Abstract
Evidence in many experimental systems supports the idea that non-uniform distributions of morphogen proteins encode positional information in developing tissues. There is also strong evidence that morphogen dispersal is mediated by cytonemes and that morphogen proteins transfer from producing to receiving cells at morphogenetic synapses that form at sites of cytoneme contacts. This essay considers some implications of this mechanism and its relevance to various contexts including large single cells such as the pre-cellular Drosophila embryo and the ciliate Stentor.
Keywords: morphogen, gradient, gene expression, cytoneme, bicoid
Although the diversity of forms in the animal and plant kingdoms may not rival the number of shapes of snowflakes, it is nevertheless vast. We understand that the apparently infinite variety of snowflakes arises from unique combinations of atmospheric temperature and humidity and properties of initiating particles, and it is accepted that the number of these combinations is unknowable. In contrast, the processes that generate shapes and patterns in biology control space and time with precision and reproducibility, despite the many uncontrollable variations in parameters that might influence them. The conceit of the cell and developmental biologist is that these processes are knowable and can be defined in ways that match conditions and context to outcome.
Starting from the premise that some form of positional information directs the development of spatial patterns (Wolpert, 2016, 1969), then we might describe our interest as seeking to understand the form in which positional information is encoded, how it distributes in space and time, and how it is interpreted. The interpretation and execution of programmed responses are issues of signal transduction and cell biology that will not be considered here. Instead I address the nature of position information and its distribution.
One of the great successes of developmental genetics has been the identification and characterization of morphogen proteins, molecules that transmit positional information in animal tissues. They are each produced by discrete sets of cells, they disseminate across adjacent regions to form concentration gradients, and as graded positional cues, they elicit concentration-dependent responses in the cells that regulate growth and patterning. They include the Wnt, Hedgehog (Hh), Bone morphogenic protein (BMP), Fibroblast growth factor (FGF), Epidermal growth factor (EGF), and Notch/Delta proteins whose structures and functions are conserved across the animal kingdom. Available evidence suggests that the Wnt, Hh, BMP, FGF, EGF and Notch/Delta signaling systems operate in most (or all) organ systems, but that they do not act systemically to script development and morphogenesis. Rather, they act locally, produced in every organ system by groups of cells (signaling centers / developmental organizers) that are designated to make a particular morphogen in an appropriate amount, place and time. Populated with multiple signaling centers that generate distinct spatial gradients, organ systems develop subject to the sum of the morphogen activities. The general model here is that these morphogen signals constitute the vocabulary of a universal language of development, and we can marvel at the elegance of a system that produces such a vast array of different forms with so few components. We can also make the following assertion: that its economy and virtuosity can only be possible because of the precision with which it can be regulated in space and time.
It has long been assumed that morphogens are released into extracellular fluid by producing cells, and that they move away by passive diffusion until they encounter receptors on receiving cells. There are a number of reasons to have reservations about this mechanism. Tissues have complex shapes, and the relative sizes and spatial relationships between signaling centers and their fields of target cells change constantly during development. Concentration gradients of morphogens presumably must reflect these evolving conditions in real time. Although mathematical simulations have been developed that mirror morphogen distributions, they are in general formulated to mathematically flat surfaces, they do not contend with the complexities of changing topologies, and they depend on unsubstantiated parameters such as extracellular fluid volume, effects of interactions with extracellular components and boundaries, and rates of synthesis and degradation (Lander, 2007). It is not apparent how unconstrained passive diffusion could generate extracellular gradients that precisely match the universe of spatial and temporal complexities that development creates.
Another possibility is that constraints exist that can shape diffusion-generated distributions of extracellular morphogens. Surface diffusion is a process by which molecules move along surfaces by jumping between nearby low affinity binding sites. The distributions of the diffusing molecules are determined by the relative stoichiometry of diffusing particles and binding sites, by the affinity of the particles for the surface components and by surface non-uniformities. Finding direct interactions between morphogen proteins and components of the extracellular matrix (ECM) and observing that signaling is depressed and morphogen movement is impaired in mutant contexts that have defective ECM have led to models that involve extracellular morphogens whose movements are constrained by adsorption to proteins at or near the cell surface (Schwank et al., 2011; Yan and Lin, 2009; Zhou et al., 2012). Although this mechanism may conceivably define the contours of morphogen distributions in extracellular space, such morphogen distributions are temporally and spatially imprecise because of the random nature of the process, and therefore may also be inadequate for the task.
There is an alternative mechanism that has strong experimental support and has been established for several systems: morphogen distributions generated by movement along cytoskeletal structures that transport the signaling proteins to designated destinations. This can be considered a “neuronal mechanism” of paracrine signaling by non-neuronal cells, and it is inherently endowed with the potential for exquisite temporal, spatial and quantitative precision that characterizes signaling at neuronal synapses. There is evidence for this mechanism in Drosophila for Hh, Decapentaplegic (a BMP homolog), FGF, Wg, EGF and Notch/Delta signaling (Bischoff et al., 2013; Cohen et al., 2010; Huang and Kornberg, 2015; Inaba et al., 2015; Rojas-Ríos et al., 2012; Roy et al., 2011), for Wnt and Notch signaling in zebrafish (Eom and Parichy, 2017; Hamada et al., 2014; Stanganello et al., 2015), and for Hh signaling in chick (Sanders et al., 2013). Both neurons and non-neuronal cells make cellular extensions (a.k.a. axons, dendrites, filopodia, cytonemes, nanotubes) that have a cytoskeletal core of actin or tubulin, and employ these organelles to send and receive signals. Cytonemes make “morphogenetic” synapses that juxtapose cytoneme tips with a target cell at distances comparable to the gaps that separate pre- and post-synaptic membranes, and the cytoneme synapses use some of the same proteins (Neuroglian, Diaphanous, Shibire (a dynamin), and Capricious (a LRR cell adhesion protein)) that are needed to make neuronal synapses. The common features of paracrine signaling by neurons and non-neuronal cells is explored in more depth elsewhere (Kornberg and Roy, 2014).
Cytonemes also navigate to their targets dependent on some of the same proteins (planar cell polarity components and heparan sulfate proteoglycans (HSPGs)) that have been implicated in axon pathfinding (Bischoff et al., 2013; Huang and Kornberg, 2016). The finding that extracellular HSPGs are required for cytonemes and cytoneme-mediated signaling is likely the reason that these regions have reduced dispersion of signaling proteins and result in reduced signaling. This explanation suggests that previous interpretations of the effects of defective ECM on signaling that invoke surface diffusion and binding of extracellular proteins to ECM proteins in the course of a random walk (Yan and Lin, 2009) may not be correct. Indeed, evidence suggests that if signaling proteins are present in the extracellular milieu, they do not signal (Roy et al., 2014). We do not yet know whether the parallels between signaling in neurons and non-neuronal cells extend to ion channels and electrical functionalities, but the homologies that have already been identified suggest that neurons and non-neuronal cells use similar mechanisms to exchange information at designated cell-cell contacts. This mode of contact-based signaling is a general one for the signaling systems that have been queried.
The premise that cytonemes are conduits that deliver morphogens directly to target cells provides a mechanism for distributing positional information across fields of cells. It does not address how cell extensions such as cytonemes navigate to their targets, how cytoneme half-life is controlled, how cytonemes generate distributions of signaling proteins, how release and uptake at the synapses is regulated, or how receptors and signaling proteins move along cytonemes. These are fascinating questions, but the fact that we do not have the answers at this time does not impugn the strength of the evidence establishing that cytonemes and other similar cell extensions transport morphogens and mediate exchange of information between cells. An important aspect of this evidence is that the contact-based cytoneme mechanism of exchanging information between cells has been established for each of the signaling systems that have been studied, suggesting that it might be universal. To query this premise, we might ask if there are known contexts whose features make this mechanism unworkable?
The Drosophila embryo, for example, develops to the blastoderm stage without cytokinesis, its nuclei multiplying in a syncytium according to a set program of 13 synchronous divisions and an orchestrated choreography that brings the nuclei simultaneously from the interior to the egg cortex at nuclear cycle 9. The egg is large (approximately 450 × 150 microns) and bean shaped, and is enveloped by a plasma membrane, but the pre-blastoderm nuclei of the early embryo are not individually encased by plasma membranes. Yet despite their syncytial state, these early stage nuclei acquire distinctive identities before they reach the embryo cortex. At nuclear stage 7 when the embryo has only 64 nuclei that are all at least 30 microns from the embryo cortex, 4-7 nuclei at about 50% egg length express the Krüppel gene (Ali-Murthy and Kornberg, 2016). With each successive nuclear division, more nuclei in this region express Krüppel and the levels of expression increase (Figure 1), but the key point is that the banded pattern of Krüppel expression originates in the syncytial interior of the embryo. This pattern of expression is dependent on a concentration gradient of the Bicoid morphogen protein that extends from the anterior end where it is highest to the posterior end where it is lowest. Thus, the early embryo is patterned along its anterior-posterior axis by a morphogen gradient when it is a syncytium that has only 64 nuclei in its interior.
Figure 1. Krüppel RNA expression and Bicoid protein in the pre-cellular Drosophila embryo.
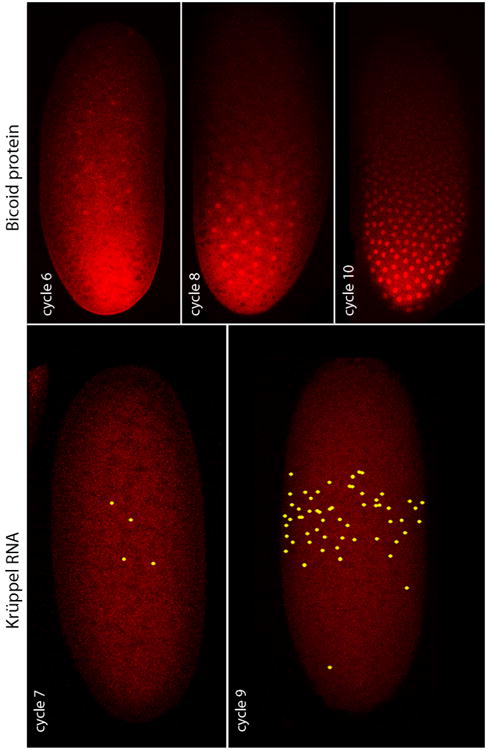
Nascent Krüppel RNA was detected by in situ hybridization in several cycle 7 nuclei and in many more cycle 9 nuclei, all centered at the approximate midpoint along the anterior-posterior axis. Bicoid protein was detected by antibody staining in a concentration gradient and concentrated in nuclei at the indicated nuclear cycles. The nuclei do not reach the embryo cortex until nuclear cycle 9. From Ali-Murthy and Kornberg, 2016
The process that generates the Bicoid concentration gradient is not known, but all detectable Bicoid protein and bicoid mRNA is localized to the anterior pole at fertilization and the gradient forms rapidly after nuclear cycle 1. In the early pre-blastoderm nuclear cycles, the distribution of Bicoid in the embryo interior forms an internal plume that has a defined shape with distinctive dorsal, ventral and lateral boundaries. Passive diffusion would seem to be incompatible with the rapidity with which this Bicoid plume forms and with its distinctive shape and well-defined borders (Ali-Murthy and Kornberg, 2016), but there are no cytonemes (plasma membrane-encased, cytoskeletal filament-containing cellular extensions) in the interior of a pre-blastoderm embryo. The question therefore arises whether despite the absence of cytonemes in the pre-blastoderm embryo, there might be common features to the way signaling proteins distribute in the unicellular, syncytial Drosophila embryo and in multicellular tissues.
Another relevant example is the ciliate Stentor coeruleus, an approximately 1mm long, single cell organism that has an exaggerated horn shape with an anchor at one end of a long tubular shaft and a large soma at the other end (Figure 2). Its cortex has patterned rows of microtubule bundles that appear as longitudinal stripes, and an oral apparatus that is reproducibly positioned relative to other cellular components. We generally associate this type of reproducibly precise patterning with multicellular tissues, yet Stentor is a single cell. Moreover, Stentor can regenerate to replace missing structures and can even rebuild its entire complex anatomy from small fragments (Slabodnick and Marshall, 2014). Although patterning and regeneration are familiar attributes of tissues, and although we normally attribute these processes to the activities of multi-cellular signaling centers and to patterned distributions of signaling proteins, Stentor has neither a classic multicellular developmental organizer or cytonemes that might distribute signaling proteins between the organizer and target cells. We know little about positional information in a ciliate, but we can speculate that these large single cells also encode positional information in spatial distributions of pattern-determining proteins. These proteins might be transcription factors that adopt a patterned distribution in the macronuclei that extend along the anterior-posterior axis, or they might be proteins that are related to the known morphogens and distribute within the cell to act locally in an autocrine manner. The relevant issue is the mechanism that distributes the pattern-determining proteins in space and time.
Figure 2. Patterned structures in Stentor.
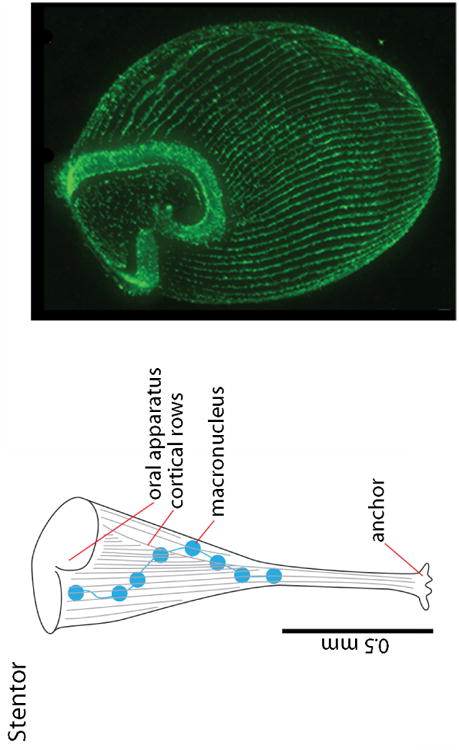
A drawing of Stentor depicts several patterned features; the fluorescence micrograph shows structures stained with anti-tubulin antibody (Slabodnick et al., 2014).
For tissues in flies, fish and chick that have been studied, morphogen proteins appear to move along cytonemes in vesicles, and it is our working assumption that these vesicles are transported along these cytoskeletal structures by molecular motors. Morphogen activity is dependent on release at pre-determined synapses. The question posed here is if there are features of this mechanism that are relevant to single cell contexts such as the pre-blastoderm Drosophila embryo and Stentor. Perhaps, the Drosophila embryo and Stentor also move the proteins that embody positional information along cytoskeletal filaments, and that in both, the process of information dispersal is dependent on and regulated by the form, direction, length, and lifetime of the filaments and by the molecular motors that service them. We might also speculate that this general mechanism could be relevant to the intercellular movement of transcription factors in plants that depend on microtubules and move through plasmodesmata that connect neighboring cells (Wu and Gallagher, 2013).
Although studies of Bicoid have not linked it to the cytoskeletal structures, we know that histological characterizations of the yolk-filled embryo interior are problematic and that the repertoire of identified cytoskeletal elements is incomplete (Cho et al., 2016). Indeed, although Bicoid appears to be constrained to the anterior end of the Drosophila egg prior to fertilization (Ali-Murthy and Kornberg, 2016), the basis for its localization has not been investigated. It is possible that Bicoid is tied by cytoskeletal tethers prior to fertilization and that it exits the anterior end after fertilization when molecular motors are induced to move cargo posteriorly. This process would presumably share properties with the cytoskeletal-based transport of morphogen-containing vesicles in cytonemes - a process that is dictated (and regulated) by the orientation and length of cytoskeletal elements and is dependent on molecular motors. It is a proposal that comes from the idea that the regulated movement of positional information in space and time has a common basis in tissues, embryos and ciliates.
Highlights.
The existence of cytonemes that distribute signaling proteins across tissues is now well established. This piece explores implications of this mechanism for single cells such as the syncytial Drosophila embryo and the ciliate Stentor.
Acknowledgments
Funding: This work was supported by grants from the National Institutes of Health (GM030637, GM105987, and GM109410)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali-Murthy Z, Kornberg TB. Bicoid gradient formation and function in the Drosophila pre-syncytial blastoderm. Elife. 2016;5 doi: 10.7554/eLife.13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff M, Gradilla AC, Seijo I, Andres G, Rodriguez-Navas C, Gonzalez-Mendez L, Guerrero I. Cytonemes are required for the establishment of a normal Hedgehog morphogen gradient in Drosophila epithelia. Nat Cell Biol. 2013;15:1269–1281. doi: 10.1038/ncb2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho A, Kato M, Whitwam T, Kim JH, Montell DJ. An Atypical Tropomyosin in Drosophila with Intermediate Filament-like Properties. Cell Rep. 2016;16:928–938. doi: 10.1016/j.celrep.2016.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Georgiou M, Stevenson NL, Miodownik M, Baum B. Dynamic Filopodia Transmit Intermittent Delta-Notch Signaling to Drive Pattern Refinement during Lateral Inhibition. Dev Cell. 2010;19:78–89. doi: 10.1016/j.devcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Eom DS, Parichy DM. A macrophage relay for long-distance signaling during postembryonic tissue remodeling. Science. 2017;355:1317–1320. doi: 10.1126/science.aal2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H, Watanabe M, Lau HE, Nishida T, Hasegawa T, Parichy DM, Kondo S. Involvement of Delta/Notch signaling in zebrafish adult pigment stripe patterning. Development. 2014;141:318–24. doi: 10.1242/dev.099804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Kornberg TB. Cells must express components of the planar cell polarity system and extracellular matrix to support cytonemes. Elife. 2016;5 doi: 10.7554/eLife.18979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Kornberg TB. Myoblast cytonemes mediate Wg signaling from the wing imaginal disc and Delta-Notch signaling to the air sac primordium. Elife. 2015;4:e06114. doi: 10.7554/eLife.06114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M, Buszczak M, Yamashita YM. Nanotubes mediate niche-stem-cell signalling in the Drosophila testis. Nature. 2015;523:329–332. doi: 10.1038/nature14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg TB, Roy S. Communicating by touch--neurons are not alone. Trends Cell Biol. 2014;24:370–376. doi: 10.1016/j.tcb.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander AD. Morpheus Unbound: Reimagining the Morphogen Gradient. Cell. 2007 doi: 10.1016/j.cell.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Rojas-Ríos P, Guerrero I, González-Reyes A. Cytoneme-mediated delivery of Hedgehog regulates the expression of bone morphogenetic proteins to maintain germline stem cells in Drosophila. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Hsiung F, Kornberg TB. Specificity of Drosophila cytonemes for distinct signaling pathways. Science. 2011;332:354–358. doi: 10.1126/science.1198949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Huang H, Liu S, Kornberg TB. Cytoneme-Mediated Contact-Dependent Transport of the Drosophila Decapentaplegic Signaling Protein. Science. 2014;343:1244624–1244624. doi: 10.1126/science.1244624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders TA, Llagostera E, Barna M. Specialized filopodia direct long-range transport of SHH during vertebrate tissue patterning. Nature. 2013;497:628–632. doi: 10.1038/nature12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwank G, Dalessi S, Yang SF, Yagi R, de Lachapelle AM, Affolter M, Bergmann S, Basler K. Formation of the long range Dpp morphogen gradient. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabodnick MM, Marshall WF. Stentor coeruleus. Curr Biol. 2014;24:R783–R784. doi: 10.1016/j.cub.2014.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabodnick MM, Ruby JG, Dunn JG, Feldman JL, DeRisi JL, Marshall WF. The Kinase Regulator Mob1 Acts as a Patterning Protein for Stentor Morphogenesis. PLoS Biol. 2014;12 doi: 10.1371/journal.pbio.1001861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanganello E, Hagemann AIH, Mattes B, Sinner C, Meyen D, Weber S, Schug A, Raz E, Scholpp S. Filopodia-based Wnt transport during vertebrate tissue patterning. Nat Commun. 2015;6:5846. doi: 10.1038/ncomms6846. [DOI] [PubMed] [Google Scholar]
- Wolpert L. Positional Information and Pattern Formation. Curr Top Dev Biol. 2016 doi: 10.1016/bs.ctdb.2015.11.008. [DOI] [PubMed] [Google Scholar]
- Wolpert L. Positional information and the spatial pattern of cellular differentiation. J Theor Biol. 1969;25:1–47. doi: 10.1016/S0022-5193(69)80016-0. [DOI] [PubMed] [Google Scholar]
- Wu S, Gallagher KL. Intact microtubules are required for the intercellular movement of the SHORT-ROOT transcription factor. Plant J. 2013;74:148–159. doi: 10.1111/tpj.12112. [DOI] [PubMed] [Google Scholar]
- Yan D, Lin X. Shaping morphogen gradients by proteoglycans. Cold Spring Harb Perspect Biol. 2009 doi: 10.1101/cshperspect.a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Lo WC, Suhalim JL, Digman MA, Gratton E, Nie Q, Lander AD. Free extracellular diffusion creates the dpp morphogen gradient of the Drosophila wing disc. Curr Biol. 2012;22:668–675. doi: 10.1016/j.cub.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]


