Abstract
Traditionally, the majority of nucleic acid amplification-based molecular diagnostic tests are done in centralized settings. In recent years, point-of-care tests have been developed for use in low-resource settings away from central laboratories. While most experts agree that point-of-care molecular tests are greatly needed, their availability as cost-effective and easy-to-operate tests remains an unmet goal. In this article, we discuss our efforts to develop a recombinase polymerase amplification reaction-based test that will meet these criteria. First, we describe our efforts in repurposing a low-cost 3D printer as a platform that can carry out medium-throughput, rapid, and high-performing nucleic acid extraction. Next, we address how these purified templates can be rapidly amplified and analyzed using the 3D printer’s heated bed or the deconstructed, low-cost thermal cycler we have developed. In both approaches, real-time isothermal amplification and detection of template DNA or RNA can be accomplished using a low-cost portable detector or smartphone camera. Last, we demonstrate the capability of our technologies using foodborne pathogens and the Zika virus. Our low-cost approach does not employ complicated and high-cost components, making it suitable for resource-limited settings. When integrated and commercialized, it will offer simple sample-to-answer molecular diagnostics.
Keywords: 3D Printers, Low-Resource Settings, Nucleic Acid Extraction, Recombinase Polymerase Amplification, Foodborne Pathogens, Zika Virus
1. Introduction
Traditionally, the majority of in vitro diagnostic (IVD) tests are done in centralized settings. In recent years, point-of-care (POC) or near-patient (NP) tests have been commercialized for use in settings where timing is critical (e.g., emergency triage), laboratory facilities are distant (physician’s offices or urgent-care facilities) or nonexistent (e.g., military settings), or resources are limited (e.g., in developing countries). It is accepted that with a good test performance, the POC/NP approach helps deliver appropriate and prompt treatments and improve clinical outcomes, thus significantly reducing expenditures in medical care.
Integration, portability, low power consumption, automation, ruggedness, and low cost are several important qualities desired by POC/NP test stakeholders in both developed and developing countries. In order for the POC/NP tests to be considered, the cost of these devices, which includes both the material and the manufacturing process, must be kept extremely low, especially in developing countries. The instrument must be portable, and the disposables must be inexpensive. However, commercially available, automated devices that can deliver low- to medium-throughput assays are currently cost prohibitive for use in POC/NP settings.
Proper sample preparation and the availability of such devices are critical to the success of reliable molecular diagnostic assays and the subsequent treatment given.[1, 2] While microfluidic devices have been demonstrated to automate sample preparation processes, [3, 4] they are not commercially available at a reasonable price, possibly because of the challenges related to system integration, complexity, manufacturability, and performance reproducibility.[5] In practice, most infectious disease applications require extraction and concentration of target nucleic acids from a sample input volume of over 500 μL to reach a suitably low limit of detection since pathogen concentration can be < 100 copies/mL. This requirement is very difficult to achieve in microfluidic devices.
Although nucleic acid (NA) purification kits and instruments are available from companies such as Invitrogen, bioMérieux, Roche, and Qiagen, these existing instruments range in cost from $15,000–$80,000. While manually operated sample preparation methods are also available, they are labor intensive and susceptible to contamination, handling variations, or user errors.[6, 7] Research communities have recognized the need for developing suitable sample preparation technologies since there are many recent reports published on new NA isolation and purification methods.[8–19]
The lack of capable devices that are low cost and can perform high-quality and consistent NA isolation is the primary limiting factor in adapting POC molecular diagnostic tests. Diagnostic tests intended for POC use should focus on being cheaper, more intuitive, and more robust to operate.[20–23] Therefore, there is an urgent need to commercialize an affordable, sensitive, and specific sample preparation device for POC use. A simple, low-cost alternative to conventional lab-based equipment would greatly reduce the barriers to providing modern medical diagnostics to low- and middle-income countries.
Consequently, we envisioned adapting a 3D printer to process NA isolation as a commercially viable means to carry out reliable NA isolation that will then lead to more reliable, sensitive, and specific diagnostics being implemented in POC/NP settings. Fig. 1 shows how we have repurposed a 3D printer to control its motion to perform NA isolation in a manner very similar to the work flow used in many magnetic particle-based NA isolation methods (such as the KingFisher mL semi-automatic nucleic acid purification system).
Fig. 1.
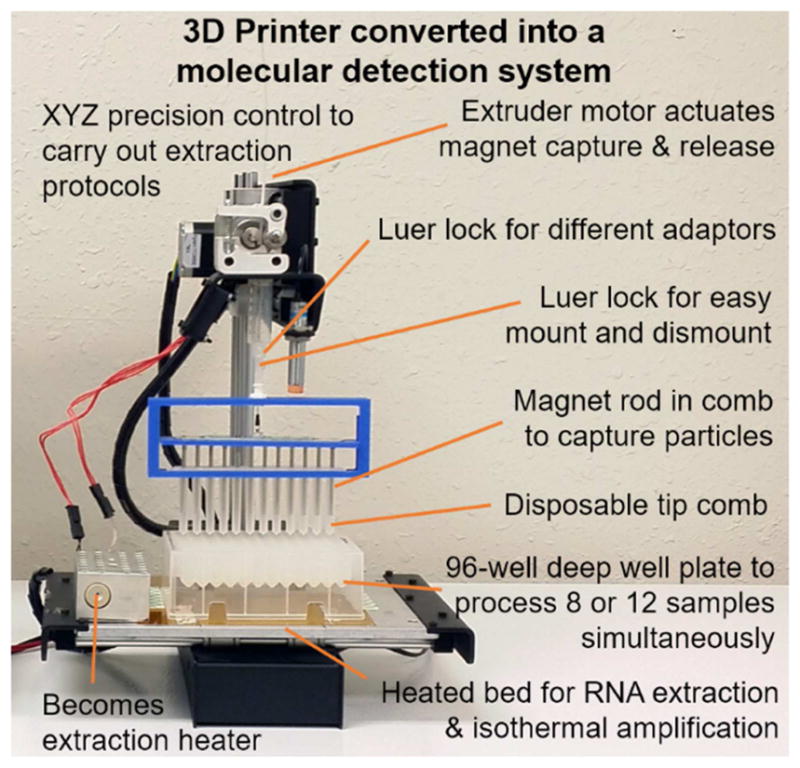
Concept of using a modified 3D printer to carry out molecular diagnostics by converting a low-cost 3D printer to perform rapid and automated NA isolation and real-time RPA amplification. The extruder of a 3D printer was removed to allow the adaptors to be mounted. The magnetic particle processor attachment (MPPA) was attached to the adaptor by a luer-lock mechanism. Its vertical and lateral movements are controlled by the Z-motor and the X and Y platform control. The disposable tip comb houses a set of magnetic rods controlled by the extruder motor. The tip comb and magnetic rods perform MP capture and resuspension via the Z-axis motor and extruder motor. There is no direct contact between the magnets, the samples, and MPs. The printer bed holds a 96-well deep-well microplate or pre-loaded cartridges. Depending on the orientation of the plate, 8 or 12 samples can be processed simultaneously. The heated bed provides elevated temperatures for isothermal amplification.
A 3D printer repurposed to perform nucleic acid extraction
We have previously reported on a method for transforming a standard 3D printer into an automated sample preparation and molecular detection device.[24, 25] We demonstrated that the device could be used to purify and detect DNA- and RNA-based pathogens, including Chlamydia trachomatis, dengue, and the Zika virus (ZIKV). Typical commercial sample preparation equipment can cost up to $25,000, with reaction times of around 40 min. Converting a 3D printer into a sample preparation device costs $750 or less and has a sample preparation processing time of 15 min.
Our previous study showed the C. trachomatis DNA was very efficiently extracted from urine samples using our method in comparison to a gold-standard spin-column-based method from Qiagen, and the NAs from both methods had similar threshold cycle (Cq) values across a range of dilutions.[24] Similar results were seen when extracting RNA from Zika virus samples, and all experiments demonstrated minimal cross-contamination.[25] The changes to the 3D printer are very simple and reversible, so the equipment can be used for 3D printing again. We note that while recombinase polymerase amplification (RPA) does not need extensive sample preparation in some reported cases, it is best if the process is high quality and highly reliable in order to produce the most sensitive and consistent results.
Need for simple nucleic acid amplification and detection
Among different IVD tests, NA-based diagnostic tests for some infectious diseases enable earlier diagnosis when compared to antibody tests.[26] The ability to amplify extremely low sample concentrations of RNA and DNA provide comparably high sensitivity and specificity.[27] Since we have already demonstrated the concept of using a 3D printer for high-quality sample preparation (DNA/RNA extractions), the next step is to simplify NA amplification and detection.
Polymerase chain reaction (PCR) is a common laboratory technique used to make copies of a particular region of DNA. Reverse transcription polymerase chain reaction (RT-PCR), a variant of PCR, is used to detect RNA. Although PCR and RT-PCR are typically used in laboratories in developed countries to amplify NAs, the need for complex equipment, staff training, electricity, and precisely timed thermal cycling limits their applicability in low-resource settings.[28]
In response, isothermal amplification techniques such as loop-mediated amplification (LAMP), helicase-dependent amplification, cross priming amplification, and RPA have been developed to amplify NAs to detectable concentrations in 15–60 min.[29, 30] For example, when combined with chemical heaters that utilize an exothermic reaction with a phase-change material that melts at an appropriate temperature (e.g., 55–65°C for LAMP and 38–42°C for RPA), these amplification techniques enable accurate diagnosis in low-resource settings where electricity is unreliable.[29, 31, 32]
RPA has an advantage over other isothermal amplification technologies because it can be performed at lower temperatures. Also, the reaction mixtures containing enzymes, nucleotides, and buffers are provided in dried pellets, which are very amenable to POC or field use. The only ingredients that need to be added are primers, a probe, and a sample. For the detection of RNAs, reverse transcriptase can be used to enable RT-RPA reactions.[33]
To power the amplification reactions, chemical-based heaters for isothermal reactions have been reported. Some have used the reaction of magnesium metal with water to produce magnesium hydroxide and hydrogen gas.[34] These galvanic, corrosion-based isothermal chemical heaters have been developed by the PATH team and the Liu laboratory.[35–37] This reaction is coupled with a solid-liquid phase-change material (PCM) with an appropriate melting point, such as palmitic acid for LAMP (phase change temperature Tf = 61.9°C), to maintain an appropriate isothermal sample temperature. Recently, Shah et al. utilized exothermic magnesium reaction with methanol functioning as both reactant and PCM, thereby eliminating the need for a separate PCM since methanol boils at 64.7°C.[38] The use of liquid-vapor phase change reduces the system’s thermal load, which may also cut the temperature ramping-up time and a potential increase in holdover time. In our study, we explored two other simple solutions that can perform RPA isothermal amplification reactions.
Using a 3D printer for isothermal amplification reactions
The first approach we took was to use the heated bed of 3D printers for isothermal amplifications. In addition to the sample preparation previously described [25], 3D printers can be converted to perform NA amplification, such as PCR or isothermal amplification. We previously explored the use of 3D printers to perform rapid “shuttle” PCR.[24] PCR reaction tubes can be transferred between two or three aluminum thermal blocks maintained at denaturing and annealing/extension temperatures (operating on the same principle as the Stratagene Robocycler). While the heated bed is maintained at a fixed temperature, the extent of heat transfer from the heated bed to the aluminum blocks can be controlled via spacer materials with different thermal conductivities or contact area with the heated bed. For isothermal amplification such as RPA, which requires only a single temperature for incubation, the heated bed can be precisely maintained at a fixed temperature (e.g., at 40°C) to perform the reactions.
Using thermoses for RPA reactions
Vintage biotech methods are not often revived, but the old-fashioned PCR technique of moving tubes between baths of heated water has now been adapted by our group, with a few adjustments, into an inexpensive thermal cycler for use in low-resource settings. In a proof-of-principle study, we showed that a PCR setup built from thermoses, soup cans, and Arduino-based microcontrollers can rival the efficiency of commercial platforms.[39, 40] The method taps into the unique thermal properties of water and has an innovative method to prevent heat loss by using oil as an insulator and a pseudo container lid for PCR amplification.
We demonstrated that the thermos thermal cycler (TTC) does not require active temperature control or continuous power supply during PCR. This unit costs $130 to build using commercial off-the-shelf items. The use of two or three vacuum-insulated, stainless-steel thermos food jars containing heated water (for denaturation and annealing/extension steps) and a layer of oil on top of the water allows for significantly stabilized temperatures in order for PCR to take place (Fig. 2A). We previously demonstrated rapid PCR and RT-PCR thermal cycling with the TTC at a rate of 15 to 30 sec per PCR cycle.[39, 40]
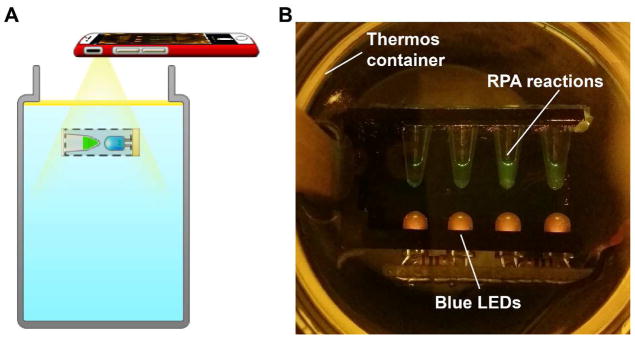
Fig. 2.
A. Device design of a thermos-based isothermal amplification device. With mineral oil as an insulator, we do not need an additional cap to seal the device for heat retention. The setup for fluorescence detection by a smartphone is also shown. An LED powered externally by a 9V battery provides the excitation under water. A phone camera can be placed above the thermos to capture the fluorescence. Low-cost theater color films were used as an emission filter. B. Photo of the reaction tubes immersed in the thermos, excited by the LEDs, and captured in ambient light (not in total darkness).
For this study, we extended the use of the TTC concept to perform RPA isothermal amplification reactions. Only a single thermos is needed for RPA reactions. The water inside the thermos, when covered by oil, can maintain a temperature at or near 40°C for over 1 hr. This means up to six 10-min RPA reactions can be performed.
While our original TTC does not have real-time detection capability for PCR, it is much easier to set up real-time fluorescence detection for use in RPA since the water temperature of the bath will only be at 40°C. For instance, the light-emitting diodes (LEDs) needed for reaction excitation underwater are easier if made waterproof. Because the reaction tubes are stationary (and not shuttled, as in PCR) and the oil layer acts as a transparent heat-retaining lid in place of an external lid covering the thermos, we can monitor the tubes’ changes in fluorescence signal much easier (Fig. 2B).
The remainder of this manuscript demonstrates that a modified 3D printer offers a viable approach to enable rapid and automated DNA and RNA extraction in a low-cost format. We show that the 3D printer can automatically process a wide concentration range of pathogens and specimens without cross-contamination. Most importantly, we demonstrate that the low-cost 3D printer and the TTC can perform RPA and real-time RPA as well as RT-RPA. Together, this proof-of-concept work demonstrates the potential in achieving rapid and high-performing point-of-care RPA molecular assays using very simple instrumentation.
2. Materials and methods
2.1 Repurposing a 3D printer into an automated NA extraction device
Modifications of the 3D printer are shown in Fig. 1. Previous studies have shown that DNA and RNA can be automatically extracted from clinical samples using the bioMérieux NucliSENS magnetic extraction reagents and a modified, consumer-grade 3D printer (Printrbot Simple, Printrbot, Inc., Lincoln, CA).[24] Advancements in the manipulation of MPs for more thorough washing were made, and the results are recorded in another publication.[25]
Automate extraction and amplification steps using G-code
To repurpose a 3D printer for molecular diagnostics, we needed to determine how to control the device to perform MP-based NA extraction and NA amplification. In 3D printing, printers are often operated using open-source host software such as Repetier-Host (Hot-World GmbH & Co., Willich, Germany) and Cura (Ultimaker B.V., Geldermalsenhe, the Netherlands) to slice the 3D model drawings and convert them into G-code, the language the 3D printer speaks. Next, either the software sends the G-code to the printer or the G-code can be copied to a secure digital card to be used by 3D printers that can be operated without a laptop. In a typical 3D printer, the G-code provides instructions, such as what the extruder nozzle temperatures should be, where the tip of the extruder will go, when to deposit melting plastic from the heated nozzle fed with plastic filament, and how much to deposit. We repurposed the 3D printer for NA extraction by writing G-codes to control the motion and many other functions (e.g., controlling temperature at the extruder and the heated bed) of the 3D printer. Using the modified 3D printer, NA was extracted with NucliSENS lysis buffer and magnetic extraction reagents (bioMérieux, Durham, NC). General steps used in our NA extraction protocols were reported previously.[24, 25]
2.2 Samples used in this work
Bacterial culture conditions
Salmonella enterica serovar Typhimurium LT2 (ATCC 19585) and Listeria monocytogenes serotype 4b (ATCC 19115) (ATCC, Manassas, VA) were cultured overnight to the exponential phase in an LB broth (Miller) medium at 37°C. S. enterica was spiked in 2% reduced fat milk at different concentrations (final concentrations of 107, 105, 103, and 0 CFU/mL), and DNA was extracted by our modified 3D printer using NucliSENS lysis buffer and magnetic extraction reagents (bioMérieux, Durham, NC). DNA was also extracted from serial tenfold dilutions of L. monocytogenes overnight culture (2.4×109 CFU/mL) using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA).
Zika virus and saliva specimens
Saliva has been investigated as an alternative sample for routine ZIKV RNA detection as ZIKV was more frequently detected in saliva when compared to blood.[41] ZIKV (a 2015 strain from Mexico) was harvested in Vero cells 4 days post-infection at a concentration of approximately 2 × 107 plaque-forming units (PFU)/mL. The ZIKV suspension was then mixed with three volumes of TRIzol Reagent (Thermo Fisher Scientific [TFS], Waltham, MA) to bring the concentration of ZIKV to 5 × 106 PFU/mL. Deidentified pooled normal human saliva (Catalog# IR100044P) were purchased from Innovative Research (Novi, MI). ZIKV RNA was extracted using our modified 3D printer as described previously.[25] In this protocol, the entire 3D printer-operated extraction process took less than 15 min (5-min lysis and RNA binding, approximately 6-min washing, and 3-min elution).
2.3 Real-time RPA and RT-RPA assays
For the detection of Salmonella, we purchased the TwistGlow® Salmonella Kit that targets the Salmonella enterica invA gene. For the detection of Listeria, we purchased the TwistAmp® exo +ListeriaM kit that targets the Listeria monocytogenes hly gene. We followed the manufacturer’s protocol to perform the RPA reactions. The incubation of the reaction tubes were performed by the 3D printer or inside the thermos.
The quantity and quality of ZIKV RNA templates extracted were evaluated in a commercial real-time PCR detection system (Bio-Rad CFX-96, Hercules, CA) using a SuperScript III Platinum One-Step Quantitative RT-PCR system (TFS, Waltham, MA), as described previously[25], or TaqPath 1-Step Multiplex Master Mix (TFS, Waltham, MA). For the Zika RT-RPA assay, we designed primers and a TwistAmp exo probe that were modified from the real-time RT-PCR primers and probe for the detection of the prM gene of ZIKV.[42] Details of primers and probe design can be found in our previous publication.[25]
Real-time RT-RPA reactions for ZIKV detection were set up using the TwistAmp exo RT kit according to manufacturer instructions (TwistDx, Cambridge, United Kingdom). Optically clear 0.2-mL low-profile PCR micro-centrifuge strips with flat caps were used in this assay. For each 12.5-μL volume reaction, 420 nM each of RPA ZV-F and RPA ZV-R primers, 120 nM of the TwistAmp exo probe, and 2 μL of the RNA template were mixed and centrifuged briefly before adding 14 mM of magnesium acetate to start the reaction.
2.4 Monitoring and recording the real-time RPA reactions
Real-time RPA assay using a benchtop instrument
In this study, real-time RPA was first performed using fluorescence detection in the FAM channel (excitation 470 nm and detection 520 nm) in an ESEQuant Tubescanner (Qiagen Lake Constance GmbH, Stockach, Germany) with a program maintaining 40°C for 15 min with a 30-sec measurement interval. Per instructions from the TwistDx’s protocol, the measurement was paused at 4.5 min to allow for reaction tubes being taken out for brief agitation and then placed back in the tube scanner to resume fluorescence measurements immediately. The threshold time (Tt) of isothermal amplification reaction was determined by using the first derivative of the fluorescent signal curve. When the first derivative became greater than or equal to 0.1, that time point was considered as the Tt of that assay.
Real-time RPA assay using the 3D printer and a USB-powered portable detector
The heated bed of a 3D printer can be used to provide the heat needed for RPA. The program used to control the 3D allows user to set the temperature and temperature probe maintains the bed’s temperature accurately (± 1°C). An aluminum block that holds eight micro-centrifuge tubes was modified to allow viewing of the reagent contents from the front (Fig. 3). The emission of the fluorescence signal was monitored by an ESElog, USB-powered fluorescence detector (Qiagen). This detector was fixed in front of the aluminum block. The heated bed of the 3D printer was programed so that the row of the tubes moved in front of the detector’s sampling lens. The back-and-forth motion of the printer bed allowed periodic sampling by the detector, so the signal intensity over time could be recorded. Alternatively, a blue LED array was placed on top of the tubes to allow fluorescent excitation of the reaction mix, and the signal intensity can be monitored by using a cell phone camera (Fig. 3C).
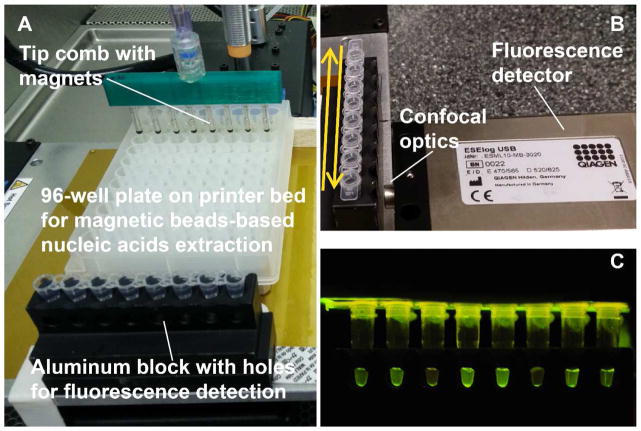
Fig. 3. Set up of 3D printer operated extraction and real-time isothermal amplification.
A. Placement of extraction plate and aluminum block for RPA incubation. B. An ESElog portable fluorescence detector placed in front of the aluminum block holding the tubes. The back-and-forth motion of the printer bed moves the tubes in front of the detection lens. C. An example photography of the RPA reactions excited by blue LED from the top of the tubes; the emission signal was captured from the side opening of the aluminum block.
Low-cost real-time RT-RPA assay using the thermos thermal cycler
NA templates extracted by the 3D printer or by traditional means were first amplified by real-time RT-PCR using a commercial real-time PCR detection system (Bio-Rad CFX-96, Hercules, CA) with SuperScript III Platinum One-Step or TaqPath qRT-PCR kits (both available from TFS, Waltham, MA). The same templates were amplified by RPA using the TTC, but only one thermos was needed as opposed to the setup mentioned in the original TTC papers.[39, 40]
We explored various heating methods for obtaining 40°C water in terms of consistency, time, and energy efficacy. One method was taken from an earlier study of ours and consisted of adding ~15 mL of oil and 520 mL of water at room temperature (~25°C) to a 24-oz Thermos-brand thermos.[40] The remaining empty space in the thermos was filled with boiling water (~155 mL) until the liquid level in the thermos reached where the double wall (and therefore best insulation) ends. This procedure allows a stable water bath at near 40°C to be created in low-resource settings. Both RPA reaction and RT-RPA reaction can be performed in the TTC thermos as long as low-temperature reverse transcriptase is used for amplification of RNAs.
The temperature was measured using K-type thermocouples (Model No. SC-TT-K-30-36, Omega Engineering, Stamford, CT) connected to a data logger thermometer (HH147U, Omega Engineering, Stamford, CT). To create a temperature probe assembly, the PCR micro-centrifuge tube cap was punctured using a 20-gauge syringe needle; the thermocouple (TC) was then inserted through the small hole and secured on the cap using a glue gun to deposit melted glue sticks. The same type of TCs was also used to record the temperature of the water baths by directly immersing the TCs into the water inside the thermoses.
Fluorescent signal detection using smartphone camera
Although the ESEQuant Tubescanner or the ESElog is a small device that can collect fluorescent signal changes over time for up to 12 tubes, it is also an expensive device (over $7,000 and $3,000 per detector, respectively). Therefore, the possibility of using a cell phone as an alternative tool to collect RPA fluorescent signals was explored. Blue LEDs (470 nm, 08R2921, Newark.com) and a cell phone (Samsung Note 3) with a plastic orange filter (Mini LED Transilluminator, I/O Rodeo, Inc., Pasadena, CA) were used to illuminate the tubes and record the signal every 30 sec (Fig. 2). Since cell phone cameras generally auto-adjust brightness and contrast, to maintain consistency during the experiment, we used a cell phone app’s locked-focus, locked-exposure, and time-lapse photography functions (Camera FV-5 installed on a Samsung Note 3 smartphone). The time-lapsed photos were analyzed visually to identify when the signal intensity of the tubes was greater than the negative control reaction’s signal.
3. Results and discussion
3.1 3D printers are fully capable of carrying out high-quality extraction
We first wanted to demonstrate that the 3D printer method can perform automatic NA extraction from saliva samples. We used the ZIKV as an example since RNA extraction is more challenging than DNA extraction.
We used a $399 3D printer (Printrbot Play) to automate magnetic bead-based (Thermo Fisher’s Dynabeads) NA extraction of up to eight samples in 13 min (5-min lysis and NA binding, 3-min washing, and 5-min elution). The input sample volume was 100 μL, and the elution buffer volume was 100 μL. Previous study showed efficient RNA extraction and detection in six orders of ZIKV concentrations spiked in human urine, and the RNA yields were similar to those extracted using the Qiagen spin-column protocol.[25] Thus, our device is capable of extracting RNA from low concentrations of ZIKV in urine. Samples as low as 5 PFU/mL (0.5 PFU per 100 μL used in the extraction) were detected by both the RT-PCR and RT-RPA assays.[25] One advantage of using the isothermal RT-RPA assay for ZIKV detection is that it requires a much shorter time to get results.[25]
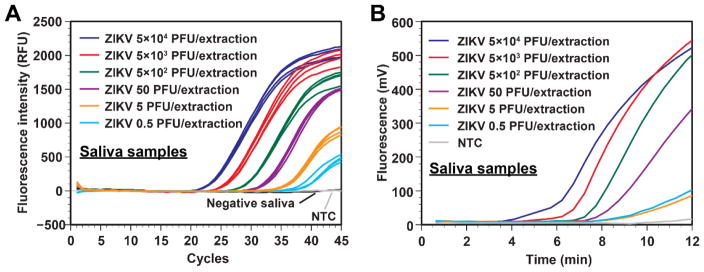
In this study, we used the same extraction protocol with ZIKV in saliva samples. Similar results in sensitivity were obtained when the templates were amplified by real-time RT-PCR (Fig. 4A). The limit of detection for RT-RPA was 0.5 PFU/extraction as the end-point fluorescence was distinguishable from the no-template control (Fig. 4B), even though the fluorescence signal curves of 5 PFU/extraction and 0.5 PFU/extraction could not be discerned.
Fig. 4.
A. Real-time RT-PCR data from serially diluted ZIKV samples spiked in human saliva processed by our 3D printer protocol. ZIKV concentrations as low as 0.5 PFU/extraction can be effectively extracted and detected by a commercial thermal cycler using real-time RT-PCR. Negative saliva samples were placed immediately next to the sample wells with 5 × 104 PFU, indicating that risk of cross-contamination is low. NTC is the no-template control. B. Real-time RT-RPA data using the same RNA templates as in panel A. ZIKV concentrations as low as 0.5 PFU/extraction can be effectively detected. NTC is the no-template control.
3.2 Real-time RPA performed on the 3D printer and ESElog
3D printer movement and scanning
S. enterica (ATCC 19585) was spiked in 2% reduced fat milk at different concentrations (final concentrations of 0, 103, 105, and 107 CFU/mL), in which 100 uL was used in each extraction, and the templates were eluted either in 100 μL elution buffer (used 2 μL template in 25 μL reaction) or directly into 25 μL master mix of Twist Diagnostics’ real-time RPA tests for Salmonella during the same extraction run. With direct elution, the capture DNA was released into a single master mix reaction, thus effectively increased the concentration of template in the reaction (not just 2 μL out of 100 μL of elution buffer). The samples were incubated together at 40°C in an aluminum block placed on the 3D printer’s heated bed, and their signal was monitored by an ESElog, USB-powered fluorescence detector (Qiagen). The aluminum block has holes drilled on the side so that LED excitation can take place above the lid and the emission signal can be measured from the side.
Fig. 5A shows the raw data of the ESElog when the tubes in the aluminum block were moved back and forth by the 3D printer’s bed in front of the fixed detector lens. The back-and-forth motion of the 3D printer bed was programmed to allow each sample to be measured for 5 sec each time, so 1 sample has at least 5 data points for every time it’s being sampled in front of the detector. We also programmed the 3D printer’s movement in a way that it quickly moves 9-mm (standard spacing in 96-well plate) to bring another tube in front of the detector for another 5 second measurement.
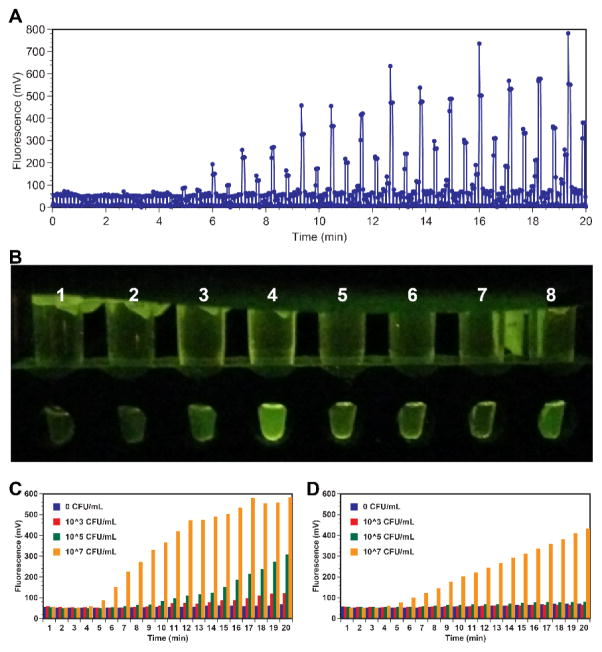
Fig. 5.
A. RPA reaction tubes were incubated and fluorescence measurement was taken every 1 sec for 20 min. Shaking of the tubes was not performed after reactions had begun. Signal growth was observed over time in tubes with high DNA template concentration. B. Photograph of the reaction tubes at the 10-min mark as taken by a cell phone. The left four tubes (1–4) were RPA reactions with the templates (from 0, 103, 105, and 107 CFU/mL samples) directly eluted into an RPA master mix. The four tubes (5 to 8) on the right are reactions (from 0, 103, 105, and 107 CFU/mL samples) with their DNA eluted into an elution buffer first, and a portion added to the RPA master mix. C. Fluorescence intensity (collected by the ESElog) of the tubes performed RPA with direct eluted templates into the master mix (25 μL). D. Fluorescence intensity (collected by the ESElog) of the tubes performed RPA with eluted templates (2 μL out of 100 μL of elution buffer).
The signal of some samples visibly increased gradually over time. Fig. 5B is an endpoint photograph that shows that the direct elution reactions produced a distinguishably higher green signal (tube 1 to 4 with 4 > 3 > 2 > 1) than those using just a 2 μL template out of a 100 μL elution buffer (tube 5 to 8 with 8 > 7 > 6 > 5). With limited testing, we were able to detect 103 CFU/mL with direct elution (100 CFU/extraction) and 105 CFU/mL (104 CFU/extraction) with a 2 μL template in 10 min. Signals from tube number 4 (107 CFU/mL) and 5 (−ve) also suggest minimal cross-contamination as they were extracted side-by-side on the 3D printer. Fig. 5C and Fig. 5D are the detected signals extracted from Fig. 5A with samples from direct elution (5C) and a portion of the total template (5D). As the reaction time increases, we can see that the signal of samples with positive template increased in portion to the template concentration. In addition, we can see that samples from direct elution reached higher overall signal than samples using just a couple μL of the entire eluted buffer. This is expected as with direct elution, all the template eluted into a single RPA reaction, effectively increased RNA concentration in each reaction.
3.3 Temperature stability of TTC for RPA runs
After successfully demonstrating the use of a 3D printer for sample preparation and RPA, we moved to demonstrate that a single thermos of TTC can be used for RPA. Fig. 6A shows representative temperature-time curves for temperature of the water inside thermoses with starting temperatures of 60 and 40°C. These two temperatures are commonly used for LAMP and RPA, respectively. The temperatures of these thermos baths were measured for 67 min. The temperature was measured using K-type thermocouples (Model No. SC-TT-K-30-36, Omega Engineering, Stamford, CT) connected to a data logger thermometer (HH147U, Omega Engineering, Stamford, CT). In each thermos, one probe was placed inside the thermos for 1 hr. Another thermometer was placed inside a PCR micro-centrifuge tube containing 25 μL of water. The tube was moved in and out of the bath around every 10 min, mimicking the step of performing multiple runs of isothermal amplification reaction within that period.
Fig. 6.
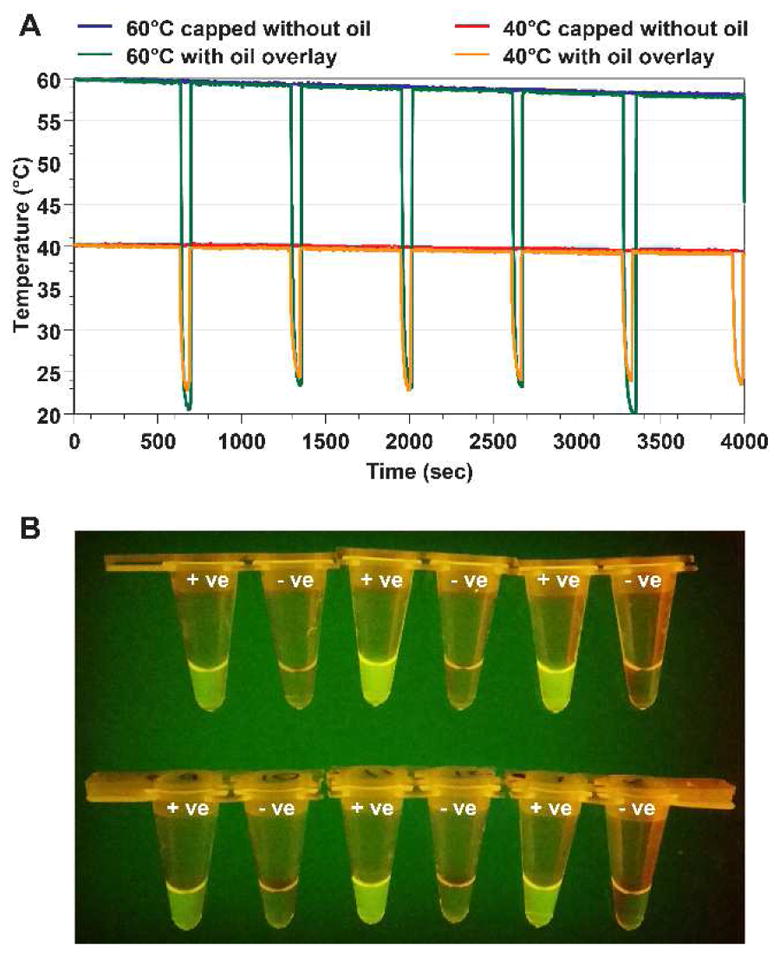
A. Characteristic temperature-time profiles that demonstrate the ability of the device to keep the temperature near 60°C and 40°C without a lid. The oil layer serves as an insulator to drastically reduce heat loss. The tubes immersed in the thermos were removed every 10 min, mimicking the time needed for an RPA run. The plots show that the temperature inside the thermos only dropped 2.1°C and 1.0°C after 66 min. B. The thermos performed six runs of RPA for the Salmonella DNA template without an active control (10 min for each run and six total runs in about 1 hr). The image shows that all six reactions produced the expected result, with positive template tubes producing increased fluorescence.
The temperature plot shows relatively little temperature change in over 1 hr (4000 sec). The temperature dropped only 2.1°C and 1.0°C for the thermoses starting at temperatures of 60 and 40°C, respectively. In addition, the motion of removing and replacing tubes (large temperature drops in the temperature profile) did not lead to a significant temperature drop. The thermoses’ ability to maintain temperatures is likely better than most chemical-based devices since switching reaction tubes will cause temperature drop, and additional time would be needed for the chamber temperature to be restored to the right reaction temperature to begin another RPA run.
Next, the thermos setup was used to carry out six consecutive RPA reactions for the detection of Salmonella DNA. A set of six replicate positive samples (in 100 μL of undiluted culture sample) and no-template control samples was prepared and then tested sequentially. Each reaction was incubated for 10 min without agitation. After the runs, the tubes were placed on top of the blue LED light box (Lonza) to view the fluorescent signal, which was recorded by a smartphone camera (Fig. 6B). The fluorescent signal of these tubes after the reaction was measured using a Tubescanner fluorescence detector, and the intensity is presented in Fig. 6B.
3.4 Comparing RPA results from the TTC and a commercial device for RPA
We demonstrated the use of single Thermos TTC for semi-quantitative analysis by amplifying samples with a serially diluted Listeria DNA template. The undiluted Listeria DNA template was estimated to have 9.6×106 genome copies per 2 μL used in the RPA reaction. As a control, the same reaction mixtures were amplified using a Qiagen ESEQuant Tubescanner benchtop device. Mixing of the tubes’ content was performed at the 4 min mark. The tubes from both devices were placed side-by-side on a blue LED gel box (Lonza) and a photo was taken (Fig. 7A). We saw that both devices can amplify the template with the same sensitivity (~9.6×102 genome copies per 2 μL). Both devices were able to amplify and deliver a positive result for the first five concentrations.
Fig. 7.
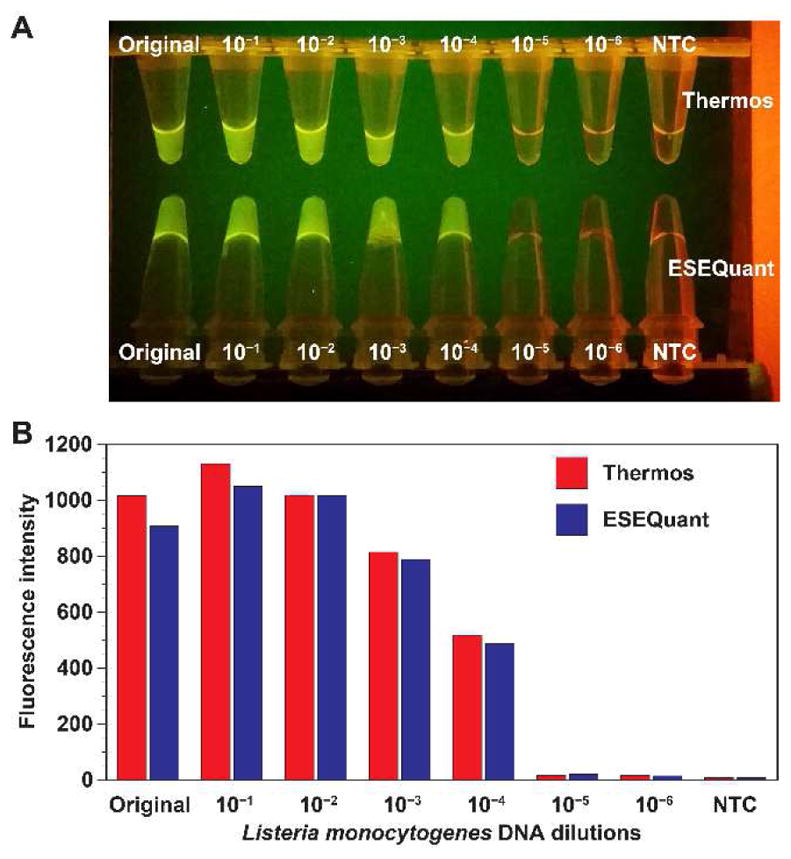
A. RPA with serially diluted Listeria DNA templates performed in Thermos vs ESEQuant Tubescanner. Mixing of the tubes were performed 4 min after the reactions have started. The image on a gel box shows that the tubes have high intensity with a higher concentration of the template. Both sets of tubes used in the TTC and the Tubescanner have similar intensity when viewed by the naked eye. The limit of detection is estimated to be 9.6×102 genome copies per 2 μL of DNA template used in the reaction. B. The endpoint fluorescence signal of RPA reaction tubes performed by the TTC (red) was measured by the Tubescanner. The signals of the RPA reaction tubes performed entirely in the Tubescanner are shown in blue.
The endpoint signal of the reaction tubes from TTC was also measured inside the Tubescanner and compared to that of the tubes processed entirely inside the Tubescanner (Fig. 7B). The incubation time for both sets were 20 min. Based on the reading of the Tubescanner detector, the endpoint signal intensities of the reactions that underwent two different incubation methods were very similar since both approaches can carry out results with similar overall sensitivity.
3.5 Real-time TTC for the real-time detection of L. monocytogenes in milk
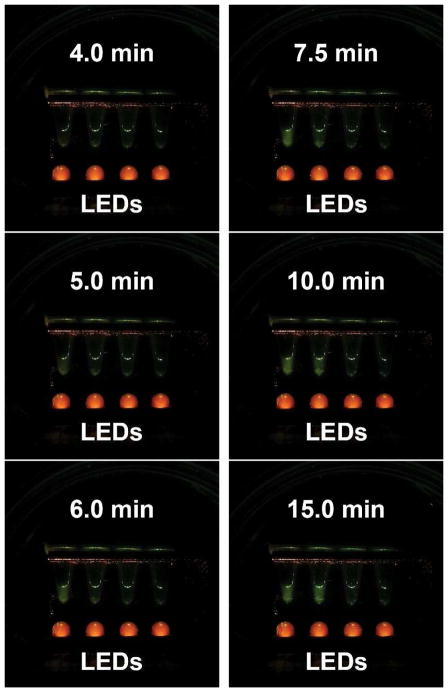
We next showed that the real-time RPA can be performed in the TTC. Listeria concentrations in milk were appropriately 2.4 × 108, 2.4 × 107, and 2.4 × 106 CFU/mL, and they were extracted manually using the spin-column method. The extracted templates were amplified by real-time RPA along with Salmonella DNA as negative template control (right to left). The tubes were incubated at 40°C in the thermos without agitation/mixing after immersion. The signal was monitored every 30 sec by a Samsung Note 3 cell phone camera. The photos taken at 4, 5, 6, 7.5, 10, and 15 min are presented in Fig. 8. The signal rose first from the most concentrated sample and became clearly visible to the naked eye and camera after 5 min, and then the next most concentrated sample’s signal rise became visible at 7.5 min. The third most concentrated Listeria sample’s signal rose at 10 min, although it was barely visible to the naked eye and camera. The signal from negative control (Salmonella DNA) remained very low. In short, the Tt of the reactions were 5, 7.5, and 10 min. This shows that monitoring signals inside a thermos with a cell phone camera is promising. As a control, the signal from a Qiagen Tubescanner showed Tt of 3.5, 4.5, and 6.5 min using the same templates (data not shown). We speculate that the slower reaction in the thermos is due to the skipping of the manufacturer’s recommended manual agitation/mixing step a few min after the initiation of the reaction in the thermos. To circumvent the need of mixing/agitation after the incubation has already begun, another researcher has recently reported that using a smaller volume of reaction without agitation during RPA run can still achieve sensitive results with minimal adverse effects on sensitivity.[43]
Fig. 8.
Underwater real-time RPA amplification of Listeria DNA. Samples from left to right: 2.4×108 Listeria CFU/mL, 2.4×107 Listeria CFU/mL, 2.4×106 Listeria CFU/mL, and Salmonella DNA as negative control. These cell phone images were taken over a period of 15 min. The images show that RPA amplification in the thermos can be monitored in real time using a smartphone above the oil layer inside the thermos. No active temperature control was needed. Since the oil layer is transparent and can act as a lid for the reaction chamber, the real-time RPA progression can also be monitored by the naked eye when an appropriate orange filter for emission viewing is used by the viewer.
4. Conclusions
This study demonstrates that a low-cost, entry level, $400 3D printer can be repurposed to perform automated high quality NA extraction; the extraction can be followed by simple methods to amplify these extracted templates using RPA isothermal amplification. Our modified 3D printer performed automated MP-based NA extraction and purification for up to 12 samples in a single run in less than 15 min. The 3D printer-operated extraction can handle samples with a 6-log difference in target concentration. We have demonstrated that the heated bed of a 3D printer can be utilized to perform RPA incubation, while the fluorescent signal from a real-time RPA assay can be monitored by a portable fluorescence detector or smartphone camera.
In addition, the simplicity of the TTC setup also allowed us to accomplish multiple RPA runs without active temperature control between runs. We also demonstrated real-time detection inside the thermos using a cell phone as a detector above the oil layer. Unlike other isothermal chemical heaters, our thermos-based device has the potential to simplify device design and manufacturability by using water and oil layer. Because of the high specific heat capacity of water and using the oil layer as a lid, multiple runs can be performed once a stable, heated bath is prepared and no active temperature control is needed. In addition, the thermos bath temperature can be changed for other isothermal amplification assays. This eliminates the need for specific PCMs and increases the likelihood of successful uptake by lowering barriers to material access and precise machining requirements. When our system is further optimized, it will have significant potential to improve access to molecular diagnostics in the developing world.
Acknowledgments
Funding was provided by the National Institutes of Health (NIH) through the Small Business Innovative Research program (NIH grants No. 1R43AI129061 and 2R44AI106239). The content is solely the responsibility of the authors and does not necessarily represent or reflect views of the NIH or the federal government. We thank Dr. Scott C. Weaver for kindly providing us the Zika virus sample.
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Note: Season Wong is the co-founder of AI Biosciences, Inc. All other authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dundas N, Leos NK, Mitui M, Revell P, Rogers BB. Comparison of Automated Nucleic Acid Extraction Methods with Manual Extraction. J Mol Diagn. 2008;10:311–316. doi: 10.2353/jmoldx.2008.070149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang YW, Sefers SE, Li H, Kohn DJ, Procop GW. Comparative Evaluation of Three Commercial Systems for Nucleic Acid Extraction from Urine Specimens. J Clin Microbiol. 2005;43:4830–4833. doi: 10.1128/JCM.43.9.4830-4833.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price CW, Leslie DC, Landers JP. Nucleic acid extraction techniques and application to the microchip. Lab on a Chip. 2009;9:2484–2494. doi: 10.1039/b907652m. [DOI] [PubMed] [Google Scholar]
- 4.Kim J, Johnson M, Hill P, Gale BK. Microfluidic sample preparation: cell lysis and nucleic acid purification. Integrative Biology. 2009;1:574–586. doi: 10.1039/b905844c. [DOI] [PubMed] [Google Scholar]
- 5.Dineva MA, Mahilum-Tapay L, Lee H. Sample preparation: a challenge in the development of point-of-care nucleic acid-based assays for resource-limited settings. The Analyst. 2007;132:1193–1199. doi: 10.1039/b705672a. [DOI] [PubMed] [Google Scholar]
- 6.Jungkind D. Automation of laboratory testing for infectious diseases using the polymerase chain reaction -- our past, our present, our future. Journal of Clinical Virology. 2001;20:1–6. doi: 10.1016/s1386-6532(00)00148-7. [DOI] [PubMed] [Google Scholar]
- 7.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 8.Sur K, McFall SM, Yeh ET, Jangam SR, Hayden MA, Stroupe SD, Kelso DM. Immiscible Phase Nucleic Acid Purification Eliminates PCR Inhibitors with a Single Pass of Paramagnetic Particles through a Hydrophobic Liquid. The Journal of Molecular Diagnostics. 2010;12:620–628. doi: 10.2353/jmoldx.2010.090190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jangam SR, Yamada DH, McFall SM, Kelso DM. Rapid, Point-of-Care Extraction of Human Immunodeficiency Virus Type 1 Proviral DNA from Whole Blood for Detection by Real-Time PCR. Journal of Clinical Microbiology. 2009;47:2363–2368. doi: 10.1128/JCM.r00092-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strotman LN, Lin G, Berry SM, Johnson EA, Beebe DJ. Facile and rapid DNA extraction and purification from food matrices using IFAST (immiscible filtration assisted by surface tension) Analyst. 2012;137:4023–4028. doi: 10.1039/c2an35506j. [DOI] [PubMed] [Google Scholar]
- 11.Berry SM, Alarid ET, Beebe DJ. One-step purification of nucleic acid for gene expression analysis via Immiscible Filtration Assisted by Surface Tension (IFAST) Lab on a Chip. 2011;11:1747–1753. doi: 10.1039/c1lc00004g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strotman L, O’Connell R, Casavant BP, Berry SM, Sperger JM, Lang JM, Beebe DJ. Selective Nucleic Acid Removal via Exclusion (SNARE): Capturing mRNA and DNA from a Single Sample. Analytical Chemistry. 2013 doi: 10.1021/ac402162r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi X, Chen C-H, Gao W, Chao S-h, Meldrum DR. Parallel RNA extraction using magnetic beads and a droplet array. Lab on a Chip. 2015;15:1059–1065. doi: 10.1039/c4lc01111b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrnes S, Fan A, Trueb J, Jareczek F, Mazzochette M, Sharon A, Sauer-Budge AF, Klapperich CM. A portable, pressure driven, room temperature nucleic acid extraction and storage system for point of care molecular diagnostics. Analytical Methods. 2013;5:3177–3184. doi: 10.1039/C3AY40162F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bordelon H, Adams NM, Klemm AS, Russ PK, Williams JV, Talbot HK, Wright DW, Haselton FR. Development of a Low-Resource RNA Extraction Cassette Based on Surface Tension Valves. ACS Applied Materials & Interfaces. 2011;3:2161–2168. doi: 10.1021/am2004009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Liu Y, Chen L, Quan S, Jiang S, Zhang D, Yang L. One Simple DNA Extraction Device and Its Combination with Modified Visual Loop-Mediated Isothermal Amplification for Rapid On-Field Detection of Genetically Modified Organisms. Analytical Chemistry. 2012;85:75–82. doi: 10.1021/ac301640p. [DOI] [PubMed] [Google Scholar]
- 17.Linnes JC, Fan A, Rodriguez NM, Lemieux B, Kong H, Klapperich CM. Paper-based molecular diagnostic for Chlamydia trachomatis. RSC advances. 2014;4:42245–42251. doi: 10.1039/C4RA07911F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandeventer PE, Weigel KM, Salazar J, Erwin B, Irvine B, Doebler R, Nadim A, Cangelosi GA, Niemz A. Mechanical Disruption of Lysis-Resistant Bacterial Cells by Use of a Miniature, Low-Power, Disposable Device. Journal of Clinical Microbiology. 2011;49:2533–2539. doi: 10.1128/JCM.02171-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi JR, Tang R, Wang S, Wan Abas WAB, Pingguan-Murphy B, Xu F. Paper-based sample-to-answer molecular diagnostic platform for point-of-care diagnostics. Biosensors and Bioelectronics. 2015;74:427–439. doi: 10.1016/j.bios.2015.06.065. [DOI] [PubMed] [Google Scholar]
- 20.Huang S, Do J, Mahalanabis M, Fan A, Zhao L, Jepeal L, Singh SK, Klapperich CM. Low cost extraction and isothermal amplification of DNA for infectious diarrhea diagnosis. PLoS One. 2013;8:e60059. doi: 10.1371/journal.pone.0060059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Connelly JT, Rolland JP, Whitesides GM. “Paper Machine” for Molecular Diagnostics. Anal Chem. 2015;87:7595–7601. doi: 10.1021/acs.analchem.5b00411. [DOI] [PubMed] [Google Scholar]
- 22.Jangam SR, Agarwal AK, Sur K, Kelso DM. A point-of-care PCR test for HIV-1 detection in resource-limited settings. Biosens Bioelectron. 2013;42:69–75. doi: 10.1016/j.bios.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 23.Yager P, Domingo GJ, Gerdes J. Point-of-care diagnostics for global health. Annu Rev Biomed Eng. 2008;10:107–144. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- 24.Chan K, Coen M, Hardick J, Gaydos CA, Wong KY, Smith C, Wilson SA, Vayugundla SP, Wong S. Low-Cost 3D Printers Enable High-Quality and Automated Sample Preparation and Molecular Detection. PLOS ONE. 2016;11:e0158502. doi: 10.1371/journal.pone.0158502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan K, Weaver SC, Wong P-Y, Lie S, Wang E, Guerbois M, Vayugundla SP, Wong S. Rapid, Affordable and Portable Medium-Throughput Molecular Device for Zika Virus. Scientific Reports. 2016;6:38223. doi: 10.1038/srep38223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel P, Bennett B, Sullivan T, Parker MM, Heffelfinger JD, Sullivan PS, Group CAS. Rapid HIV screening: missed opportunities for HIV diagnosis and prevention. Journal of Clinical Virology. 2012;54:42–47. doi: 10.1016/j.jcv.2012.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hadgu A, Dendukuri N, Hilden J. Evaluation of nucleic acid amplification tests in the absence of a perfect gold-standard test: a review of the statistical and epidemiologic issues. Epidemiology. 2005;16:604–612. doi: 10.1097/01.ede.0000173042.07579.17. [DOI] [PubMed] [Google Scholar]
- 28.Rohrman BA, Leautaud V, Molyneux E, Richards-Kortum RR. A lateral flow assay for quantitative detection of amplified HIV-1 RNA. PLoS One. 2012;7:e45611. doi: 10.1371/journal.pone.0045611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zanoli LM, Spoto G. Isothermal amplification methods for the detection of nucleic acids in microfluidic devices. Biosensors. 2012;3:18–43. doi: 10.3390/bios3010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu G, Hu L, Zhong H, Wang H, Yusa S-i, Weiss TC, Romaniuk PJ, Pickerill S, You Q. Cross priming amplification: mechanism and optimization for isothermal DNA amplification. Scientific reports. 2012;2 doi: 10.1038/srep00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waters RA, Fowler VL, Armson B, Nelson N, Gloster J, Paton DJ, King DP. Preliminary validation of direct detection of foot-and-mouth disease virus within clinical samples using reverse transcription loop-mediated isothermal amplification coupled with a simple lateral flow device for detection. PloS one. 2014;9:e105630. doi: 10.1371/journal.pone.0105630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lillis L, Lehman D, Singhal MC, Cantera J, Singleton J, Labarre P, Toyama A, Piepenburg O, Parker M, Wood R. Non-instrumented incubation of a recombinase polymerase amplification assay for the rapid and sensitive detection of proviral HIV-1 DNA. PloS one. 2014;9:e108189. doi: 10.1371/journal.pone.0108189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Euler M, Wang Y, Nentwich O, Piepenburg O, Hufert FT, Weidmann M. Recombinase polymerase amplification assay for rapid detection of Rift Valley fever virus. Journal of Clinical Virology. 2012;54:308–312. doi: 10.1016/j.jcv.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 34.Taub IA, Roberts W, LaGambina S, Kustin K. Mechanism of Dihydrogen Formation in the Magnesium-Water Reaction⊥. The Journal of Physical Chemistry A. 2002;106:8070–8078. [Google Scholar]
- 35.Singleton J, Osborn JL, Lillis L, Hawkins K, Guelig D, Price W, Johns R, Ebels K, Boyle D, Weigl B. Electricity-free amplification and detection for molecular point-of-care diagnosis of HIV-1. PloS one. 2014;9:e113693. doi: 10.1371/journal.pone.0113693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C, Mauk MG, Hart R, Qiu X, Bau HH. A self-heating cartridge for molecular diagnostics. Lab on a Chip. 2011;11:2686–2692. doi: 10.1039/c1lc20345b. [DOI] [PubMed] [Google Scholar]
- 37.LaBarre P, Hawkins KR, Gerlach J, Wilmoth J, Beddoe A, Singleton J, Boyle D, Weigl B. A simple, inexpensive device for nucleic acid amplification without electricity—toward instrument-free molecular diagnostics in low-resource settings. PloS one. 2011;6:e19738. doi: 10.1371/journal.pone.0019738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shah KG, Guelig D, Diesburg S, Buser J, Burton R, LaBarre P, Richards-Kortum R, Weigl B. Design of a New Type of Compact Chemical Heater for Isothermal Nucleic Acid Amplification. PLoS ONE. 2015;10:e0139449. doi: 10.1371/journal.pone.0139449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan K, Wong PY, Yu P, Hardick J, Wong KY, Wilson SA, Wu T, Hui Z, Gaydos C, Wong SS. A Rapid and Low-Cost PCR Thermal Cycler for Infectious Disease Diagnostics. PLoS ONE. 2016;11:e0149150. doi: 10.1371/journal.pone.0149150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong G, Wong I, Chan K, Hsieh Y, Wong S. A Rapid and Low-Cost PCR Thermal Cycler for Low Resource Settings. PLoS ONE. 2015;10:e0131701. doi: 10.1371/journal.pone.0131701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Musso D, Roche C, Nhan TX, Robin E, Teissier A, Cao-Lormeau VM. Detection of Zika virus in saliva. J Clin Virol. 2015;68:53–55. doi: 10.1016/j.jcv.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 42.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerging infectious diseases. 2008;14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lillis L, Siverson J, Lee A, Cantera J, Parker M, Piepenburg O, Lehman DA, Boyle DS. Factors influencing Recombinase polymerase amplification (RPA) assay outcomes at point of care. Molecular and Cellular Probes. 2016;30:74–78. doi: 10.1016/j.mcp.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]