Abstract
Introduction
Glioblastoma (GBM) generates a varied immune response and understanding the immune microenvironment may lead to novel immunotherapy treatments modalities. The goal of this study was to evaluate the expression of immunologic markers of potential clinical significance in primary versus recurrent GBM and assess the relationship between these markers and molecular characteristics of GBM.
Methods
Human GBM samples were evaluated and analyzed with immunohistochemistry for multiple immunobiologic markers (CD3, CD8, FoxP3, CD68, CD163, PD1, PDL1, CTLA4, CD70). Immunoreactivity was analyzed using Aperio software. Degree of strong positive immunoreactivity within the tumor was compared to patient and tumor characteristics including age, gender, MGMT promoter methylation status, and ATRX, p53, and IDH1 mutation status. Additionally, the TCGA database was used to perform similar analysis of these factors in GBM using RNA-seq by expectation-maximization (RSEM).
Results
Using odds ratios, IDH1 mutated GBM had statistically significant decreased expression of CD163 and CD70 and a trend for decreased PD1, CTLA4, and Foxp3. ATRX-mutated GBMs exhibited statistically significant increased CD3 immunoreactivity, while those with p53 mutations were found to have significantly increased CTLA4 immunoreactivity. The odds of having strong CD8 and CD68 reactivity was significantly less in MGMT methylated tumors. No significant difference was identified in any immune marker between the primary and recurrent GBM, nor was a significant change in immunoreactivity identified among age intervals. TCGA analysis corroborated findings related to the differential immune profile of IDH1 mutant, p53 mutant, and MGMT unmethylated tumors.
Conclusion
Immunobiologic markers have greater association with the molecular characteristics of the tumor than with primary/recurrent status or age.
Keywords: immune infiltrates, tumor infiltrating lymphocytes, immunobiologic markers, high grade glioma, glioblastoma
Introduction
Glioblastoma (GBM) is associated with a dismal prognosis despite aggressive measures with maximal safe resection, fractionated radiation and chemotherapy. Most importantly, many novel agents that have had promising results in pre-clinical and early human trials have failed in larger randomized phase III trials [1–3]. Part of these failures can be attributed to the remarkable heterogeneity within GBM tumors[4]. This heterogeneity results in a robust ability[5] for cancer cells to evade therapy through multiple redundant pathways that can be up-regulated to preserve growth and invasion in the face of targeted therapeutic disruption. This robustness of GBM continues to hamper the development of effective therapeutics for invasive glial tumors. The attempts to advance promising cancer immunotherapies in the treatment of GBM will undoubtedly face similar developmental challenges. Recently matured late stage immunotherapy trials in GBM that have failed to meet clinical study endpoints highlight these challenges[6–8].
Immunotherapeutic efforts are well underway with various approaches including cellular and peptide vaccines, T cell transfer or immune checkpoint blockade. The efficacy of these strategies will most likely be dependent on patient and tumor-related factors that will determine ability to mount an effective immune response against tumor cells that drive growth, invasion and resistance. Others have shown that tumor biology is associated with variable immune infiltrates when comparing GBM to other gliomas[9, 10]. As our understanding increases of molecular characterization of GBM, the interplay between tumor immune infiltrates and molecular markers is paramount.
Our group previously published that intra-tumoral T cell infiltrates were associated with survival outcomes in glioma patients[11]. To build on our previous work, the goal of this study was to evaluate the expression of immunologic markers of potential clinical significance in primary versus recurrent GBM and assess the relationship between these markers and clinically relevant molecular characteristics of GBM.
Methods
Human GBM pathologic specimen analysis
Thirty-eight primary and twelve cases of recurrent glioblastoma were collected from the Florida Center for Brain Tumor Research (FCBTR) for immunohistochemical analysis, including four paired primary and recurrent cases. IHC staining of paraffin-embedded, formalin-fixed tissue blocks was performed using polymer method. Briefly, the sections were steamed for 30 min in antigen retrieval citra buffer (Biogenex, Freemont, CA) for antigen retrieval. The primary antibodies were monoclonal mouse anti-FOXP3 (14-4776; eBioscience), CD70 (CD27L, sc-365539, Santa Cruz), CTLA-4 (sc-376016, Santa Cruz), PD-L1 (ab58810, Abcam), PD1 (ab52587, Abcam), CD163 (cm353ak, Biocare). A board-certified neuropathologist evaluated each case utilizing Aperio software for analysis[11] (Fig 1). Evaluation was limited to only tumor-rich areas in order to avoid nonspecific staining, such as that seen in regions of necrosis. Additionally, the software was calibrated to only recognize cells with strong positive immunoreactivity.
Figure 1. Examples of IHC staining and Aperio analysis.
A) Representations of positive IHC staining for each immune expression marker including CD70, CTLA4, FoxP3, PD-L1, PD-1 and CD163. B) With each case, only tumor-rich areas (yellow encircled region) were selected for Aperio analysis to avoid non-specific staining such as necrotic regions (star) and plasma (arrow). C) Screenshot of Aperio analysis demonstrating quantification of cell staining. The red arrow shows the area of the tumor chosen for the analysis. The red circle shows the cells that stained positively for the marker and were counted by the Aperio software.
For each immune marker, the percentage of strongly positive cells over all nucleated cells was calculated for each case. Using these values, the overall average percentage of positive cells for all cases was calculated in addition to the averages for all primary and recurrent cases. Additionally, the percentage of macrophages (CD68 and CD163), CTLA4, CD8+ T cells, and FoxP3+ T cells was also calculated for each case and averaged. Reporting of PD1 values was an exception due to low overall staining. In this case, positivity was assigned a value from 0–3, with 0 representing no detectable immunoreactivity and 3 representing very strong immunoreactivity as previously described[11]. Averages were calculated similarly to the other immune markers.
Using odds ratios, patterns of immunoreactivity were compared between patient characteristics such as age and gender, as well as tumor characteristics including IDH1, ATRX, and p53 mutation status and MGMT promoter methylation status. The immunoreactivity patterns of the paired primary and recurrent cases were also compared to each other.
Statistical Analysis
The proportion of cells that stained strongly positive for each of 9 markers was calculated and analyzed as a response variable. Initially, logistic regression was used to model the expected log-odds of strongly positive cell staining as a linear function of combinations of 7 covariates of interest: primary vs recurrent tumor, age, gender, and presence/absence of ATRX, P53, IDH1, and MGMT. Fitted models demonstrated significant overdispersion of errors, so proportions were logit-transformed and modeled directly as continuous responses using linear regression. Graphical assessment of residual errors from these models appeared reasonably normal.
Because the MGMT status covariate had roughly 60% of values missing, two parallel analyses were carried out. One analysis considered all of the study sample observations and excluded MGMT from consideration as a potential predictor. The other analysis only included observations with non-missing MGMT status values. Best models with 4 or fewer covariates (MGMT excluded) or 2 or fewer covariates (MGMT included) in the model were determined for each marker among the family of all possible models using the small-sample Akaike Information Criterion (AICc). The predictors in these models were then used to adjust estimated odds ratios for each predictor of each marker. These odds ratios are summarized in tables that include 95% confidence intervals and p-values for a Wald test that the odds ratios differ from 1. Forest plots contrasting the 9 marker odds ratios for each predictor were also generated.
TCGA Analysis
Primary (n=146) and recurrent (n=19) GBM patients mRNA gene expression data and clinical information were culled from TCGA GBM RNAseqv2 dataset (currently available in UCSC repository, https://genome-cancer.ucsc.edu/proj/site/hgHeatmap/). Patients’ IDH1, ATRX, TP53 gene mutation were derived from TCGA GBM gene-level non-silent somatic mutation (broad) dataset of UCSC repository. Patient’s MGMT promoter methylation prediction were based on supplementary table from Bady et al.’s publication[12]. Patients mRNA expression, survival outcomes, TCGA/WHO subtypes, gene mutation and MGMT promoter methylation data were mapped into one cohort by Subioplatform. This merged cohort was then bifurcated into mutation/wild-type or methylated/unmethylated groups, respectively. Normalized, log-transformed RSEM gene expression levels for each immune marker were analyzed using linear regression, in a manner similar to that used for strong cell staining responses from the FCBTR dataset. Differences between mutated and wild-type strata (IDH1, ATRX, p53) or between methylated and unmethylated strata (MGMT) were expressed as adjusted RSEM mean ratios and displayed using forest plots.
We used Cox proportional hazards regression[13] to assess the effect of TCGA immune marker gene expression on overall survival in GBM patients. For each immune marker, gene expression levels were treated as a continuous predictor. Patients were stratified by wild-type or mutated (IDH1, ATRX, p53) or by methylated or unmethylated forms for MGMT. In each immune marker model, we included the expression levels of the marker, a stratifying variable, and a term representing interaction between the marker and the stratifying variable. We used restricted cubic splines[13] to assess possible non-linearity in the association of immune markers with survival. We used our fitted Cox models to estimate hazard ratios (HR) along with 95% confidence intervals and Wald tests for the null hypothesis HR=1. We estimated median survival for selected expression levels of each immune marker and used these estimates to generate expected median survival trend curves over the range of observed expression levels.
Results
Table 1 summarizes the demographic and percent of samples expressing ATRX, p53, IDH-1 and MGMT methylation status in the human GBM samples from the FCBTR. Overall, 61% of samples were from male patients and 18.4% of samples were IDH-1 mutated. About half of the samples had no ATRX, p53 or IDH-1 mutation (Table 2). The most common mutation patterns were isolated p53 mutation (12.5%), p53 and IDH-1 mutation (12.5%) and an isolated ATRX mutation (10.42%). The presence of these genetic mutations in primary and recurrent samples is shown in Table 3. No statistically significant differences in gender, ATRX mutation, p53 mutation, IDH-1 mutation or MGMT promoter methylation status were found between primary and recurrent GBM samples. The immune marker expression in primary and recurrent tumors is shown in Table 4. PD-L1, CD163 and CD68 had the highest percent positivity. Not surprisingly, recurrent GBM samples were more likely to be from younger patients. Otherwise, immune marker expression did not differ between primary and recurrent samples. No significant difference in the expression of immune cell markers was identified between primary and recurrent GBM, including markers for lymphocytes (CD3, CD8, FoxP3) and macrophages (CD68 and CD163) (Suppl Fig 1). The lymphocyte and macrophage values between paired cases from the same patient (4 cases) demonstrated no significant differences. Differing age groups also did not correlate with differences in the immune profiles of GBMs (Suppl Fig 2).
Table 1.
Clinical and molecular characteristics of GBM samples included in the study.
| Variable | n | n total | Percent |
|---|---|---|---|
| Recurrent | 11 | 49 | 22.45 |
| Male | 30 | 49 | 61.22 |
| ATRX mutated | 9 | 49 | 18.37 |
| P53 mutated | 14 | 48 | 29.17 |
| IDH1 mutated | 9 | 49 | 18.37 |
| MGMT methylated | 7 | 20 | 35 |
The GBM samples used for the study were mostly newly diagnosed GBM and from male patients. As expected a fraction of patients had ATRX, p53 and/or IDH-1 mutated tumors. The majority of samples were MGMT unmethylated.
Table 2.
The combination patterns of genetic mutations in all GBM samples.
| ATRX mutated | p53 mutated | IDH-1 mutated | Frequency | Percent % |
|---|---|---|---|---|
| N | N | N | 27 | 56.25 |
| N | N | Y | 1 | 2.08 |
| N | Y | N | 6 | 12.50 |
| N | Y | Y | 6 | 12.50 |
| Y | N | N | 5 | 10.42 |
| Y | N | Y | 1 | 2.08 |
| Y | Y | N | 1 | 2.08 |
| Y | Y | Y | 1 | 2.08 |
The combination of mutations are shown.
Table 3.
Primary versus recurrent GBM sample categorical variables.
| Variable | Group | n | n total | Percent % | p value |
|---|---|---|---|---|---|
| Male | Primary | 23 | 38 | 60.53 | 1.0 |
| Recurrent | 7 | 11 | 63.64 | ||
| ATRX mutated | Primary | 7 | 38 | 18.42 | 1.0 |
| Recurrent | 2 | 11 | 18.18 | ||
| p53 mutated | Primary | 10 | 37 | 27.03 | 0.7075 |
| Recurrent | 4 | 11 | 36.36 | ||
| IDH1 mutated | Primary | 5 | 38 | 13.16 | 0.1786 |
| Recurrent | 4 | 11 | 36.36 | ||
| MGMT methylated | Primary | 6 | 19 | 31.58 | 0.3500 |
| Recurrent | 1 | 1 | 100 |
The patient gender, the clinically relevant mutations (ATRX, p53, IDH-1) and MGMT methylation status are shown by primary or recurrent tumor status. No statistically significant differences were found between the primary or recurrent samples in these variables.
Table 4.
Primary versus recurrent GBM sample continuous variables.
| Variable | Group | n | Mean (yrs or % positive staining) |
SD | p value |
|---|---|---|---|---|---|
| Age | Primary | 38 | 61.05 | 10.67 | 0.0009 |
| Recurrent | 11 | 43.36 | 12.71 | ||
| CD3 | Primary | 37 | 0.35 | 0.44 | 0.9416 |
| Recurrent | 11 | 0.33 | 0.35 | ||
| CD8 | Primary | 36 | 0.12 | 0.14 | 0.1937 |
| Recurrent | 11 | 0.30 | 0.44 | ||
| CD68 | Primary | 38 | 5.20 | 3.39 | 0.7120 |
| Recurrent | 11 | 5.46 | 2.06 | ||
| CD163 | Primary | 38 | 6.12 | 6.40 | 0.7658 |
| Recurrent | 11 | 5.56 | 3.72 | ||
| FOXP3 | Primary | 37 | 0.35 | 0.47 | 0.9279 |
| Recurrent | 10 | 0.37 | 0.47 | ||
| PDL1 | Primary | 37 | 8.11 | 7.66 | 0.4508 |
| Recurrent | 11 | 7.42 | 4.34 | ||
| CTLA4 | Primary | 38 | 0.43 | 0.59 | 0.9525 |
| Recurrent | 11 | 0.27 | 0.23 | ||
| CD70 | Primary | 34 | 0.69 | 1.38 | 0.3401 |
| Recurrent | 6 | 0.82 | 0.79 | ||
| PD1 | Primary | 38 | 1.42 × 105 | 3.95 × 105 | 0.4888 |
| Recurrent | 11 | 0.38 × 105 | 0.75 × 105 |
The patient age and percent positive cell expression of immune markers are shown in primary and recurrent samples. The primary samples were associated with older patients compared to recurrent samples. No significant differences in immune marker expression was found between primary and recurrent samples.
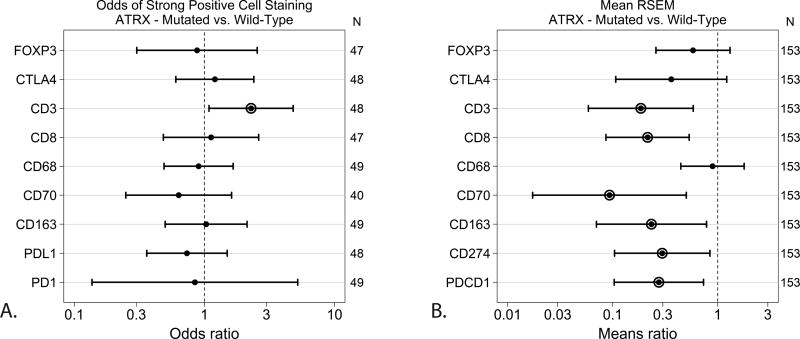
These samples were analyzed for mutation status and immune marker expression. The cases that were positive for IDH1 mutation showed significantly lower odds of expressing CD163 (macrophages) (p=0.0464) and CD70 (on lymphocytes and tumor cells) (p=0.0428), and also showed a trend of decreased expression of PD1, CTLA4 (immune checkpoints), and Foxp3 (regulatory T cells) (Fig 2a). The TCGA RNA-sequencing analysis similarly demonstrated that IDH1 mutated tumors had significantly lower means ratios of CD163 (p=0.0005), CD70 (p=0.0094) as well as CD3 (T cells) (p=0.0044), CD8 (cytotoxic T cells) (p=0.0044), PDCD1 (gene for PD-1) (p=0.0057) and CD274 (gene for PD-L1) (p=0.0192) (Fig 2b). ATRX-mutated GBM in the human GBM samples exhibited significantly increased odds of CD3 immunoreactivity (p=0.0306) (Fig 3a). These results were different in the TCGA analysis which demonstrated decreased means ratio of CD3 (p=0.0043), CD8 (p=0.0011), CD70 (p=0.0060), CD163 (p=0.0135), CD274 (gene for PD-L1) (p=0.0229), and PDCD1 (gene for PD-1) (p=0.0101) in ATRX mutated tumors (Fig 3b). GBM with p53 mutations were found to have increased odds of expressing CTLA4 (p=0.0225) (Fig 4a). The TCGA data did not demonstrate statistically significant differences between p53 mutated or wildtype tumors (Fig 4b). MGMT promoter methylated GBM cases were found to have significantly lower odds of expressing CD8 (p=0.0489) and CD68 (p=0.0192) than MGMT unmethylated tumors (Fig 5a). The TCGA analysis showed that MGMT methylated tumors had less CTLA4 (p=0.0207), CD70 (p=0.0423) and PDCD1 (gene for PD-1) (p=0.0044) with only a trend for decreased CD68 (Fig 5b).
Figure 2. Effect of Mutated versus Wild-Type IDH1 on Immune Marker Expression.
A. GBM samples were tested for immune marker expression by IHC and compared based on IDH-1 mutant status. CD70 (p=0.0428) and CD163 (p=0.0464) were significantly lower in IDH-1 mutated tumors shown as an odds ratio of less than 1. B. TCGA analysis of GBM samples revealed that CD3 (p=0.0044), CD8 (p=0.0044), CD70 (p=0.0094), CD163 (p=0.0005), CD274 (gene for PD-L1) (p=0.0192), and PDCD1 (gene for PD-1) (p=0.0057) were lower in IDH1 mutated tumors.
Circled odds ratio or mean ratio point estimates differ significantly from one at α=0.05.
Figure 3. Effect of Mutated versus Wild-Type ATRX on Immune Marker Expression.
A. GBM samples were tested for immune marker expression by IHC and compared based on ATRX mutant status. CD3 was significantly higher in ATRX mutated tumors (p=0.0306) shown as an odds ratio of greater than 1. B. TCGA analysis of GBM samples revealed that CD3 (p=0.0043), CD8 (p=0.0011), CD70 (p=0.0060), CD163 (p=0.0135), CD274 (gene for PD-L1) (p=0.0229), and PDCD1 (gene for PD-1) (p=0.0101) were lower in ATRX mutated tumors.
Circled odds ratio or mean ratio point estimates differ significantly from one at α=0.05.
Figure 4. Effect of Mutated versus Wild-Type P53 on Immune Marker Expression.
A. GBM samples were tested for immune marker expression by IHC and compared based on p53 mutant status. CTLA4 was significantly higher in p53 mutated tumors (p=0.0225) shown as an odds ratio of greater than 1. B. TCGA analysis of GBM samples revealed no significant differences in mutant and wildtype p53 GBM tumors.
Circled odds ratio or mean ratio point estimates differ significantly from one at α=0.05.
Figure 5. Effect of MGMT Promoter Methylation on Immune Marker Expression.
A. GBM samples were tested for immune marker expression by IHC and compared based on MGMT promoter methylation status. CD8 (p=0.0489) and CD68 (p=0.0192) were significantly lower in MGMT promoter methylated tumors shown as an odds ratio of less than 1. B. TCGA analysis of GBM samples revealed decreased CTLA4 (p=0.0207), CD3 (p=0., CD8, CD70 (p=0.0423) and PDCD1 (gene for PD-1) (p=0.0044) in MGMT promoter methylated tumors.
Circled odds ratio or mean ratio point estimates differ significantly from one at α=0.05.
Using the TCGA database, mutation of ATRX, IDH1, and p53 correlated with molecular subtype of GBM (Fig 6). ATRX or IDH1 mutated tumors were more likely to be proneural[14], p53 mutated tumors were either proneural or mesenchymal. MGMT promoter methylated tumors fell into all four molecular subtypes. Subsequently, the association of immune marker RNA expression with overall survival was analyzed. The factors that were found to be significant on IHC analysis were used to determine survival associations using the TCGA dataset. CD8 expression was significantly associated with survival in GBM. MGMT unmethylated tumors had decreased survival with very low or very high expression of CD8, while methylated GBM had increased survival with low CD8 expression (Fig 7a–b). This finding will require further exploration. Other markers were associated with trends in survival but these did not reach statistical significance.
Figure 6. GBM Mutation Status and Molecular Subtype in TCGA Dataset.
TCGA data analysis shows molecular subtype of GBM and presence of ATRX, IDH1, p53 mutation or MGMT methylation status. The red bars indicate mutation or promoter methylation and the black bars represent wildtype or promoter unmethylated.
Figure 7. Effect of CD8A expression level on median overall survival in GBM patients.
A. TCGA analysis demonstrating correlation between CD8A expression and survival in GBM tumors that are MGMT promoter unmethylated. B. TCGA analysis demonstrating correlation between CD8A expression and survival in GBM tumors that are MGMT promoter methylated.
The red line indicates the Cox model estimate of median overall survival (OS) before consideration of immune marker expression. The solid black line shows the expected median OS when the immune marker expression level is included as a predictor in the Cox model. The displayed P-value tests whether the immune marker hazard ratio from the Cox model differs significantly from one.
Discussion
In this study, we identified a significant correlation of immune marker expression with mutational profiles of GBM. However, immune marker expression between primary and recurrent GBM were not significantly different. The importance of molecular characteristics of GBM to predict outcome is reflected in the latest World Health Organization (WHO) classification of glioma. The 2016 WHO classification incorporates both histological and molecular characteristics of the tumor given the increasing evidence that molecular signatures predict tumor behavior[15, 16]. We have now found that these molecular signatures may also be important in immune mechanisms related to tumor control, growth, resistance and ultimately to how a tumor responds to immunotherapy. Our data shows that IDH-1 mutated tumors which are known to have a better prognosis were less likely to express immune checkpoint receptors, monocyte markers and regulatory T cells (Treg) markers which are known to be immunosuppressive. Alternatively, ATRX positive tumors had discordant results with the IHC evaluation demonstrating an increase in CD3+ T cell infiltration, and the TCGA analysis demonstrating less CD3+ and CD8+ T cells. Tumors with p53 mutations were more likely to express immune checkpoint receptor CTLA4. We also found that MGMT unmethylated tumors had increased CD8+ T cell and macrophage infiltrates. Our data also demonstrate the only statistically significant effects on survival is the expression of CD8 in MGMT unmethylated and methylated tumors. Others have described improved survival in GBM patients who have more CD8+ T cell infiltrates[17]. Our data demonstrated that MGMT unmethylated tumors did not have improved survival with increased CD8+ expression. The other immune markers did not have significant association with survival based on gene expression analysis of the tumors by RNA sequencing. Additionally, the expression of immune markers measured by IHC within molecular subgroups of GBM differed from the transcriptional expression measured by TCGA. This discordance is most likely multi-factorial and related to the difference in measuring protein expression versus transcriptional expression, sensitivity of the assays employed for measuring quantitative differences, and the power of the studies driven by differences in sample size. However, both TCGA and IHC analyses demonstrated that immunologic differences within the tumor microenvironment are notable between molecular subtypes of GBM tumors and our studies demonstrate that molecular subtype rather than age of patient or primary versus recurrent disease is more important in driving immunologic heterogeneity. These studies suggest that molecular subtype of malignant gliomas should be incorporated in the analysis of response to immunotherapy and be considered in the development of novel immune therapeutics for this patient population.
Other groups have described association of markers of immune function with molecular characteristics of gliomas. In a study of 976 gliomas in the Chinese Glioma Genome Atlas and the Cancer Genome Atlas (TCGA), investigators found a correlation between PD-L1 expression and mesenchymal subtype[18]. They also found these tumors were more likely to be infiltrated with T cells and macrophages compared to PD-L1 negative tumors. These data provide the rationale for potentially stratifying patients enrolled in immunotherapy trials to maximize benefit. This strategy could also avoid the likelihood of a negative trial, which has significantly limited advances in GBM therapeutic efforts. The definition of immune markers expression groups could also be used as a surrogate for molecular subtypes of glioma. Determining mesenchymal versus proneural subtypes require genomic-level analysis which can be costly, time-intensive and is not utilized clinically. Other groups have described using immunohistochemistry (IHC) methods as a surrogate biomarker of certain molecular subtypes of glioma[19]. Therefore, the methods we describe here have the potential to be used to subclassify gliomas based on immune infiltrates, and be used to determine which patients could benefit from certain immunotherapeutic options.
The limitations of this study are small sample size and a limited analysis of each sample of a tumor that is known for heterogeneity. Additionally, the PD-L1 immunostain does not have consensus for interpretation of positive versus negative. There were very few recurrent samples compared to primary tumor samples. Despite these limitations, this work demonstrates that the immune microenvironment of GBM is associated with molecular subtype, which may have implications for treatment.
Conclusion
Primary GBM and recurrent GBM samples demonstrated similar immune infiltrates. However certain genetic markers such as IDH1, p53, ATRX and MGMT methylation status correlated with immune infiltrates. These findings may have implications for immunotherapy moving forward.
Supplementary Material
Circled odds ratio point estimates differ significantly from one at α=0.05.
Circled odds ratio point estimates differ significantly from one at α=0.05.
Acknowledgments
This work was funded by the Florida Center for Brain Tumor Research. We would like to acknowledge Barbara Frentzen for her work in obtaining samples for analysis through the FCBTR.
References
- 1.Stupp R, Hegi ME, Gorlia T, Erridge SC, Perry J, Hong YK, Aldape KD, Lhermitte B, Pietsch T, Grujicic D, Steinbach JP, Wick W, Tarnawski R, Nam DH, Hau P, Weyerbrock A, Taphoorn MJ, Shen CC, Rao N, Thurzo L, Herrlinger U, Gupta T, Kortmann RD, Adamska K, McBain C, Brandes AA, Tonn JC, Schnell O, Wiegel T, Kim CY, Nabors LB, Reardon DA, van den Bent MJ, Hicking C, Markivskyy A, Picard M, Weller M. Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071-22072 study): a multicentre, randomised, open-label, phase 3 trial. The lancet oncology. 2014;15:1100–1108. doi: 10.1016/s1470-2045(14)70379-1. [DOI] [PubMed] [Google Scholar]
- 2.Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, Carpentier AF, Hoang-Xuan K, Kavan P, Cernea D, Brandes AA, Hilton M, Abrey L, Cloughesy T. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370:709–722. doi: 10.1056/NEJMoa1308345. [DOI] [PubMed] [Google Scholar]
- 3.Weathers SP, Han X, Liu DD, Conrad CA, Gilbert MR, Loghin ME, O'Brien BJ, Penas-Prado M, Puduvalli VK, Tremont-Lukats I, Colen RR, Yung WK, de Groot JF. A randomized phase II trial of standard dose bevacizumab versus low dose bevacizumab plus lomustine (CCNU) in adults with recurrent glioblastoma. Journal of neuro-oncology. 2016;129:487–494. doi: 10.1007/s11060-016-2195-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suva ML, Regev A, Bernstein BE. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science (New York, NY) 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitano H. Cancer as a robust system: implications for anticancer therapy. Nature reviews Cancer. 2004;4:227–235. doi: 10.1038/nrc1300. [DOI] [PubMed] [Google Scholar]
- 6.Inman S. Rintega (Rindopepimut) misses survival endpoint in phase 3 glioblastoma trial. [Accessed 5/12/17 2017];Cure. 2016 [Google Scholar]
- 7.Green J. ImmunoCellular Therapeutics Phase II Study Demonstrates That Glioblastoma Patients Live Longer Without Disease Progression When Treated With ICT-107. Immunocellular therapeutics Ltd. 2013 http://investors.imuc.com/releasedetail.cfm?ReleaseID=813442.
- 8.Abernathy A. Bristol-Myers Squibb Announces Results from CheckMate-143, a Phase 3 Study of Opdivo (nivolumab) in Patients with Glioblastoma Multiforme. Bristol-Myers Squibb. 2017 https://news.bms.com/press-release/bmy/bristol-myers-squibb-announces-results-checkmate-143-phase-3-study-opdivo-nivoluma.
- 9.Yang I, Han SJ, Sughrue ME, Tihan T, Parsa AT. Immune cell infiltrate differences in pilocytic astrocytoma and glioblastoma: evidence of distinct immunological microenvironments that reflect tumor biology. J Neurosurg. 2011;115:505–511. doi: 10.3171/2011.4.jns101172. [DOI] [PubMed] [Google Scholar]
- 10.Hewedi IH, Radwan NA, Shash LS, Elserry TH. Perspectives on the immunologic microenvironment of astrocytomas. Cancer management and research. 2013;5:293–299. doi: 10.2147/cmar.s48942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sayour EJ, McLendon P, McLendon R, De Leon G, Reynolds R, Kresak J, Sampson JH, Mitchell DA. Increased proportion of FoxP3+ regulatory T cells in tumor infiltrating lymphocytes is associated with tumor recurrence and reduced survival in patients with glioblastoma. Cancer immunology, immunotherapy : CII. 2015;64:419–427. doi: 10.1007/s00262-014-1651-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bady P, Sciuscio D, Diserens AC, Bloch J, van den Bent MJ, Marosi C, Dietrich PY, Weller M, Mariani L, Heppner FL, McDonald DR, Lacombe D, Stupp R, Delorenzi M, Hegi ME. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta neuropathologica. 2012;124:547–560. doi: 10.1007/s00401-012-1016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harell FE. Regression modeling strategies: with applications to linear models, logistic and ordinal regression, and survival analysis. Springer; New York: 2015. [Google Scholar]
- 14.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 17:98–110. doi: 10.1016/j.ccr.2009.12.020. doi:S1535-6108(09)00432-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louis DN, Perry A, Burger P, Ellison DW, Reifenberger G, von Deimling A, Aldape K, Brat D, Collins VP, Eberhart C, Figarella-Branger D, Fuller GN, Giangaspero F, Giannini C, Hawkins C, Kleihues P, Korshunov A, Kros JM, Beatriz Lopes M, Ng HK, Ohgaki H, Paulus W, Pietsch T, Rosenblum M, Rushing E, Soylemezoglu F, Wiestler O, Wesseling P. International Society Of Neuropathology--Haarlem consensus guidelines for nervous system tumor classification and grading. Brain pathology (Zurich, Switzerland) 2014;24:429–435. doi: 10.1111/bpa.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta neuropathologica. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 17.Kmiecik J, Poli A, Brons NH, Waha A, Eide GE, Enger PO, Zimmer J, Chekenya M. Elevated CD3+ and CD8+ tumor-infiltrating immune cells correlate with prolonged survival in glioblastoma patients despite integrated immunosuppressive mechanisms in the tumor microenvironment and at the systemic level. Journal of neuroimmunology. 2013;264:71–83. doi: 10.1016/j.jneuroim.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, Zhang C, Liu X, Wang Z, Sun L, Li G, Liang J, Hu H, Liu Y, Zhang W, Jiang T. Molecular and clinical characterization of PD-L1 expression at transcriptional level via 976 samples of brain glioma. Oncoimmunology. 2016;5:e1196310. doi: 10.1080/2162402x.2016.1196310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conroy S, Kruyt FA, Joseph JV, Balasubramaniyan V, Bhat KP, Wagemakers M, Enting RH, Walenkamp AM, den Dunnen WF. Subclassification of newly diagnosed glioblastomas through an immunohistochemical approach. PloS one. 2014;9:e115687. doi: 10.1371/journal.pone.0115687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Circled odds ratio point estimates differ significantly from one at α=0.05.
Circled odds ratio point estimates differ significantly from one at α=0.05.