Abstract
All eukaryotes require the transition metal, iron, a redox active element that is an essential cofactor in many metabolic pathways, as well as an oxygen carrier. Iron can also react to generate oxygen radicals such as hydroxyl radicals and superoxide anions, which are highly toxic to cells. Therefore, organisms have developed intricate mechanisms to acquire iron as well as to protect themselves from the toxic effects of excess iron. In fungi and plants, iron is stored in the vacuole as a protective mechanism against iron toxicity. Iron storage in the vacuole is mediated predominantly by the vacuolar metal importer Ccc1 in yeast and the homologous transporter VIT1 in plants. Transcription of yeast CCC1 expression is tightly controlled primarily by the transcription factor Yap5, which sits on the CCC1 promoter and activates transcription through the binding of Fe–S clusters. A second mechanism that regulates CCC1 transcription is through the Snf1 signaling pathway involved in low-glucose sensing. Snf1 activates stress transcription factors Msn2 and Msn4 to mediate CCC1 transcription. Transcriptional regulation by Yap5 and Snf1 are completely independent and provide for a graded response in Ccc1 expression. The identification of multiple independent transcriptional pathways that regulate the levels of Ccc1 under high iron conditions accentuates the importance of protecting cells from the toxic effects of high iron.
Keywords: Aft1, Ccc1, Fe, S clusters, Iron homeostasis, Snf1, Vacuolar transporter and Yap5
Introduction
Iron is an essential nutrient for all eukaryotes because it plays a role in a wide variety of cellular oxidation–reduction reactions such as cellular respiration, DNA replication and repair, lipid biosynthesis and oxygen transport, however, although iron is essential, it can also be toxic due to its ability to generate reactive oxygen species, which can damage nucleic acids, lipids and proteins. Both the necessity and toxicity of iron underscores the importance of acquiring iron and localizing it to subcellular compartments where it is utilized or stored. As there is no mechanism for regulated iron excretion, intracellular iron levels are regulated by acquisition and storage. Alterations in these parameters can result in iron-limited growth or iron toxicity and death. Saccharomyces cerevisiae has evolved complex mechanisms to obtain iron and to protect itself from the toxic effects of excess cellular iron [for review see (Martinez-Pastor et al. 2017; Outten and Albetel 2013; Philpott et al. 2012)]. Fungi and plants store iron in the vacuole to protect themselves from toxicity as they do not express the cytosolic iron-binding protein ferritin (Bode et al. 1995; Gollhofer et al. 2014; Kim et al. 2006; Li et al. 2001; Portnoy et al. 2000; Singh et al. 2007; Szczypka et al. 1997; Urbanowski and Piper 1999). Iron storage in the vacuole also assists in maintaining cytosolic iron levels. When cytosolic iron levels decrease, iron can be efficiently exported from the vacuole by specific iron transporters. In S. cerevisiae, those transporters include Smf3 and the high affinity iron transport system Fet5/Fth1 (Portnoy et al. 2000; Singh et al. 2007; Urbanowski and Piper 1999). When cytosolic iron levels are replete, iron is imported into the vacuole to protect from iron toxicity as well as to store iron for use under iron-limited growth conditions. Iron import into the vacuole occurs primarily through the membrane transporter Ccc1 or its homologue VIT1 in plants (Chen and Kaplan 2000; Gollhofer et al. 2014; Kim et al. 2006; Lapinskas et al. 1996; Li et al. 2001). The absence of Ccc1 results in increased sensitivity to iron toxicity and cell death. In yeast, Ccc1 levels are tightly regulated by transcription (Li et al. 2008, 2011, 2012, 2017; Rietzschel et al. 2015) and by mRNA degradation (Puig et al. 2005, 2008). In this review, we will focus on the transcriptional regulation of iron sensing in S. cerevisiae and how Ccc1 levels are controlled by the high iron-sensing transcription factor Yap5 and Snf1 kinase signaling to the stress transcription factors Msn2 and Msn4.
Iron sensing in S. cerevisiae
Under iron-limiting conditions, the low iron transcription factors Aft1 and Aft2 regulate the expression of a large group of genes (> 25 genes) collectively termed the “low iron regulon”. These genes encode for proteins involved in plasma membrane iron acquisition, vacuolar iron export, mitochondrial iron import and iron–sulfur cluster synthesis along with proteins involved in iron metabolism [for review see (Martinez-Pastor et al. 2017; Outten and Albetel 2013)]. These transcription factors, Aft1 and Aft2, predominantly respond to the levels of mitochondrial iron-sulfur (Fe–S) cluster biogenesis and not to cytosolic iron levels. When mitochondrial Fe–S cluster synthesis is diminished, cytosolic Aft1 and Aft2 are translocated to the nucleus and bind to target DNA resulting in transcription of the “low iron regulon”. While Aft1 and Aft2 are the major low-iron sensing transcription factors, mathematical modeling of yeast iron acquisition suggests that there may be other transcription factors that respond to changes in cytosolic iron levels (Cockrell et al. 2014).
Transcriptional regulation of CCC1
Notably, under low iron conditions there is little transcription of the vacuolar iron importer Ccc1. The lack of CCC1 transcription ensures that cytosolic and mitochondrial proteins have maximal access to iron. Further, two Aft1-target genes, CTH1 and CTH2, are responsible for the low iron induced degradation of CCC1 mRNA (Puig et al. 2005, 2008). Under iron-replete conditions, the transcription factor Yap5 acts to increase CCC1 transcription resulting in increased iron import into the vacuole (Li et al. 2008; Pimentel et al. 2012). Yap5 is a member of the basic leucine zipper transcription regulators of which there are eight in fungi (Fernandes et al. 1997; Reinke et al. 2013). Yap5 is localized to the nucleus and occupies a consensus Yap binding site in the CCC1 promoter regardless of cellular iron levels (Li et al. 2008). Yap5-mediated CCC1 transcription is activated under iron excess and two different studies suggest that activation of transcription is mediated through sensing Fe–S clusters (Li et al. 2012; Rietzschel et al. 2015). Rietzschel et al. specifically determined that Yap5-dependent CCC1 transcriptional activation is mediated through the binding of two 2Fe–2S clusters to cysteine-rich domains in the activation domain of Yap5, inducing a conformational change that is suggested to activate CCC1 transcription.
The absence of Yap5 greatly reduces CCC1 transcription, however, iron-responsive CCC1 transcription still occurs. Cells deleted for YAP5 have higher iron tolerance than cells deleted for CCC1. These results suggested that there were other transcriptional mechanisms that regulate CCC1 expression in iron-replete conditions. Yeast have developed several stress-activated signaling networks to deal with deprivation or excess nutrients conditions (Ho and Gasch 2015; Nishida-Aoki et al. 2015; Yadav et al. 2016). Recently, a role for the low-glucose sensor Snf1 kinase complex [for review see (Conrad et al. 2014)] in regulating CCC1 transcription has been identified (Li et al. 2017). Under iron-replete conditions, the absence of Yap5 and Snf1 has an additive effect on CCC1 transcription with the concomitant decrease in Ccc1 protein levels, increased sensitivity to iron toxicity and poor growth. This additive effect suggests that Yap5 and the Snf1 complex act independently to activate CCC1 transcription (Fig. 1). Indeed, Snf1 activation is independent of Fe–S cluster synthesis and does not affect the transcription of TYW1, another target of the Yap5-high iron regulon (Li et al. 2011). Iron-replete conditions can be sensed as a stress on cells and in fact, the general stress transcription factor Msn4 is upregulated in response to high iron (Du et al. 2012). Li et al. show that the Snf1-mediated activation of CCC1 transcription works partially through Msn2/Msn4, but the Snf1 complex also activates further CCC1 transcription through an unknown mechanism (Li et al. 2017).
Fig. 1.
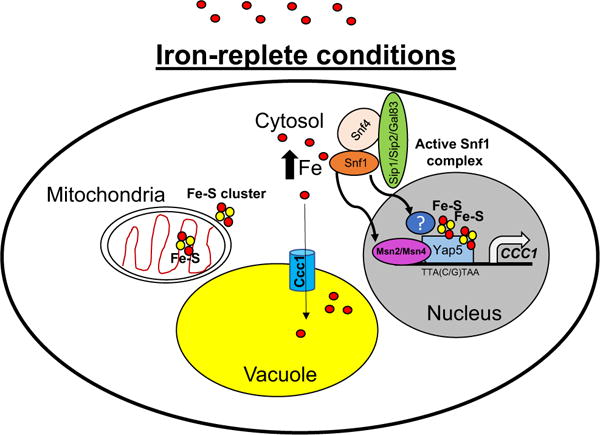
Model of transcriptional regulation of CCC1 expression. Under iron-replete conditions, sufficient Fe–S clusters are synthesized in the mitochondria and nuclear localized Yap5 binds 2 2Fe–2S clusters to activate transcription. The high iron also acts by some unknown mechanism to activate the Snf1-complex (Snf1/Snf4/Sip1/Sip2/Gal83), which acts through the transcription factors Msn2/Msn4 and another mechanism (blue question mark circle) to induce CCC1 transcription
Conclusions
There is a clear balance that must be maintained between cytosolic iron and mitochondrial iron levels depending upon iron utilization. Dysregulation of this balance results in iron toxicity and cell death. Studies have shown that removing cytosolic iron either by sequestration in the vacuole or in the mitochondria can act as a protective mechanism against iron toxicity (Chen and Kaplan 2000; Li et al. 2001; Lin et al. 2011). Ccc1 plays a key role in this balance by importing iron into the vacuole. The levels of Ccc1 are determined by an equilibrium between regulating CCC1 transcription under iron-replete conditions and CCC1 mRNA degradation under iron-limited conditions. Transcriptionally, CCC1 expression is regulated through two known mechanisms including the high iron sensing transcription factor Yap5 and the Snf1-kinase stress pathway signal to transcription factors Msn2 and Msn4, however, additional studies are needed to determine other “signals” and transcription factors that contribute to increased CCC1 transcription under high iron conditions.
Acknowledgments
The authors express their appreciation to Dr. Jerry Kaplan for critically reading the manuscript. We apologize to those authors whose work we were unable to cite due to space limitations. This work is supported by the National Institutes of Health Grant DK030534 to D.M.W.
Footnotes
Compliance with Ethical Standards
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
References
- Bode HP, Dumschat M, Garotti S, Fuhrmann GF. Iron sequestration by the yeast vacuole. A study with vacuolar mutants of Saccharomyces cerevisiae. Eur J Biochem. 1995;228:337–342. [PubMed] [Google Scholar]
- Chen OS, Kaplan J. CCC1 suppresses mitochondrial damage in the yeast model of Friedreich’s ataxia by limiting mitochondrial iron accumulation. J Biol Chem. 2000;275:7626–7632. doi: 10.1074/jbc.275.11.7626. [DOI] [PubMed] [Google Scholar]
- Cockrell A, McCormick SP, Moore MJ, Chakrabarti M, Lindahl PA. Mossbauer, EPR, and modeling study of iron trafficking and regulation in Deltaccc1 and CCC1-up Saccharomyces cerevisiae. Biochemistry. 2014;53:2926–2940. doi: 10.1021/bi500002n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2014;38:254–299. doi: 10.1111/1574-6976.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Cheng W, Li WF. Expression profiling reveals an unexpected growth-stimulating effect of surplus iron on the yeast Saccharomyces cerevisiae. Mol Cells. 2012;34:127–132. doi: 10.1007/s10059-012-2242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes L, Rodrigues-Pousada C, Struhl K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol. 1997;17:6982–6993. doi: 10.1128/mcb.17.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollhofer J, Timofeev R, Lan P, Schmidt W, Buckhout TJ. Vacuolar-iron-transporter1-like proteins mediate iron homeostasis in Arabidopsis. PLoS One. 2014;9:e110468. doi: 10.1371/journal.pone.0110468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho YH, Gasch AP. Exploiting the yeast stress-activated signaling network to inform on stress biology and disease signaling. Curr Genet. 2015;61:503–511. doi: 10.1007/s00294-015-0491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Punshon T, Lanzirotti A, Li L, Alonso JM, Ecker JR, Kaplan J, Guerinot ML. Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science. 2006;314:1295–1298. doi: 10.1126/science.1132563. [DOI] [PubMed] [Google Scholar]
- Lapinskas PJ, Lin SJ, Culotta VC. The role of the Saccharomyces cerevisiae CCC1 gene in the homeostasis of manganese ions. Mol Microbiol. 1996;21:519–528. doi: 10.1111/j.1365-2958.1996.tb02561.x. [DOI] [PubMed] [Google Scholar]
- Li L, Chen OS, McVey Ward D, Kaplan J. CCC1 is a transporter that mediates vacuolar iron storage in yeast. J Biol Chem. 2001;276:29515–29519. doi: 10.1074/jbc.M103944200. [DOI] [PubMed] [Google Scholar]
- Li L, Bagley D, Ward DM, Kaplan J. Yap5 is an iron-responsive transcriptional activator that regulates vacuolar iron storage in yeast. Mol Cell Biol. 2008;28:1326–1337. doi: 10.1128/MCB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Jia X, Ward DM, Kaplan J. Yap5 protein-regulated transcription of the TYW1 gene protects yeast from high iron toxicity. J Biol Chem. 2011;286:38488–38497. doi: 10.1074/jbc.M111.286666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Miao R, Bertram S, Jia X, Ward DM, Kaplan J. A role for iron–sulfur clusters in the regulation of transcription factor Yap5-dependent high iron transcriptional responses in yeast. J Biol Chem. 2012;287:35709–35721. doi: 10.1074/jbc.M112.395533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kaplan J, Ward DM. The glucose sensor Snf1 and the transcription factors Msn2 and Msn4 regulate transcription of the vacuolar iron importer gene CCC1 and iron resistance in yeast. J Biol Chem. 2017;292:15577–15586. doi: 10.1074/jbc.M117.802504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Li L, Jia X, Ward DM, Kaplan J. Genetic and biochemical analysis of high iron toxicity in yeast: iron toxicity is due to the accumulation of cytosolic iron and occurs under both aerobic and anaerobic conditions. J Biol Chem. 2011;286:3851–3862. doi: 10.1074/jbc.M110.190959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Perea-Garcia A, Puig S. Mechanisms of iron sensing and regulation in the yeast Saccharomyces cerevisiae. World J Microbiol Biotechnol. 2017;33:75. doi: 10.1007/s11274-017-2215-8. [DOI] [PubMed] [Google Scholar]
- Nishida-Aoki N, Mori H, Kuroda K, Ueda M. Activation of the mitochondrial signaling pathway in response to organic solvent stress in yeast. Curr Genet. 2015;61:153–164. doi: 10.1007/s00294-014-0463-9. [DOI] [PubMed] [Google Scholar]
- Outten CE, Albetel AN. Iron sensing and regulation in Saccharomyces cerevisiae: ironing out the mechanistic details. Curr Opin Microbiol. 2013;16:662–668. doi: 10.1016/j.mib.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott CC, Leidgens S, Frey AG. Metabolic remodeling in iron-deficient fungi. Biochim Biophys Acta. 2012;1823:1509–1520. doi: 10.1016/j.bbamcr.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel C, Vicente C, Menezes RA, Caetano S, Carreto L, Rodrigues-Pousada C. The role of the Yap5 transcription factor in remodeling gene expression in response to Fe bioavailability. PLoS One. 2012;7:e37434. doi: 10.1371/journal.pone.0037434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy ME, Liu XF, Culotta VC. Saccharomyces cerevisiae expresses three functionally distinct homologues of the nramp family of metal transporters. Mol Cell Biol. 2000;20:7893–7902. doi: 10.1128/mcb.20.21.7893-7902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig S, Askeland E, Thiele DJ. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell. 2005;120:99–110. doi: 10.1016/j.cell.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Puig S, Vergara SV, Thiele DJ. Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab. 2008;7:555–564. doi: 10.1016/j.cmet.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke AW, Baek J, Ashenberg O, Keating AE. Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science. 2013;340:730–734. doi: 10.1126/science.1233465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietzschel N, Pierik AJ, Bill E, Lill R, Muhlenhoff U. The basic leucine zipper stress response regulator Yap5 senses high-iron conditions by coordination of [2Fe–2S] clusters. Mol Cell Biol. 2015;35:370–378. doi: 10.1128/MCB.01033-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Kaur N, Kosman DJ. The metalloreductase Fre6p in Fe-efflux from the yeast vacuole. J Biol Chem. 2007;282:28619–28626. doi: 10.1074/jbc.M703398200. [DOI] [PubMed] [Google Scholar]
- Szczypka MS, Zhu Z, Silar P, Thiele DJ. Saccharomyces cerevisiae mutants altered in vacuole function are defective in copper detoxification and iron-responsive gene transcription. Yeast. 1997;13:1423–1435. doi: 10.1002/(SICI)1097-0061(199712)13:15<1423::AID-YEA190>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Urbanowski JL, Piper RC. The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J Biol Chem. 1999;274:38061–38070. doi: 10.1074/jbc.274.53.38061. [DOI] [PubMed] [Google Scholar]
- Yadav KK, Singh N, Rajasekharan R. Responses to phosphate deprivation in yeast cells. Curr Genet. 2016;62:301–307. doi: 10.1007/s00294-015-0544-4. [DOI] [PubMed] [Google Scholar]


