20min), and GFP or GFP-tagged lamin A was immunoprecipitated using anti-GFP antibody and Dynabeads protein G (Invitrogen). Immunoprecipitates were resolved via SDS-PAGE and transferred to polyvinylidene fluoride membranes (Bio-Rad, Hercules, CA) for immunoblotting. Where indicated, data were quantified by densitometry using ImageJ version 1.4.3.67.
LAP2-binding protein identification by tandem mass spectrometry
Proteins were eluted by incubating the immunoprecipitates in 50μL of 0.1M glycine (pH2.5) with shaking (10min, 22°C), then addition of an equal volume of 1M Tris (pH8) prior to trypsin digestion and analysis by LC-MS/MS.
Statistical analysis
For continuous data, the two-tailed Student’s t-test or the Mann-Whitney U test was used to assess statistical significance. For categorical data, a two-tailed Fisher’s exact test was used.
Results
Study participants
Thirty-seven (37) twin and sibling pairs underwent intake history, serologic evaluation, MRI, and lamina-related gene sequencing. Within this group of 74 subjects (Table 1), 21 met criteria for NAFLD (MRI-PDFF ≥5% without another apparent cause of hepatic steatosis). This NAFLD group consisted of five concordant twin pairs (4 monozygotic, 1 dizygotic), one concordant sibling pair, and 9 members of discordant twin (4 monozygotic, 3 dizygotic) or sibling (2) pairs. This NAFLD group was compared with 53 subjects (Controls) without NAFLD (19 concordant twin pairs, 3 concordant sibling-sibling pairs, and 9 members of discordant twin or sibling pairs). Cases and Controls differed in body mass index, hemoglobin A1c, alanine aminotransferase (ALT) levels, and MRI scores (Table 1). Age, sex, race, and ethnicity showed no significant differences between the two groups.
Table 1.
Characteristics of twin and sibling cohort (37 pairs, n=74 subjects). S.D., standard deviation; BMI, body mass index; Hgb, hemoglobin.
| Parameter | Cases (n = 21) [with NAFLD] | Controls (n = 53) [without NAFLD] | P-value |
|---|---|---|---|
| Age (S.D.) | 52.1 (19.6) | 46.1 (20.6) | 0.26 |
| % Male | 57 | 32 | 0.07 |
| % Caucasian | 67 | 51 | 0.30 |
| % Hispanic | 24 | 15 | 0.27 |
| BMI (S.D.) | 32.3 (5.0) | 27.2 (5.4) | < 0.001 |
| Hgb A1c (S.D.) | 6.1 (0.7) | 5.7 (0.4) | 0.001 |
| ALT (S.D.) | 30 (18) | 21 (12) | 0.01 |
| MRI-PDFF (S.D.) | 10.7 (5.4) | 2.6 (0.9) | < 0.0001 |
Identification of variants in lamina-related genes
Given the frequency of hepatic steatosis and steatohepatitis among patients with lipodystrophy syndromes caused by LMNA mutations (20,21) and the abnormal nuclear morphology in some individuals with metabolic syndrome (22), we hypothesized that variants might be found in genes encoding lamins, lamin-related proteins, or nuclear lamina-associated proteins in patients with NAFLD. To address this, we performed exon-directed sequencing of ten genes encoding lamins, lamin-binding proteins (including two transcription factors involved in lipid homeostasis that bind lamins), and lamin-processing enzymes (LMNA/LMNB2/ZMPSTE24/ICMT/FNTA/FNTB/TMPO/BANF1/SREBF1/SREBF2, Table S3). Sequencing was performed with an amplicon-based platform, achieving an average read coverage of 1100x across the 10 gene target with ~400 variants identified across all samples. Of these, <10% were predicted to result in an amino acid change in the encoded protein; these were confirmed via Sanger sequencing (Fig. S2).
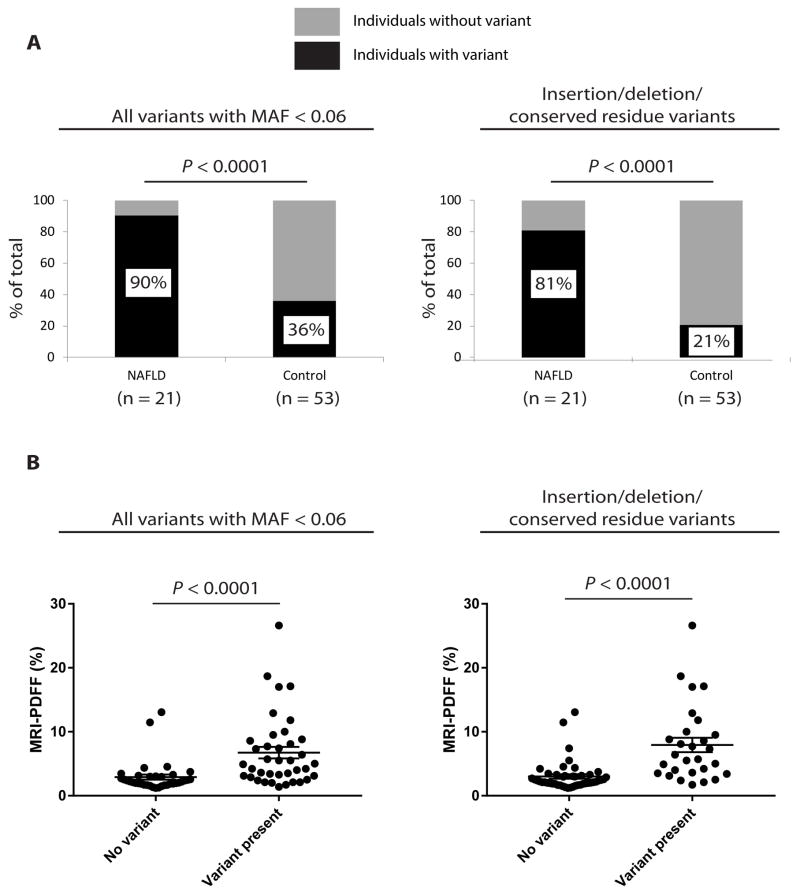
Twelve unique variants in four genes were validated (Table 2). Of these, SREBF2 G595A was previously found to have a minor allele frequency (MAF) in the general population of 0.41 and, therefore, was excluded from further analyses; the remainder were novel or have MAF < 0.06 (24). Among these, novel insertion variants in TMPO and SREBF2 were identified in two individuals with NAFLD. The TMPO insertion was also present in the monozygotic twin without NAFLD, while the SREBF2 insertion was not present in the unaffected sibling. While no single genetic variant was significantly associated with NAFLD in this cohort after Bonferroni correction, collectively the nuclear lamina genetic variants were found preferentially in study participants with NAFLD. The majority of the patients with NAFLD (19 of 21, 90%) were heterozygous for a variant in at least one lamina-related gene, versus 19 of 53 (36%) controls (P<0.0001) (Fig. 1A, left panel). When only variants predicted to result in insertion/deletion or change in a conserved amino acid were included (Fig. 1A, right panel), 17 of 21 NAFLD patients (81%) carried such a variant, compared to 11 of 53 (21%) controls (P<0.0001). Similarly, the hepatic fat content (quantitated by MRI-PDFF) was compared in individuals without a lamina-related variant versus subjects with a variant (Fig. 1B). The MRI-PDFF in the former group was 2.91±0.41% versus 6.74±0.90% in the latter group (mean±SEM;P<0.0001); when only variants predicted to result in an insertion/deletion or change in a conserved residue were included, the findings were similar: 3.02±0.34% for subjects without versus 7.93±1.12% for subjects with a variant (P<0.0001) (Fig. 1B). Comparable results were obtained when subgroup analysis of the twin pairs (Fig. S3), monozygotic twin pairs (Fig. S4A), dizygotic twin pairs (Fig. S4B), and non-twin sibling pairs (not shown) was performed separately. Similarly, when an additional 24 subjects (15 NAFLD cases, 9 controls) who were recruited as part of the same study, but not part of a twin or sibling pair, were included in the analysis, the conclusions were unchanged: 23 of 36 cases had a variant versus 24 of 62 controls (P=0.02); 20 of 36 cases had an insertion/deletion or conserved residue variant versus 14 of 62 controls (P=0.002) (Fig. S5).
Table 2.
Confirmed coding variants. Minor allele frequency (MAF) cited in far right column is derived from the 1000 Genomes Project (reference 24). Ins, insertion; fs, frameshift.
| Gene | Nucleotide change | Amino acid change | Conserved residue | Number of cases with variant | Number of controls with variant | Minor allele frequency |
|---|---|---|---|---|---|---|
| ZMPSTE24 | C > T | L438F | Yes | 2 | 0 | 0.001 |
| TMPO | InsA | T99fs | N/A | 1 | 1 | Not identified |
| TMPO | G > A | R274K | No | 0 | 1 | 0.002 |
| TMPO | C > G | T317S | Yes | 0 | 1 | 0.030 |
| TMPO | C > G | Q599E | Yes | 11 | 7 | 0.06 |
| TMPO | C > T | R690C | Yes | 1 | 0 | 0.011 |
| SREBF1 | C > T | V610M | Yes | 1 | 1 | 0.007 |
| SREBF2 | Ins (24bp) | S72Ins (in-frame) | N/A | 1 | 0 | Not identified |
| SREBF2 | G > A | R371K | Yes | 0 | 2 | 0.001 |
| SREBF2 | G > C | G595A | No | 16 | 23 | 0.41 |
| SREBF2 | G > C | R860S | No | 6 | 7 | 0.06 |
| SREBF2 | G > A | R1080Q | Yes | 1 | 0 | 0.001 |
Figure 1.
Variants in lamina-related genes were predominantly found in individuals with NAFLD in a cohort of twin and sibling pairs. (A) Left panel: Percentages of individuals (y-axis) with and without genetic variants are shown for both NAFLD cases and Controls (includes all coding variants with minor allele frequency < 0.06). Right panel: Percentages of individuals (y-axis) with and without a variant resulting in insertion/deletion or change in a conserved residue are shown for NAFLD cases and Controls. Fisher’s exact test was used to assess statistical significance at a threshold of P<0.05. (B) Left panel: Scatter plot of liver fat content (assessed by MRI-PDFF) of individuals without and with a lamina-related genetic variant (includes all coding variants with minor allele frequency < 0.06). Right panel: Scatter plot of liver fat content (assessed by MRI-PDFF) of individuals without and with a lamina-related genetic variant (includes variants resulting in insertion/deletion or change in a conserved residue). Error bars represent standard error of the mean. Mann-Whitney U test was used to assess statistical significance at a threshold of P<0.05.
In addition to traditional MRI, some study participants underwent MR elastography (MRE) to assess the extent of hepatic fibrosis. Within the group of 37 twin/sibling pairs (n=74 subjects), 20 of the 21 subjects with NAFLD underwent MRE. Of these, 18 of 20 (90%) had a variant in a lamina-related gene, but this high number prevented a meaningful assessment of the effect of variants on MRE-stiffness in the NAFLD group. Within the other group of 24 subjects who were not part of a twin/sibling pair (referenced above), there were 15 subjects with NAFLD, of whom 13 underwent MRE. When these 13 were combined with the 20 twins and siblings with NAFLD (n=33 total subjects with NAFLD who underwent MRE), there was a trend toward those having significant or advanced fibrosis (MRE-stiffness >3.66 kPa or >4.11 kPa, respectively (27)) being more likely to carry lamina-related variants compared to controls (P=0.2 and 0.06, respectively) (Fig. S6).
Effect of PNPLA3 I148M genotype
A single nucleotide polymorphism (C>G) in PNPLA3, which encodes patatin-like phospholipase domain containing 3, resulting in an I148M change, is a well-described susceptibility allele for NAFLD (28,29). To address whether there was any interaction between PNPLA3 genotype and the presence of lamina-related variants in our cohort, the PNPLA3 I148M polymorphism was genotyped in the 37 twin and sibling pairs. Notably, presence of one or two G alleles was not significantly associated with NAFLD in our cohort (odds ratio conferred by having at least one G allele=2.48, 95% confidence interval 0.88–7.00, P=0.09), similar to what was previously noted (7). In contrast, the NAFLD odds ratio associated with a lamina variant was 17.0 (95% confidence interval 3.6–81.0, P<0.001), and the effect of a lamina variant was highest in subjects lacking a PNPLA3 G allele (n=40): MRI-PDFF 2.40±0.17% in subjects without a lamina variant versus 5.57±0.87% in those with lamina variants (P<0.001) (Fig. S7A). Among those carrying at least one G allele (n=34), the effect of a lamina-related variant was also significant: MRI-PDFF 4.08±1.25% in those subjects without a variant versus 7.49±1.37% in those with variants (P=0.02). Similar findings were observed in subjects lacking a PNPLA3 G allele when only the 31 twin pairs were examined (Fig. S7B), although within this subgroup the effect of a lamina variant was not as prominent among subjects carrying a PNPLA3 G allele (P=0.06).
Multiple variants in TMPO
Several variants were identified in TMPO (Table 2) and were over-represented in subjects with NAFLD (13 of 21) compared to controls (10 of 53), a difference which remained significant after Bonferroni correction (P=0.0006 by Fisher’s exact test, with a threshold of P<0.005 given that ten genes were tested). TMPO encodes six isoforms (α,β,γ,δ,ε,ζ) of lamina-associated polypeptide 2 (LAP2) that are produced via alternative splicing (Fig. S8). The longest isoform is known as LAP2α and was previously found to interact with lamin A/C via its carboxy terminus (30). Notably, the majority of the variants we identified in TMPO were in the portion of the gene that is unique to the α-isoform. The lone exception was a novel single base pair insertion (c.287_288insA) which was identified in one set of monozygotic 41 year-old twins (one twin with NAFLD and one without) and is located in the amino-terminal region common to all isoforms. This insertion is predicted to cause a frameshift and premature stop codon after Thr 99, resulting in truncation of all LAP2 isoforms.
Two variants of LAP2 impair interaction with lamin A
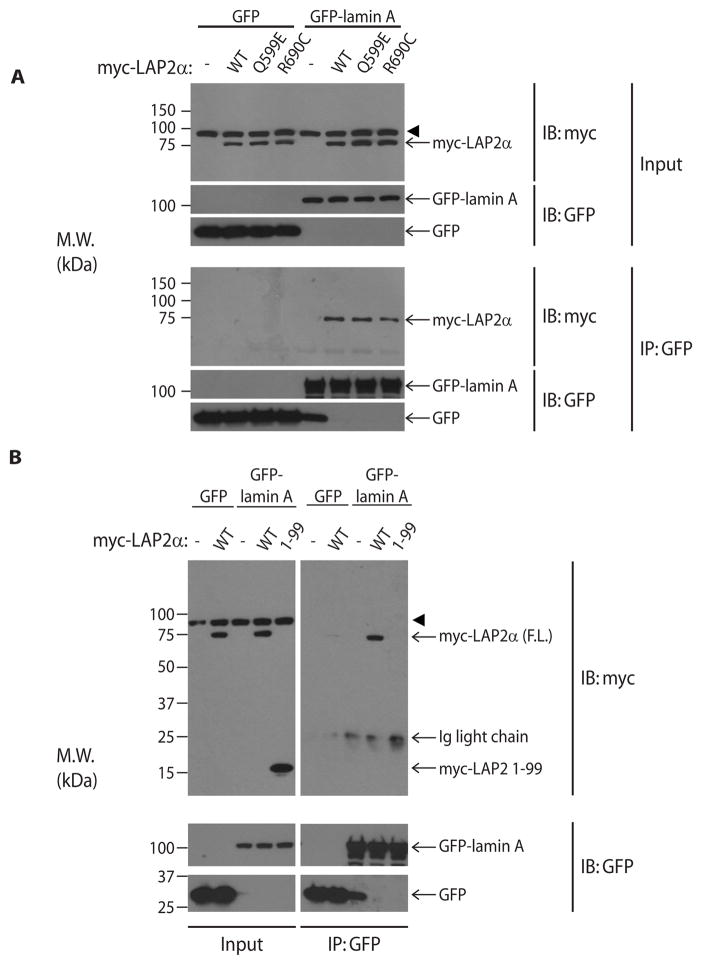
The α-isoform of LAP2 is largely found in the nucleoplasm (rather than directly associated with the inner nuclear membrane) and binds and stabilizes nucleoplasmic lamin A (30–32). Previous studies have narrowed the lamin-binding domain of LAP2α to residues 601–694 (Fig. S8) (30). Hence, the novel truncated variant of LAP2 (LAP2 1-99) lacks the lamin-binding domain. In addition, one other variant in TMPO that was found in a patient with NAFLD (R690C) results in a charge change in a conserved residue and was previously reported to affect lamin binding in vitro (33). Interestingly, this variant was previously identified in a family with hereditary cardiomyopathy but has not been linked to liver disease (33). To address whether these variants of LAP2α affect binding to lamin A, we co-expressed GFP-tagged human lamin A with wild-type or mutant myc-tagged LAP2α in Huh7 cells and performed co-immunoprecipitation using anti-GFP antibody. GFP-tagged lamin A, but not GFP alone, readily co-precipitated wild-type LAP2α (Fig. 2A). In agreement with a previous report showing reduced binding to the lamin A tail in vitro (33), LAP2α R690C exhibited a modest, though not statistically significant (quantitation via densitometry of three independent experiments – data not shown), reduction in co-precipitation with lamin A (Fig. 2A), while a second variant of LAP2α (Q599E) located outside the lamin-binding region co-precipitated with lamin A similarly to wild-type LAP2α. In contrast, truncation of LAP2 at amino acid 99 completely abrogated binding to lamin A, despite similar expression to wild-type LAP2α (Fig. 2B).
Figure 2.
Variants of LAP2 found in NAFLD patients interfere with binding to lamin A. (A) Huh7 cells were co-transfected with myc-tagged wild-type (WT) or variant LAP2α and GFP-tagged lamin A, followed by immunoprecipitation using an antibody directed to the GFP tag. Immunoprecipitates were resolved on SDS-PAGE, and precipitated proteins were visualized after immunoblotting with antibodies to myc or the GFP tag. (B) Huh7 cells were co-transfected with GFP-tagged lamin A and WT LAP2α or its truncated variant (LAP2 1-99). Lamin A was immunoprecipitated, followed by visualization as in panel A. Ig, immunoglobulin; M.W., apparent molecular weight; kDa, kilodaltons; IP, immunoprecipitation; IB, immunoblot. Arrowhead indicates a non-specific band recognized by the myc antibody.
Full-length, but not truncated, LAP2α forms a soluble, detergent-resistant, high molecular weight species
Previous work demonstrated that the carboxy-terminal portion of LAP2α can dimerize or trimerize in vitro and that multimeric LAP2α migrates as a high molecular species on semi-native SDS-PAGE (34,35). Given that lamin A forms intra- and intermolecular disulfide bonds (36), we hypothesized that multimeric species of LAP2α might also be disulfide-linked. To address this, we expressed myc-tagged LAP2α in Huh7 cells, then performed SDS-PAGE under semi-native, denaturing/non-reducing, and denaturing/reducing conditions. Similar to prior findings (35), >50% of LAP2α migrated as high-molecular weight species (>250 kDa) under semi-native conditions (not shown), with identical results under denaturing non-reducing conditions (Fig. S9A). In contrast, under reducing conditions, >90% of LAP2α migrated as a single species (~75 kDa) consistent with monomeric LAP2α (Fig. S9A). These data suggest that a substantial proportion of LAP2α can exist as disulfide-linked multimeric complexes.
We hypothesized that truncated LAP2 1-99 might differ from wild-type LAP2α in its ability to form high-molecular-weight species, as the truncated protein lacks cysteines. In contrast to full-length LAP2α, LAP2 1-99 formed no disulfide-linked high-molecular-weight species (Fig. S9A). Notably, the LAP2α R690C variant migrated as high-molecular-weight species similar to wild-type LAP2α, consistent with a prior report (35). In addition, >50% of high-molecular-weight LAP2α species were Triton-soluble, which did not vary between wild-type LAP2α and the R690C variant. In contrast, LAP2 1-99, which did not form detectable high-molecular-weight species in the Triton-soluble fraction, formed β-mercaptoethanol-resistant insoluble high-molecular-weight forms not seen with wild-type LAP2α or any point-variant tested (Fig. S9B). Of note, although the high-molecular-weight smear formed by truncated LAP2 1-99 appears more prominent after β-mercaptoethanol treatment (Fig. S9B), quantitation of four independent experiments showed no significant difference in the relative amount of high-molecular-weight species with or without β-mercaptoethanol treatment.
Truncation of LAP2 disrupts its nuclear localization and increases lipid accumulation compared to full-length LAP2α
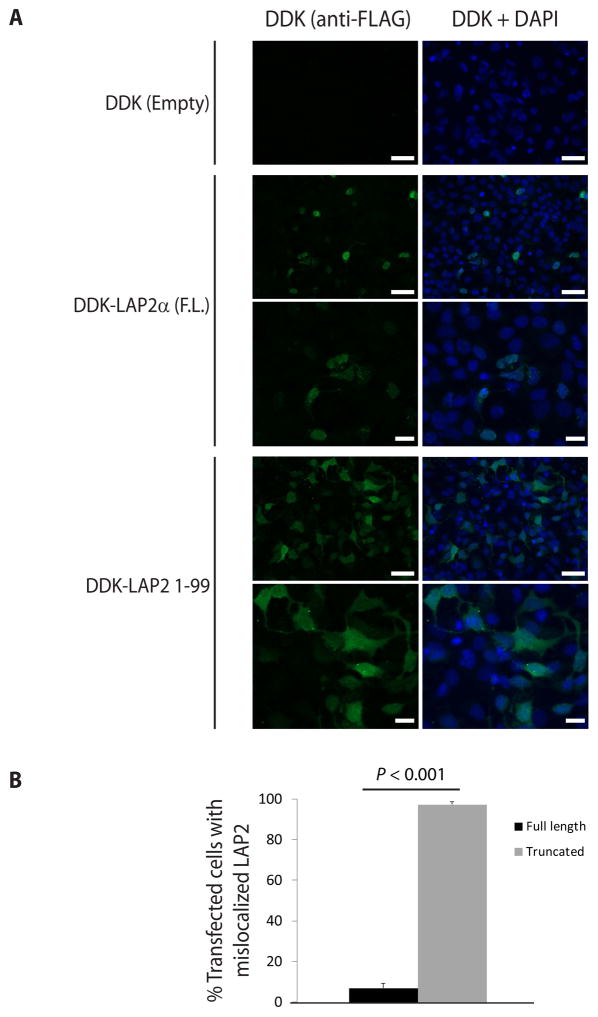
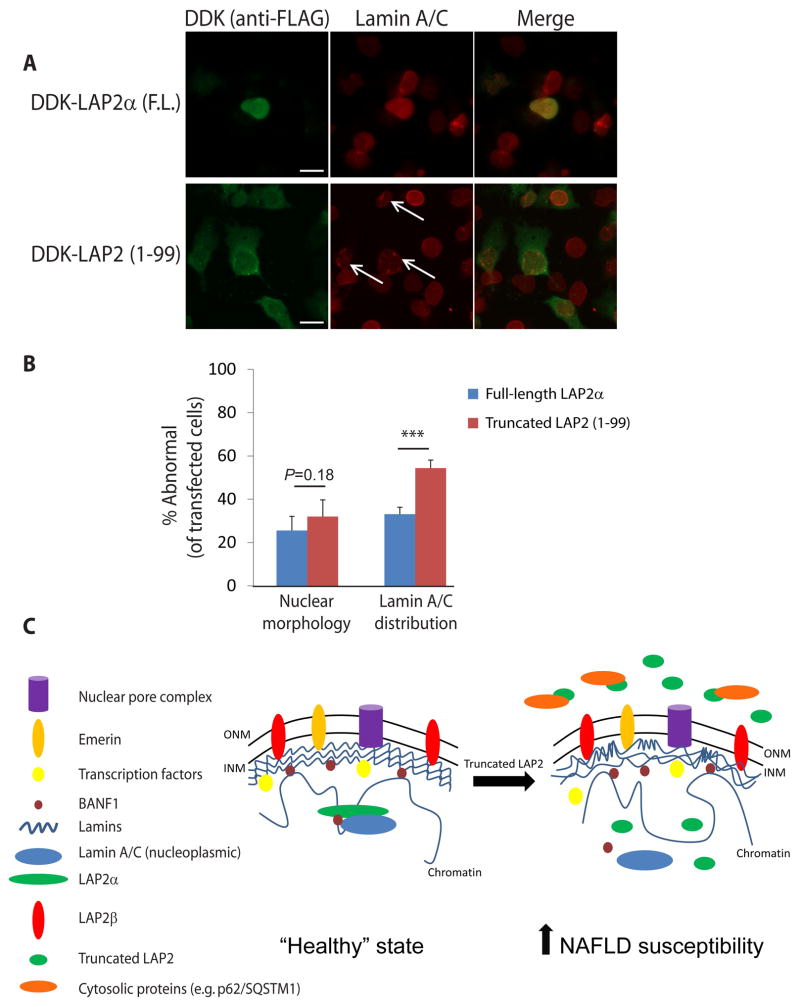
In prior studies of LAP2 in cultured cells, a truncated protein containing amino acids 1–187 (generated to study LAP2 domains) lost exclusive targeting to the nucleus, suggesting that the mature protein region required for nuclear localization is C-terminal to residue 187 (37). Hence, we reasoned that LAP2 1-99 would also be mislocalized and addressed this by examining DDK-tagged full-length LAP2α or LAP2 1-99 localization in Huh7 cells. As expected, full-length LAP2α localized exclusively to the nucleus, while LAP2 1-99 was found throughout the nucleus and cytoplasm under identical conditions (Fig. 3). Similar results were obtained in transfected human lung adenocarcinoma (A549) cells and baby hamster kidney (BHK) cells (data not shown).
Figure 3.
The truncated variant of LAP2 is mislocalized in transfected cells. (A) Huh7 cells were transfected with full-length (F.L.) LAP2α or truncated LAP2 (1-99) with DDK tag, or empty vector. Cells were then fixed, and LAP2 was visualized by indirect immunofluorescence using anti-FLAG antibody (which recognizes the DDK tag). Scale bar: 50 μm (lower magnification images: first, second, and fourth rows), 20 μm (higher magnification images: third and fifth rows). (B) Cells transfected as in panel A were scored according to whether LAP2 was specifically localized to the nucleus or mislocalized throughout the cell (represented as percent of transfected cells with mislocalized LAP2). Data were derived from counting 9–12 high-power fields from three independent experiments. Error bars represent the standard error of the mean. Student’s t test was used to determine statistical significance at a threshold of P<0.05.
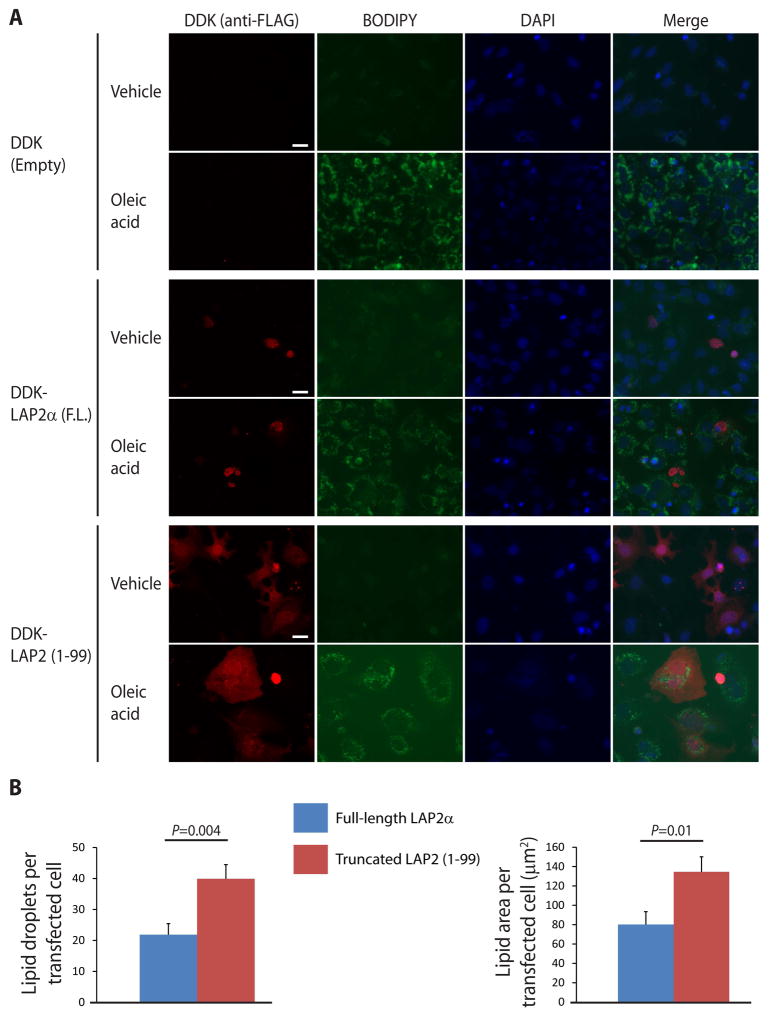
To address whether LAP2 1-99 might contribute to NAFLD development by facilitating lipid accumulation in hepatocytes, we treated Huh7 cells expressing full-length LAP2α or LAP2 1-99 with oleic acid and performed lipid staining. After overnight incubation with oleic acid, lipid droplet accumulation in cells transfected with truncated LAP2 (1-99) was similar to that in adjacent untransfected cells (quantitation showed no statistically significant difference between these two groups of cells), but was significantly greater than in cells transfected with full-length LAP2α (Fig. 4).
Figure 4.
Truncated LAP2 causes increased lipid accumulation in transfected cells. (A) Huh7 cells were transfected with DDK-tagged full-length (F.L.) LAP2α or truncated LAP2 (1-99) or empty vector, then treated with 500 μM oleic acid or vehicle (isopropanol) in serum-free medium overnight. After fixation, transfected LAP2 was visualized by indirect immunofluorescence using anti-FLAG antibody. Lipid droplets were stained with BODIPY 493/593 as described in Materials and Methods. Representative images are shown for each condition; scale bar, 20 μm. (B) Lipid accumulation was quantitated as described in Materials and Methods for >10 high-power fields for each condition, and the data shown are representative of 3 independent experiments. Error bars represent standard error of the mean. Student’s t test was used to determine statistical significance at a threshold of P<0.05.
Truncated LAP2 has multiple unique interaction partners in Huh7 cells
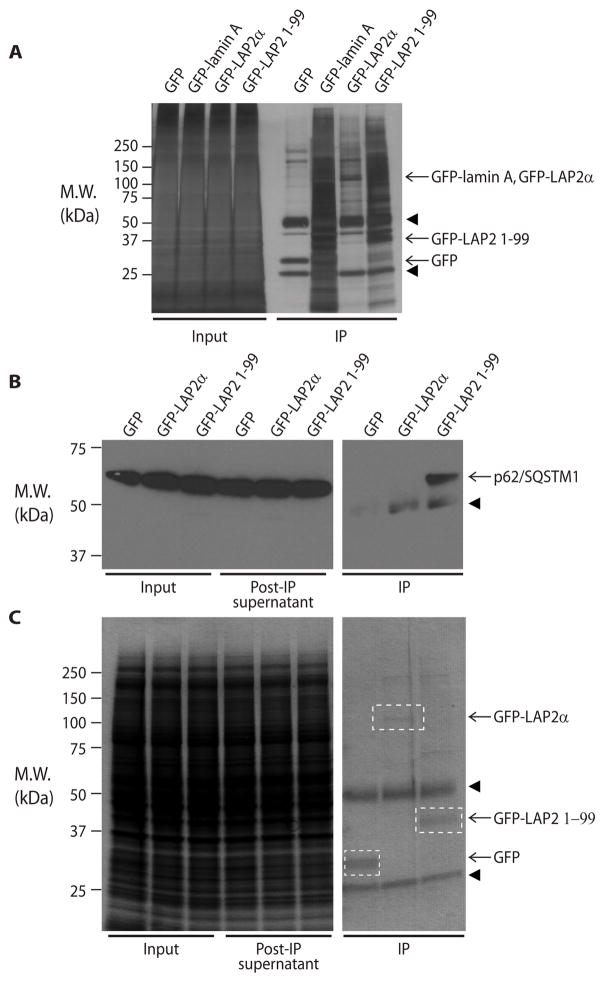
Given that LAP2 1-99 exhibited unique biochemical properties compared to full-length LAP2α (Fig. 2, Fig. S9), and was mislocalized in cultured cells (Fig. 3) and led to increased lipid accumulation compared to full-length LAP2α in oleic acid-treated cells (Fig. 4), we hypothesized that truncated LAP2 might exhibit unique protein-protein interactions compared to the full-length protein. To address this, we immunoprecipitated GFP-tagged LAP2α or LAP2 1-99 from Huh7 cell lysates and carried out mass spectrometry analysis of the immunoprecipitates and associated endogenous proteins (Fig. 5A). Numerous unique interacting partners for truncated LAP2 1-99 as compared to full-length protein were detected, many of which were cytoplasmic proteins (Table S4). We selected the cytoplasmic ubiquitin-binding protein p62/SQSTM1 for validation because it ranked first among 235 identified LAP2-associated proteins after sorting by the ratio of spectra obtained with truncated LAP2 1-99 compared to full-length LAP2α, then by percent coverage, and because it was reported to regulate lipogenesis in mouse liver (38). Notably, endogenous p62/SQSTM1 interacted preferentially with LAP2 1-99 as determined by co-immunoprecipitation of GFP-tagged LAP2 followed by immunoblotting with anti-p62 antibody (Fig. 5B,C).
Figure 5.
Truncated LAP2 has multiple unique cytoplasmic interaction partners, including p62/SQSTM1. (A) Huh7 cells were transfected with GFP-tagged LAP2α or truncated LAP2 (1-99), followed by immunoprecipitation using an anti-GFP antibody; empty vector (GFP only) and GFP-tagged lamin A were included as controls. The immunoprecipitates and the input cell lysates were visualized by silver staining. (B) Huh7 cells were transfected with GFP alone, GFP-tagged LAP2α, or truncated LAP2 (1-99), followed by immunoprecipitation of LAP2 using anti-GFP antibody. Co-precipitated p62/SQSTM1 was visualized by immunoblotting. (C) Immunoprecipitated GFP and GFP-tagged proteins were visualized by Coomassie staining for the samples shown in panel B. M.W., apparent molecular weight; kDa, kilodaltons; IP, immunoprecipitates. Arrowheads indicate antibody heavy (~50 kDa) and light (~25 kDa) chains.
Expression of truncated LAP2 alters endogenous lamin distribution in Huh7 cells
Given the unique biochemical properties of truncated LAP2 (Fig. 2,3,5; Fig. S9), its effect on lipid accumulation in cells (Fig. 4), and prior reports demonstrating a role for LAP2α in regulating lamin A/C organization (31,32), we hypothesized that LAP2 truncation might lead to altered lamin A/C organization. To address this, we expressed wild-type LAP2α and LAP2 1-99 in Huh7 cells and examined endogenous lamin A/C distribution (Fig. 6A). Although LAP2 1-99 did not affect nuclear shape at the level of resolution we tested, more abnormal punctate and globular lamin A/C staining was noted in nuclei of cells transfected with LAP2 1-99 as compared to full-length LAP2α (Fig. 6B). A comparable effect was noted for B-type lamins under the same conditions (Fig. S10).
Figure 6.
Truncated LAP2 causes altered lamin A/C distribution. (A) Huh7 cells were transfected with DDK-tagged full-length (F.L.) LAP2α or truncated LAP2 (1-99). After fixation, transfected LAP2 and endogenous lamin A/C were visualized by immunofluorescence using anti-FLAG and anti-lamin A/C antibodies, respectively. Representative high-magnification images are shown; nuclei of transfected cells with abnormal lamin A/C staining (punctate/globular) are highlighted by arrows. Scale bar, 20 μm. (B) Nuclear morphology and lamin A/C distribution in cells transfected with full-length LAP2α or truncated LAP2 (1-99) were scored in a blinded fashion from three independent experiments (3–6 fields/condition/experiment, >85 total nuclei/condition). Error bars represent standard error of the mean. Student’s t test was used to determine statistical significance; ***, P<0.001. (C) Schematic of alterations predicted to occur due to expression of LAP2 1-99, thereby predisposing to NAFLD via ectopic protein-protein interactions (e.g., LAP2 1-99 with p62/SQSTM1), altered lamin A/C distribution, chromatin reorganization, and increased lipid accumulation in hepatocytes. ONM, outer nuclear membrane; INM, inner nuclear membrane.
Discussion
Large-scale genome-wide association studies (GWAS) in individuals with NAFLD have identified several susceptibility loci (8,39), though these loci cannot account for the high degree of heritability suggested by twin and familial aggregation studies (7,8,10,11). Given that a subset of patients with mutations in LMNA develop lipodystrophy, metabolic syndrome, and NAFLD (20,21), as noted in mice that lack lamin A/C expression in hepatocytes (40), we reasoned that other variants in genes encoding lamin-related and lamina-associated proteins might be found in patients with NAFLD, and that rare variants that escape detection via GWAS might be identified by candidate gene sequencing approaches.
Here we report a set of variants, several of which are novel, in genes encoding lamina-related proteins in a cohort of twins and siblings with NAFLD. The majority result in a single amino acid change, while two are insertion variants (each of which was identified in one patient). Collectively these variants conferred a significantly increased risk of NAFLD within this cohort (odds ratio 17.0, P<0.001). In our patient cohort, the PNPLA3 I148M polymorphism did not have a significant NAFLD effect, as noted previously (7), likely due to the small size of the cohort in relation to the frequency of this variant in the general population. Importantly, the effect of the lamina-related variants was most prominent among subjects with wild-type PNPLA3 (CC genotype), but the interaction of lamina-related variants with PNPLA3 genotypes will need to be assessed in a larger population.
Among the variants we identified, one set of twins with NAFLD carried a point mutation in ZMPSTE24 (L438F), which encodes a protease involved in lamin A processing, that was previously identified in individuals with metabolic syndrome (22,41). Notably, over one-third of the variants were in TMPO, most of which were located in the portion of the gene that is unique to the α-isoform of the encoded protein (LAP2). Of these, one was previously reported to cause dilated cardiomyopathy in one family (33) but has not previously been associated with metabolic disease. No disease association has previously been described for any of the other variants in TMPO, including the truncation at Thr99. The NAFLD patient with LAP2 1-99 has a monozygotic twin with normal MRI-PDFF, underlining the importance of environmental factors in NAFLD and suggesting that this truncation may represent a predisposition rather than a high-penetrance cause of NAFLD. However, the possibility of other NAFLD patients carrying TMPO variants will need to be investigated, and it is not possible to make definitive genotype-phenotype conclusions because of the small numbers and the lack of longitudinal and clinical/lifestyle information, as the involved twins were lost to follow-up after their initial enrollment in 2012 (Table S5).
The dramatic effects of truncated LAP2 1-99 in transfected cells, including its altered intracellular distribution, alteration of endogenous lamin distribution, and increased lipid droplet accumulation, support a role for this variant in predisposition to liver disease. Truncated LAP2 robustly co-precipitated endogenous p62/SQSTM1, which regulates lipogenic gene expression in mouse liver (38), but the functional importance of this and the other unique protein-protein interactions identified via mass spectrometry remain to be defined. In addition, the effects of truncated LAP2 on both A- and B-type lamins (Fig. 6, Fig. S10) are noteworthy given previous studies showing that Lmnb1 and Lmnb2 depletion in hepatocytes did not lead to misshapen nuclei as determined by liver tissue staining but did lead to nuclear blebbing of cultured hepatocytes (42). It is unknown whether these mice develop spontaneous hepatosteatosis upon aging or have increased susceptibility to fibrosis upon high fat feeding as was noted in hepatocyte-specific Lmna-null mice (40). However, liver involvement was reported in a patient with acquired partial lipodystrophy due to LMNB2 mutation (23,43). In addition, alterations of lamins A/C and B1/B2 occur in the context Mallory-Denk body formation and porphyria-associated liver injury (44,45).
Limitations of our findings include the relatively small number of participants which precludes statistical significance for any individual variant. Nevertheless, the fact that we identified several variants, even in our small cohort, underscores the potential importance of the nuclear lamina in NAFLD and the need for further study. Another limitation is that non-invasive assessment of liver disease was used rather than liver biopsy. While the accuracy of MRI in assessing hepatic steatosis is well-established (46), the lack of histologic data precludes definitive conclusions about liver disease severity. Still, among study participants who underwent MRE, there was a trend toward higher MRE-stiffness among those subjects carrying lamina-related variants (Fig. S6). This suggests the possibility that these variants might contribute to liver disease progression but such a conclusion requires study in larger cohorts.
The data presented here have a number of implications for the study of nuclear lamina function, lamina-related disease, and NAFLD. Our findings underscore the importance of proper localization and interaction of lamins and lamin-binding proteins at the nuclear lamina. Prior studies in cell culture systems and mice have illustrated the role of LAP2α-lamin interaction in regulating the stability, localization, and function of lamin A/C (31,32). Our findings are the first to suggest a potential link of TMPO variants with liver disease, possibly via impaired interaction with lamin A. In this context, one of the TMPO variants identified herein, the R690C variant, was previously reported to cause dilated cardiomyopathy (33), or to serve as a genetic modifier in a family with dilated cardiomyopathy (47), without the description of liver disease. Identification of this variant in an individual with NAFLD underscores the variability in laminopathy phenotypes and the likelihood that genetic modifiers influence both the severity of disease and the affected organ(s). In addition, extrapolation of the number of patients in our small cohort with variants (19 of 21) in the ten genes sequenced raises the possibility that they might be relatively common among patients with familial NAFLD. We also posit that some patients with nonfamilial NAFLD might have an unrecognized laminopathy, which may have future therapeutic implications as drugs are developed to target laminopathies (48). Further study in larger patient cohorts will be needed to clarify the frequency and relative contributions of variants in lamina-related genes in patients with NAFLD. Taken together, our data suggest several mechanisms by which lamina-related variants such as truncated LAP2 1-99 might promote susceptibility to NAFLD via inappropriate interaction with cytoplasmic proteins including p62/SQSTM1, altered lamin distribution (which could alter chromatin organization (49) and cause downstream transcriptional changes (50)), and promotion of lipid accumulation in hepatocytes (Fig. 6C).
Supplementary Material
Acknowledgments
Financial Support: The work was supported by National Institutes of Health (NIH) grant R01 DK47918 (M.B.O.); NIH training grant T32 DK094775 (G.F.B.); American Liver Foundation (ALF) Liver Scholar Award (G.F.B.); and by institutional grant P30 DK034933 to the University of Michigan.
The authors would like to thank Dr. Robert Lyons and Jeanne Geskes of the University of Michigan DNA Sequencing core for their assistance with next-generation DNA sequencing, and members of the Omary laboratory for helpful discussions and critical review of the manuscript.
Abbreviations
- ALT
alanine aminotransferase
- DAPI
4′,6-diamidino-2-phenylindole
- GFP
green fluorescent protein
- IF
intermediate filament proteins
- LAP2
lamina-associated polypeptide 2
- MAF
minor allele frequency
- MRE
magnetic resonance elastography
- MRI
magnetic resonance imaging
- MRI-PDFF
MRI-determined proton-density fat-fraction
- NAFLD
nonalcoholic fatty liver disease
- PAGE
polyacrylamide gel electrophoresis
- TMPO
thymopoietin
Footnotes
Potential conflict of interest: None to report.
References
Author names in bold designate co-first authorship.
- 1.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology. 2012;55:2005–2023. doi: 10.1002/hep.25762. [DOI] [PubMed] [Google Scholar]
- 2.Michelotti GA, Machado MV, Diehl AM. NAFLD, NASH and liver cancer. Nat Rev Gastroenterol Hepatol. 2013;10:656–665. doi: 10.1038/nrgastro.2013.183. [DOI] [PubMed] [Google Scholar]
- 3.Sattar N, Forrest E, Preiss D. Non-alcoholic fatty liver disease. BMJ. 2014;349:g4596. doi: 10.1136/bmj.g4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marengo A, Jouness RI, Bugianesi E. Progression and Natural History of Nonalcoholic Fatty Liver Disease in Adults. Clin Liver Dis. 2016;20:313–324. doi: 10.1016/j.cld.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 5.Wong RJ, Aguilar M, Cheung R, Perumpail RB, Harrison SA, Younossi ZM, Ahmed A. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology. 2015;148:547–555. doi: 10.1053/j.gastro.2014.11.039. [DOI] [PubMed] [Google Scholar]
- 6.Sanyal AJ, Chalasani N, Kowdley KV, McCullough A, Diehl AM, Bass NM, et al. Pioglitazone, vitamin E, or placebo for nonalcoholic steatohepatitis. N Engl J Med. 2010;362:1675–1685. doi: 10.1056/NEJMoa0907929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loomba R, Schork N, Chen CH, Bettencourt R, Bhatt A, Ang B, et al. Heritability of Hepatic Fibrosis and Steatosis Based on a Prospective Twin Study. Gastroenterology. 2015;149:1784–1793. doi: 10.1053/j.gastro.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anstee QM, Seth D, Day CP. Genetic Factors That Affect Risk of Alcoholic and Nonalcoholic Fatty Liver Disease. Gastroenterology. 2016;150:1728–1744. e1727. doi: 10.1053/j.gastro.2016.01.037. [DOI] [PubMed] [Google Scholar]
- 9.Zarrinpar A, Gupta S, Maurya MR, Subramaniam S, Loomba R. Serum microRNAs explain discordance of non-alcoholic fatty liver disease in monozygotic and dizygotic twins: a prospective study. Gut. 2016;65:1546–1554. doi: 10.1136/gutjnl-2015-309456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grove JI, Austin M, Tibble J, Aithal GP, Verma S. Monozygotic twins with NASH cirrhosis: cumulative effect of multiple single nucleotide polymorphisms? Ann Hepatol. 2016;15:277–282. doi: 10.5604/16652681.1193726. [DOI] [PubMed] [Google Scholar]
- 11.Cui J, Chen CH, Lo MT, Schork N, Bettencourt R, Gonzalez MP, et al. Shared genetic effects between hepatic steatosis and fibrosis: A prospective twin study. Hepatology. 2016;64:1547–1558. doi: 10.1002/hep.28674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Omary MB. “IF-pathies”: a broad spectrum of intermediate filament-associated diseases. J Clin Invest. 2009;119:1756–1762. doi: 10.1172/JCI39894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butin-Israeli V, Adam SA, Goldman AE, Goldman RD. Nuclear lamin functions and disease. Trends Genet. 2012;28:464–471. doi: 10.1016/j.tig.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun J, Groppi VE, Gui H, Chen L, Xie Q, Liu L, Omary MB. High-Throughput Screening for Drugs that Modulate Intermediate Filament Proteins. Methods Enzymol. 2016;568:163–185. doi: 10.1016/bs.mie.2015.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burke B, Stewart CL. The nuclear lamins: flexibility in function. Nat Rev Mol Cell Biol. 2013;14:13–24. doi: 10.1038/nrm3488. [DOI] [PubMed] [Google Scholar]
- 16.Worman HJ, Fong LG, Muchir A, Young SG. Laminopathies and the long strange trip from basic cell biology to therapy. J Clin Invest. 2009;119:1825–1836. doi: 10.1172/JCI37679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davidson PM, Lammerding J. Broken nuclei--lamins, nuclear mechanics, and disease. Trends Cell Biol. 2014;24:247–256. doi: 10.1016/j.tcb.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor MR, Fain PR, Sinagra G, Robinson ML, Robertson AD, Carniel E, et al. Natural history of dilated cardiomyopathy due to lamin A/C gene mutations. J Am Coll Cardiol. 2003;41:771–780. doi: 10.1016/s0735-1097(02)02954-6. [DOI] [PubMed] [Google Scholar]
- 19.Van Esch H, Agarwal AK, Debeer P, Fryns JP, Garg A. A homozygous mutation in the lamin A/C gene associated with a novel syndrome of arthropathy, tendinous calcinosis, and progeroid features. J Clin Endocrinol Metab. 2006;91:517–521. doi: 10.1210/jc.2005-1297. [DOI] [PubMed] [Google Scholar]
- 20.Guenantin AC, Briand N, Bidault G, Afonso P, Bereziat V, Vatier C, et al. Nuclear envelope-related lipodystrophies. Semin Cell Dev Biol. 2014;29:148–157. doi: 10.1016/j.semcdb.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 21.Ajluni N, Meral R, Neidert AH, Brady GF, Buras E, McKenna B, et al. Spectrum of disease associated with partial lipodystrophy: lessons from a trial cohort. Clin Endocrinol (Oxf) 2017;86:698–707. doi: 10.1111/cen.13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dutour A, Roll P, Gaborit B, Courrier S, Alessi MC, Tregouet DA, et al. High prevalence of laminopathies among patients with metabolic syndrome. Hum Mol Genet. 2011;20:3779–3786. doi: 10.1093/hmg/ddr294. [DOI] [PubMed] [Google Scholar]
- 23.Omary MB. Intermediate filament proteins of digestive organs: physiology and pathophysiology. Am J Physiol Gastrointest Liver Physiol. 2017;312:G628–G634. doi: 10.1152/ajpgi.00455.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.1000 Genomes Project Consortium. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Jian X, Boerwinkle E. dbNSFP v2. 0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34:E2393–2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malhi H, Bronk SF, Werneburg NW, Gores GJ. Free fatty acids induce JNK-dependent hepatocyte lipoapoptosis. J Biol Chem. 2006;281:12093–12101. doi: 10.1074/jbc.M510660200. [DOI] [PubMed] [Google Scholar]
- 27.Singh S, Venkatesh SK, Wang Z, Miller FH, Motosugi U, Low RN, et al. Diagnostic performance of magnetic resonance elastography in staging liver fibrosis: a systematic review and meta-analysis of individual participant data. Clin Gastroenterol Hepatol. 2015;13:440–451. e446. doi: 10.1016/j.cgh.2014.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Speliotes EK, Butler JL, Palmer CD, Voight BF, et al. GIANT Consortium, MIGen Consortium. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 2010;52:904–912. doi: 10.1002/hep.23768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dechat T, Korbei B, Vaughan OA, Vlcek S, Hutchison CJ, Foisner R. Lamina-associated polypeptide 2alpha binds intranuclear A-type lamins. J Cell Sci. 2000;113(Pt 19):3473–3484. doi: 10.1242/jcs.113.19.3473. [DOI] [PubMed] [Google Scholar]
- 31.Naetar N, Korbei B, Kozlov S, Kerenyi MA, Dorner D, Kral R, et al. Loss of nucleoplasmic LAP2alpha-lamin A complexes causes erythroid and epidermal progenitor hyperproliferation. Nat Cell Biol. 2008;10:1341–1348. doi: 10.1038/ncb1793. [DOI] [PubMed] [Google Scholar]
- 32.Gesson K, Vidak S, Foisner R. Lamina-associated polypeptide (LAP)2alpha and nucleoplasmic lamins in adult stem cell regulation and disease. Semin Cell Dev Biol. 2014;29:116–124. doi: 10.1016/j.semcdb.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor MR, Slavov D, Gajewski A, Vlcek S, Ku L, Fain PR, et al. Thymopoietin (lamina-associated polypeptide 2) gene mutation associated with dilated cardiomyopathy. Hum Mutat. 2005;26:566–574. doi: 10.1002/humu.20250. [DOI] [PubMed] [Google Scholar]
- 34.Bradley CM, Jones S, Huang Y, Suzuki Y, Kvaratskhelia M, Hickman AB, et al. Structural basis for dimerization of LAP2alpha, a component of the nuclear lamina. Structure. 2007;15:643–653. doi: 10.1016/j.str.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Snyers L, Vlcek S, Dechat T, Skegro D, Korbei B, Gajewski A, et al. Lamina-associated polypeptide 2-alpha forms homo-trimers via its C terminus, and oligomerization is unaffected by a disease-causing mutation. J Biol Chem. 2007;282:6308–6315. doi: 10.1074/jbc.M605782200. [DOI] [PubMed] [Google Scholar]
- 36.Pekovic V, Gibbs-Seymour I, Markiewicz E, Alzoghaibi F, Benham AM, Edwards R, et al. Conserved cysteine residues in the mammalian lamin A tail are essential for cellular responses to ROS generation. Aging Cell. 2011;10:1067–1079. doi: 10.1111/j.1474-9726.2011.00750.x. [DOI] [PubMed] [Google Scholar]
- 37.Vlcek S, Just H, Dechat T, Foisner R. Functional diversity of LAP2alpha and LAP2beta in postmitotic chromosome association is caused by an alpha-specific nuclear targeting domain. EMBO J. 1999;18:6370–6384. doi: 10.1093/emboj/18.22.6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popineau L, Morzyglod L, Carre N, Cauzac M, Bossard P, Prip-Buus C, et al. Novel Grb14-Mediated Cross Talk between Insulin and p62/Nrf2 Pathways Regulates Liver Lipogenesis and Selective Insulin Resistance. Mol Cell Biol. 2016;36:2168–2181. doi: 10.1128/MCB.00170-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwan R, Brady GF, Brzozowski M, Weerasinghe SV, Martin H, Park M-J, et al. Hepatocyte-specific deletion of mouse lamin A/C leads to male-selective steatohepatitis. Cell Mol Gastroenterol Hepatol. 2017 doi: 10.1016/j.jcmgh.2017.06.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galant D, Gaborit B, Desgrouas C, Abdesselam I, Bernard M, Levy N, et al. A Heterozygous ZMPSTE24 Mutation Associated with Severe Metabolic Syndrome, Ectopic Fat Accumulation, and Dilated Cardiomyopathy. Cells. 2016:5. doi: 10.3390/cells5020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang SH, Jung HJ, Coffinier C, Fong LG, Young SG. Are B-type lamins essential in all mammalian cells? Nucleus. 2011;2:562–569. doi: 10.4161/nucl.2.6.18085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao J, Li Y, Fu X, Luo X. A Chinese patient with acquired partial lipodystrophy caused by a novel mutation with LMNB2 gene. J Pediatr Endocrinol Metab. 2012;25:375–377. doi: 10.1515/jpem-2012-0007. [DOI] [PubMed] [Google Scholar]
- 44.Zatloukal K, Denk H, Spurej G, Hutter H. Modulation of protein composition of nuclear lamina. Reduction of lamins B1 and B2 in livers of griseofulvin-treated mice. Lab Invest. 1992;66:589–597. [PubMed] [Google Scholar]
- 45.Singla A, Griggs NW, Kwan R, Snider NT, Maitra D, Ernst SA, et al. Lamin aggregation is an early sensor of porphyria-induced liver injury. J Cell Sci. 2013;126:3105–3112. doi: 10.1242/jcs.123026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reeder SB, Sirlin CB. Quantification of liver fat with magnetic resonance imaging. Magn Reson Imaging Clin N Am. 2010;18:337–357. doi: 10.1016/j.mric.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zaragoza MV, Fung L, Jensen E, Oh F, Cung K, McCarthy LA, et al. Exome Sequencing Identifies a Novel LMNA Splice-Site Mutation and Multigenic Heterozygosity of Potential Modifiers in a Family with Sick Sinus Syndrome, Dilated Cardiomyopathy, and Sudden Cardiac Death. PLoS One. 2016;11:e0155421. doi: 10.1371/journal.pone.0155421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muchir A, Worman HJ. Targeting Mitogen-Activated Protein Kinase Signaling in Mouse Models of Cardiomyopathy Caused by Lamin A/C Gene Mutations. Methods Enzymol. 2016;568:557–580. doi: 10.1016/bs.mie.2015.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barton LJ, Soshnev AA, Geyer PK. Networking in the nucleus: a spotlight on LEM-domain proteins. Curr Opin Cell Biol. 2015;34:1–8. doi: 10.1016/j.ceb.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vidak S, Kubben N, Dechat T, Foisner R. Proliferation of progeria cells is enhanced by lamina-associated polypeptide 2alpha (LAP2alpha) through expression of extracellular matrix proteins. Genes Dev. 2015;29:2022–2036. doi: 10.1101/gad.263939.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.