Abstract
Post-translational modifications are biologically important and wide-spread modulators of protein function. Although methods for detecting the presence of specific modifications are becoming established, approaches for quantifying their mol modification/mol protein stoichiometry are less well developed. Here we introduce a ratiometric, label-free, targeted liquid chromatography tandem mass spectroscopy-based method for estimating Lys and Arg methylation stoichiometry on post-translationally modified proteins. Methylated Lys and Arg were detected with limits of quantification at low fmol and with linearity extending from 20 – 5000 fmol. This level of sensitivity allowed estimation of methylation stoichiometry from microgram quantities of various proteins, including those derived from either recombinant or tissue sources. The method also disaggregated total methylation stoichiometry into its elementary mono-, di-, and tri-methylated residue components. In addition to being compatible with kinetic experiments of protein methylation, the approach will be especially useful for characterizing methylation states of proteins isolated from cells and tissues.
Keywords: Post-translational modification, protein methylation, mass spectrometry, label-free
Introduction
Protein methylation is a post-translational modification implicated in the control of gene expression and other functions [1]. Although extensively characterized in the context of nuclear core histone proteins, the majority of methylation occurs on non-histone proteins [2]. For example, the microtubule-associated protein tau is methylated on ≥17 Lys and Arg residues when isolated from mammalian brain [3, 4]. “Bottom-up” proteomic approaches indicate that the tau methylation signature varies with disease state, and is positioned to modulate tau aggregation propensity and turnover [3, 5]. However, the full functional implications of these observations remain unknown in part because biological effects are mediated by modification stoichiometry, and bottom-up approaches alone capture site distribution but not occupancy. Characterization of complex methylation substrates such as tau would benefit from a “top-down” method [6] capable of capturing overall stoichiometry in terms of quantity (mol methyl equivalents per mol of protein) and quality (chemical form) of methylation under various biological conditions.
The classic top-down approach for characterizing bulk methylation stoichiometry from an ensemble of intact proteins is amino acid analysis, which leverages the unique stability of Lys [7], Arg [7, 8], and their methylated derivatives 1meK, 2meK, 3mek, 1meR and 2meR (composed of both ADMA, NG,NG′-Dimethyl-L-arginine; and SDMA, NG,NG′-Dimethyl-L-arginine) upon acid hydrolysis [9, 10]. In contrast, other Lys and Arg derivatives are unstable and reduce to the parent amino acids under these conditions. For example, the most prevalent stable mammalian Lys modifications yield amide linkages through acylation (e.g., acetylation, etc; [11]) and isopeptide linkages through conjugation with ubiquitin-like proteins [12], both of which readily hydrolyze in parallel with peptide bonds. Similarly, ADP-ribosylation of Arg, which is mediated by N,O-acetal linkages, also is acid labile. As a result, it has been possible to estimate Lys and Arg methylation stoichiometries for specific proteins including calmodulin [13] and myelin basic protein (MBP) [14–17]. Nonetheless, limitations of amino acid analysis have hampered its general application. First, simultaneous separation of Lys, Arg, and all of their methylated derivatives by liquid chromatography is problematic [10], and so methylation stoichiometry has been reported primarily for proteins of low methylation complexity such as calmodulin and MBP (calmodulin contains 3meK and no other methylated residue [13], whereas MBP methylation is limited to 1meR and 2meR [14–17]). Second, amino acid analysis requires derivitization for quantification, which yields poor sensitivity with classic colorimetric agents such as ninhydrin [18]. Replacement with fluorometric detection greatly improves sensitivity [9, 10], but even mid-pmol quantification is not adequate for characterizing proteins isolated in low abundance from tissues. Moreover, common derivitization agents such as o-pthalaldehyde (OPA) yield Lys adducts that dimerize and quench [19], thereby lowering detection sensitivity for this amino acid still further. Alternative top-down approaches are confined to samples where the complexity of methylation is modest [20].
Here we introduce a targeted mass spectrometric approach for quantifying protein Lys and Arg methylation stoichiometry. The method leverages the established stability of methylated amino acids to acid hydrolysis, but then uses LC-MS/MS to quantify analytes with high accuracy and sensitivity. Because targeted precursor/product ion pairs are detected by a triple-quadrupole mass spectrometer operated in multiple reaction monitoring (MRM) mode, the approach yields simultaneous and sensitive detection of even complex analyte mixtures. The final assay successfully quantified the methylation stoichiometry of multiply modified, tissue-derived proteins.
Experimental
Materials
Recombinant human 2N4R tau was prepared as described previously [3]. All other reagents were obtained from commercial vendors, including Sigma-Aldrich (St Louis, MO) for calmodulin from bovine testes (P1431), MBP from bovine brain (M1891), unfractionated whole histone from calf thymus (H9250), PVDF membrane (IPVH00005), 1meR (M7033), ADMA (D4268), SDMA (D0390), and all unmodified L-amino acids, and Chem-Impex Intl (Wood Dale, IL) for amino acids 1meK, 2meK and 3meK.
Reductive tau methylation
Recombinant human 2N4R tau was reductively methylated with NaBH3CN and formaldehyde as described previously [3, 21]. Reactions were quenched by addition of glycine after 0, 7, 15, 30 and 60 min incubation. Non-methylated controls were processed identically, except that formaldehyde was omitted from the reaction.
SDS PAGE and blotting
Tau proteins were electrophoresed through 8% acrylamide gels, then transferred (100 V for 1 h at 4°C) to PVDF membranes in Transfer Buffer A (25 mM Tris, 0.2 mM glycine, 10% methanol). PVDF membranes were then stained (50% Methanol, 7% Acetic Acid, 0.1% Coomassie Brilliant Blue) for 30 min at room temperature with agitation, and then destained (50% methanol, 7% acetic acid) for ~10 min at room temperature with agitation until the staining pattern was visible. After membranes dried at room temperature, Coomassie-blue stained bands were excised using a razor blade (2 × 6 mm slices) and stored at −20°C until used.
MBP, unfractionated histones, and calmodulin were subjected to SDS-PAGE as above, except that electrophoresis was performed through 15% acrylamide gels. MBP and unfractionated histones were then transferred (100 V for 1 h at 4°C) to PVDF membranes in Transfer Buffer B (25 mM Tris-HCl, 0.2 mM glycine, 20% methanol), whereas calmodulin was transferred (100 V for 1 h at 4°C) to PVDF in Transfer Buffer C (25 mM Tris, 0.2 mM glycine, 2 mM CaCl2, 20% methanol) as reported previously [22]. Staining and band excision were performed in the same way as described above for tau proteins.
Acid hydrolysis
Excised PVDF membranes were placed in glass tubes containing 1 mL 6 M HCl and purged with nitrogen as described previously [23]. Samples were then sealed and incubated for 24 h at 125°C. Following hydrolysis, samples were dried under nitrogen at 60°C and then stored at −20°C until LC-MS/MS analysis.
LC-MS/MS quantification
Dried hydrolysates were resuspended in 250 μL of 10 mM HCl, vortexed, filtered (0.2 μm Pall nanosep Mf operated at 14,000g for 10 min at 4°C), and finally prepared for LC by 10-fold dilution with water in glass vials. LC separations were performed on a Hypercarb column (100 × 2.1 mm, 5 μm pore; Thermo Fisher Scientific) operated at 0.2 mL/min using a mobile phase prepared from mixtures of Solvent A (acetonitrile containing 0.1% formic acid) and Solvent B (water containing 0.1% formic acid). The gradient expressed in terms of %-Solvent A was: 0–1 min, 0%; 1–1.1 min, 20%; 1.1–2.7 min, 45%; 2.7–5 min, 60%; 5–5.1 min 90%; 5.1–7 min, 90%; 7–7.1 min, 0%; 7.1–10 min, 0% [24].
Mass spectra were acquired on a triple-quadrupole QTRAP 5500 (AB Sciex) using turbo spray ionization at 2.5 kV in positive ion mode and MRM of parent and characteristic product ions (the transitions monitored are listed in Table 1). The curtain gas (nitrogen) and the collision-activated dissociation were set to 30 psi and medium, respectively. The MS was set to have a dwell time of 35 ms. Analyst 1.6.1 software was used to acquire and process all data.
Table 1.
Mass spectrometry parameters and calibration of amino acids.
| Amino Acid | Precursor ion | Product ion | Retention timea | DPb | EPb | CEb | CXPb | Linear range | r2 | LoDc | LoQc |
|---|---|---|---|---|---|---|---|---|---|---|---|
| (mass) | (mass) | (min ± SD) | (volts) | (volts) | (volts) | (volts) | (fmol) | (fmol) | (fmol) | ||
| Lysine and derivatives | |||||||||||
| Lys | 147 | 84 | 1.16 ± 0.01 | 40 | 10 | 24 | 12 | 20 – 5000 | 0.995 | 1.30 | 4.35 |
| 1meK | 161 | 84 | 1.27 ± 0.02 | 70 | 10 | 23 | 10 | 20 – 5000 | 0.998 | 0.56 | 1.85 |
| 2meK | 175 | 84 | 1.39 ± 0.07 | 70 | 10 | 25 | 10 | 20 – 5000 | 0.999 | 0.51 | 1.69 |
| 3meK | 189 | 84 | 1.46 ± 0.08 | 70 | 10 | 29 | 10 | 20 – 5000 | 0.999 | 0.42 | 1.41 |
| Arginine and derivatives | |||||||||||
| Arg | 175 | 70 | 3.27 ± 0.01 | 60 | 10 | 33 | 10 | 20 – 5000 | 0.999 | 0.20 | 0.65 |
| 1meR | 189 | 70 | 3.27 ± 0.01 | 50 | 10 | 31 | 8 | 20 – 5000 | 0.999 | 0.56 | 1.85 |
| 2meR | 203 | 70 | 3.26 ± 0.01 | 46 | 10 | 33 | 10 | 20 – 5000 | 0.999 | 0.24 | 0.80 |
Mean ± SD of eight biological replicates collected over a six-month period
DP; declustering potential; EP, entrance potential; CE, collision energy; CSP, collision cell exit potential as optimized by Analyst 1.6.1 software.
Limits of detection (LoD) and quantification (LoQ) were extrapolated from linear regression analysis as signal-to-background ratios of 3:1 and 10:1, respectively [30].
Analytical methods
To capture modification quality, the relative proportion (PKX) of all peptidyl-Lys residues in the form of 1meK, 2meK or 3meK (XmeK) was calculated ratiometrically by the equation:
| (1) |
The stoichiometry of total peptidyl-Lys methylation (i.e., mol methyl groups per mol protein; SK) was then calculated by summing the relative proportion of each methyl-Lys species (XPKX) and multiplying it by the number of peptidyl-Lys residues (N) expressed in the target analyte:
| (2) |
Similarly, the relative proportion (PRX) of Arg methylation was calculated ratiometrically from mol amounts of all methyl-Arg species (XmeR) by the equation:
| (3) |
The stoichiometry of peptidyl-Arg methylation (i.e., mol methyl groups per mol protein) was calculated by summing the relative proportion of each methyl-Arg species (XPRX) and multiplying it by the number of peptidyl-Arg residues (N) expressed in the target analyte:
| (4) |
All methylation time series were modeled as simple exponential processes as described previously [25, 26]:
| (5) |
where kapp and ymax are the rate constant and maximum stoichiometry of methylation, respectively.
Statistics
Stoichiometry data were calculated as the mean ± SD of three biological replicates unless otherwise stated. Groups were compared with Student’s t-test for single values and one-sample t-test for relative values. The probability (p) of differences between estimated parameters (kapp and ymax) was assessed by z-test:
| (6) |
where x1 ± sx1 and x2 ± sx2 are the pair of estimates ± SE being compared, and z is the 1−α point of the standard normal distribution using and JMP 13.1 (SAS Institute, Cary, NC). The null hypothesis was rejected for all statistical tests at p < 0.05.
Results
LC-MS/MS quantification of methylated amino acids
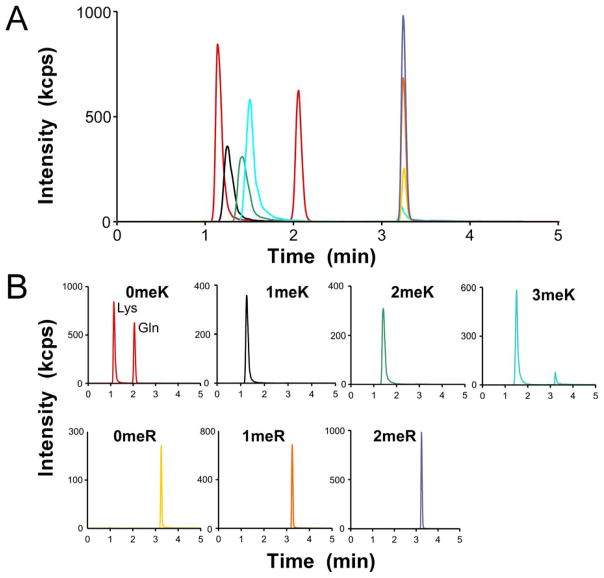
To establish assay conditions, unmodified Arg, Lys, the Lys isobar Gln, and methylated amino acids 1meK, 2meK, 3meK, 1meR, ADMR and SDMR were mixed together and subjected to LC-MS/MS. LC separation leveraged a porous graphitic carbon stationary phase developed with an acetonitrile gradient in 0.1% formic acid as the mobile phase. These conditions were used because they had previously been optimized for separation of the 20 naturally-occurring L-amino acids [27]. All nine test amino acids eluted within the gradient (Fig. 1) where they were resolved by a combination of retention time and fragmentation pattern (Table 1). For example, Lys and its isobar, Gln, generated identical precursor/product transitions, but were easily resolved from each other on the basis of retention time (reported in [28] and confirmed in Fig. 1). In contrast, Arg and its methylated derivatives migrated identically during LC, but were resolved from each other by their distinct precursor/product transitions (Fig. 1). As a result, it was possible to simultaneously detect Lys, Arg, and their methylated derivatives in the mixture. However, as reported previously [29], isobaric dimethylated arginine derivatives ADMA and SDMA were not distinguishable under these conditions. For this reason, dimethyl arginine (i.e., 2meR) was captured and reported as the sum of SDMA and ADMA.
Fig. 1.
Ion chromatograms for amino acid standards detected by multiple reaction monitoring. (A) Total ion count of mixed amino acid standards Lys (3.75 pmol), Gln (3.75 pmol), methyl-Lys (1meK, 2meK, 3meK; 1.5 pmol each), Arg (750 fmol), and methyl-Arg (1meR, 1.5 pmol; 2meR, 0.75 pmol each of ADMA and SDMA), where each color represents the measured ion intensity of an individual transition precursor/product ion pair (see Table 1). (B) LC-MS/MS chromatograms of individual amino acid standards extracted from Panel A. The separation of all amino acids standards was completed within 10 min.
To determine detection sensitivity [30], calibration graphs relating total area under the LC curve (i.e., signal) to the amount of each analyte were generated. These showed excellent linearity from mid fmol to low pmol levels, with correlation coefficients >0.995 and with corresponding limits of detection ranging from 0.2 – 1.3 fmol and limits of quantification ranging between 0.8 – 4.4 fmol (Table 1). Together these data demonstrate the feasibility of quantifying mixtures of Lys, Arg, and their methylated derivatives in the mid-fmol – low pmol range by LC/MS-MS.
Method validation with tau protein
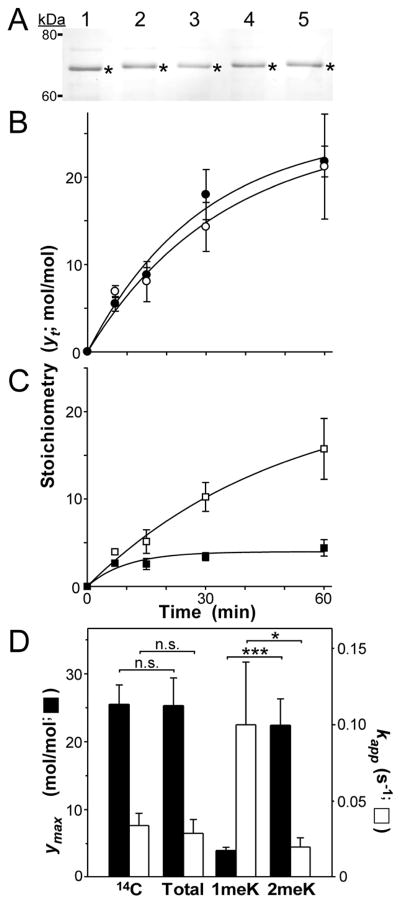
Application of the above detection method to protein hydrolysates provides a route toward estimating relative molar stoichiometries of peptidyl Lys and Arg methylation. To validate this approach, proteins with established methylation stoichiometries were separated by SDS-PAGE, immobilized by blotting onto PVDF filters, hydrolyzed to amino acids with concentrated HCl, then assayed for methylated amino acid content by the LC/MS-MS method described above. First we examined human recombinant 2N4R tau, which we previously showed could be reductively methylated on Lys residues with 14C-formaldehyde to yield estimates of methylation stoichiometry [3]. When quantified over reaction time course, 2N4R tau methylation stoichiometry was found to increase monotonically to ~22 mol methyl/mol tau protein by 60 min of incubation [3]. By replacing 14C-formaldehyde with unlabeled formaldehyde and subjecting the samples to bottom-up proteomic analysis, it was shown that all detectable methyl incorporation at all analyzed time points was in the form of 1meK and 2meK [3]. We used these unlabeled samples (taken after 0, 7, 15, 30 and 60 min reductive methylation) to validate the ratiometric LC/MS-MS approach. When subjected to SDS-PAGE, all five samples migrated identically with intact 2N4R tau (Fig. 2A). Following transfer to PVDF membrane and acid hydrolysis, only Lys, 1meK, 2meK and Arg were identified in the samples by LC/MS-MS, consistent with reported proteomic results [3] and the established selectivity of reductive methylation for Lys residues [21]. The stoichiometry of Lys methylation (SK) was then calculated from measured quantities of Lys, 1meK and 2 meK, the known Lys composition of human 2N4R tau (44 Lys residues; [31]) and eqns. 1 and 2. Comparison of these estimated stoichiometries with those determined previously by 14C-labeling showed excellent concordance between the two methods (Fig. 2B), with the null hypothesis being accepted at all time points (triplicate determinations).
Fig. 2.
Quantification of Lys methylation stoichiometry in tau protein. (A) recombinant human 2N4R tau that was reductively methylated for 0 (lane 1), 7 (lane 2), 15 (lane 3), 30 (lane 4) or 60 (lane 5) min was subjected to SDS-PAGE and Coomassie blue staining. Bands excised for hydrolysis and LC-MS/MS analysis are marked by asterisks. (B) Time course of methylation stoichiometry. Solid circles represent total methylation stoichiometry (in units of mol methyl/mol tau) as a function of time (yt) determined through radiolabeling with [14C]formaldehyde (n = 3) whereas hollow circles represent total methylation stoichiometry determined using LC-MS/MS (n = 3). Solid lines represent best fit of each time series with Eqn. 5. (C) Time course of methylation stoichiometry, where methylation stoichiometry determined by LC-MS/MS in Panel B was disaggregated into its 1meK (solid squares) and 2meK (hollow squares) components. Solid lines represent best fit of each time series with Eqn. 5. (D) Replot of data shown in Panels B and C, where ymax corresponds to the calculated plateau stoichiometry (in units of mol methyl/mol tau) and and kapp corresponds to the pseudo-first order rate constant for each time series. Key: 14C ([14C] formaldehyde labeling), Total (total methylation stoichiometry determined by LC/MS-MS), 1meK (1meK component of LC/MS-MS stoichiometry), and 2meK (2meK component of LC/MS-MS stoichiometry). *, p < 0.05; ***; p < 0.001; n.s., p > 0.05 on z-test (eqn. 6).
These data indicate that LC/MS-MS could replace 14C-labeling for quantifying tau methylation stoichiometry. However, the approach provided additional information by disaggregating methylation stoichiometry into its qualitative 1meK and 2meK components. These components appeared together early in the time series, but differed in their subsequent kinetics, with 1meK plateauing early while 2meK continued to increase over time (Fig. 2C). By 60 min, most methylation stoichiometry was in the form of 2meK (compare Fig. 2 panels B and C). To quantify these observations, stoichiometry data were modeled as simple first-order approaches to plateau by eqn. 5. Both 14C-labeling and LC/MS-MS stoichiometry data produced statistically similar estimates of pseudo-first order rate constant kapp and total plateau methylation stoichiometry ymax (Fig. 2D), demonstrating the utility of the LC/MS-MS assay for kinetic analysis. Disaggregating these total stoichiometry data into their 1meK and 2meK components revealed that the kapp for 2N4R tau mono-methylation was faster than that for dimethylation, but plateaued at lower levels (Fig. 2CD). As a result, 2meK was the predominant contributor to total stoichiometry at later time points (Fig. 2D). These data were consistent with the established kinetics of reductive methylation, where monomethylation must precede addition of a second methyl group [21]. Together these data illustrate the utility of LC/MS-MS detection for quantification of Lys protein methylation stoichiometry and its dissociation into elementary components.
Application to tissue-derived proteins
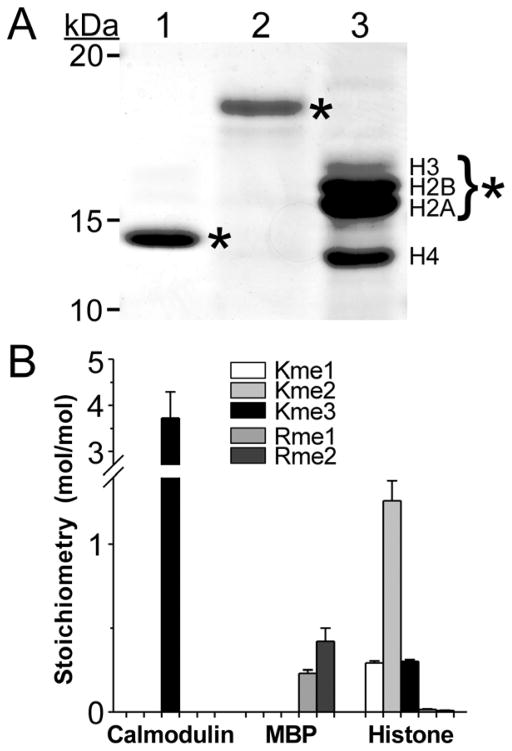
Because of its sensitive and label-free method of detection, the LC/MS-MS approach is ideally suited for analysis of low abundance, tissue-derived samples. To test this utility, three tissue-derived methyl-protein samples were analyzed. The first of these was bovine calmodulin, which when isolated from beef brain or testes is reported to contain one 3meK residue at position 115 and no other Lys or Arg methylations [13]. When subjected to SDS-PAGE, bovine calmodulin electrophoresed as a 14 kDa species (Fig. 3A). After gel-purified calmodulin was acid hydrolyzed and assayed by LC-MS/MS, only 3meK was detected (Fig. 3B). On the basis of the amino acid composition of calmodulin [13], a stoichiometry of 3.72 ± 0.57 mol methyl/mol calmodulin was calculated (Fig. 3B). This value corresponded to 1.24 ± 0.19 mol 3meK/mol calmodulin, consistent with the single 3meK modification site established by protein chemistry methods (null hypothesis accepted, p = 0.16).
Fig. 3.
Quantification of methylation stoichiometry in protein standards. (A) Coomassie-blue stained SDS-PAGE gel of input protein standards calmodulin (lane 1, 1 μg), MBP (lane 2, 2 μg) and mixed histones (lane 3, 3 μg). Bands excised for hydrolysis and LC-MS/MS analysis are marked by asterisks. The excised histone bands consisted of core histones H2A, H2B and H3. (B) Methylation stoichiometry determined by LC-MS/MS and eqns. 1–4. Bar height indicates contributions of 1meK, 2meK, 3meK, 1meR and 2meR to mol methyl/mol protein in each sample (triplicate determination) whereas error bars represent the standard deviation.
The second sample was MBP, which reportedly contains 1meR and 2meR (but not methyl Lys) when isolated from bovine brain [14–17]. Upon SDS-PAGE separation, bovine MBP electrophoresed as a ~18 kDa species (Fig. 3A). After acid hydrolysis and LC-MS/MS analysis, 1meR and 2meR were the only detectable methyl amino acids (Fig. 3B). On the basis of the amino acid composition of MBP, a stoichiometry of 0.65 ± 0.14 mol methyl/mol MBP was calculated (Fig. 3B) composed of ~0.2 mol each of 1meR and 2meR per mol MBP. These values were consistent with previous estimates made on the basis of amino acid analysis and colorimetric detection (0.18 – 0.80 mol 1meR/mol MBP and 0.12 – 0.31 mol 2meR/mol MBP; [14–17]), demonstrating that the utility of the ratiometric approach for estimating protein methylation stoichiometry also extends to methyl-Arg.
Finally, mixed histones were assayed to gauge the feasibility of detecting broad-spectrum methylation occurring within one sample. Histones are subject to diverse post-translational modifications, and have been reported to contain 1meK, 2meK and 3meK as well as 1meR and 2meR residues [32, 33]. When analyzed by SDS-PAGE, mixed histones prepared from calf thymus separated into constituent core H2A, H2B, H3, and H4 components (Fig. 3A). After H2A, H2B, and H3 were excised as a mixture and subjected to acid hydrolysis, LC-MS/MS analysis successfully confirmed 1meK, 2meK, 3meK, 1meR and 2meR as being simultaneously present in the sample (Fig. 3B). Estimates of stoichiometry in this case were averaged over the composition of the mixture (H2A, H2B and H3) and therefore not reflective of any single species. However, the observed rank order stoichiometry of 2meK > 1meK was consistent with previously reported characterizations of arginine-rich histones by amino acid analysis [34, 35]. These results indicate that the LC-MS/MS method is able to detect and quantify both Lys and Arg methylation when present simultaneously in a qualitatively complex sample.
Discussion
These data indicate that an LC-MS/MS approach can reliably detect mid-fmol quantities of methylated Lys and Arg yielding estimates of their stoichiometries in proteins. Its overall precision and accuracy are sufficient to replace radio-labeling in kinetic analyses of protein Lys reductive methylation. It may also be of use in characterizing the kinetics of Lys and Arg methyltransferases [36]. However, its greatest utility is its compatibility with a broad range of low abundance, tissue-derived protein samples, its ability to disaggregate bulk Arg/Lys methylation stoichiometry into its individual mono-, di-, and tri-methylated residue components, and its compatibility with diverse sample sources owing to its leveraging of SDS-PAGE for final sample preparation. Despite these advantages, accurate ratiometric quantification of tissue protein methylation stoichiometry will depend on three conditions. First, as with other methods, the analyte must be of high purity. While not attainable for all protein samples, the incorporation of an SDS-PAGE isolation step provides a convenient and powerful means of maximizing input sample purity. Second, ratiometric analysis requires full-length protein or at least precise knowledge of Lys and Arg composition. In the case of tau protein, this was easily achieved by using recombinant protein of defined isoform composition. Full-length, post-translationally modified tau proteins of known isoform composition also can be isolated from mammalian brain tissue and separated by SDS-PAGE [37, 38], and so these will likely be amenable to analysis as well. In contrast, the tau proteins that accumulate in human cerebral spinal fluid can become extensively proteolyzed, resulting in complex peptide mixtures ([39, 40]). Accurate quantification of methylation stoichiometry in this pool would be challenging. Finally, the ratiometric approach requires that diverse post-translational modifications on Lys and Arg residues other than methylation break down to unmodified amino acids after acid hydrolysis. This condition is largely met for stable Lys modifications, which apart from specific hydroxylated derivatives associated with collagen [41], and certain acid-stable advanced glycation end products associated with aging and metabolic disease [42], are dominated by acid-sensitive acylations. Arg too is subjected to hydrolysable PTMs. However, Arg also can be modified by peptidyl Arg deiminases that convert peptidyl-Arg residues to citrulline [43], which upon hydrolysis yields ornithine rather than Arg. As a result, the denominator for eqn. 3 will be underestimated when applied to citrullinated analytes, resulting in overestimation of methylation stoichiometry. However, the stoichiometry of citrullination in vivo is low [44]. Indeed, we found that ratiometric quantification of bovine MBP methyl-Arg stotichiometry was consistent with previously reported analyses despite the presence of five citrullination sites on this analyte [45]. Because ornithine also is amenable to LC-MS/MS analysis [28], it may be possible to create a ratiometric assay for quantification of citrullination stoichiometry in future.
In addition to Lys and Arg residues, proteins can be methylated on their N-termini, on His residues, and on certain carboxylic acid groups (reviewed in [46]). Because carboxymethylated residues are not stable to acid hydrolysis (for example [47]), a targeted ratiometric approach similar to the one disclosed here for methylated Lys and Arg would not be appropriate for these analytes. However, both His- and N-terminal methylation are acid stable [48, 49], and so it may be possible to target these methyl-residues for bulk quantification using our methods.
In summary, the method disclosed herein will complement existing bottom-up proteomic identification of Lys and Arg protein methylation sites by yielding estimates of overall methylation stoichiometry and quality. The approach will facilitate study of methylation kinetics as well as placing observations of in vivo methylation into a more quantitative context.
Acknowledgments
We thank the Targeted Metabolomics Laboratory at The Ohio State University for access to LC–MS/MS equipment (funded by the Translational Plant Sciences Targeted Investment in Excellence). This work was supported by Public Health Service grants NS77441 and AG54018.
Abbreviations used
- 1meK, 2meK and 3meK
Nε-(methyl)-, Nε-(dimethyl)-, and Nε-(trimethyl)-L-lysine, respectively
- 1meR
NG-Methyl-L-arginine
- ADMA
NG,NG′-Dimethyl-L-arginine
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- MBP
myelin basic protein
- MRM
multiple reaction monitoring
- PTM
post-translational modification
- PVDF
polyvinylidene fluoride
- SDMA
NG,NG′-Dimethyl-L-arginine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications - writers that read. EMBO reports. 2015;16:1467–1481. doi: 10.15252/embr.201540945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biggar KK, Li SS. Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol. 2015;16:5–17. doi: 10.1038/nrm3915. [DOI] [PubMed] [Google Scholar]
- 3.Funk KE, Thomas SN, Schafer KN, Cooper GL, Liao Z, Clark DJ, Yang AJ, Kuret J. Lysine methylation is an endogenous post-translational modification of tau protein in human brain and a modulator of aggregation propensity. Biochem J. 2014;462:77–88. doi: 10.1042/BJ20140372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris M, Knudsen GM, Maeda S, Trinidad JC, Ioanoviciu A, Burlingame AL, Mucke L. Tau post-translational modifications in wild-type and human amyloid precursor protein transgenic mice. Nat Neurosci. 2015;18:1183–1189. doi: 10.1038/nn.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas SN, Funk KE, Wan Y, Liao Z, Davies P, Kuret J, Yang AJ. Dual modification of Alzheimer’s disease PHF-tau protein by lysine methylation and ubiquitylation: a mass spectrometry approach. Acta Neuropathol. 2012;123:105–117. doi: 10.1007/s00401-011-0893-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coorssen JR, Yergey AL. Proteomics Is Analytical Chemistry: Fitness-for-Purpose in the Application of Top-Down and Bottom-Up Analyses. Proteomes. 2015;3:440–453. doi: 10.3390/proteomes3040440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore S, Stein WH. Chromatographic determination of amino acids by the use of automatic recording equipment. Methods Enzymol. 1963;6:819–831. [Google Scholar]
- 8.Murray K, Rasmussen PS, Neustaedter J, Luck JM. The Hydrolysis of Arginine. J Biol Chem. 1965;240:705–709. [PubMed] [Google Scholar]
- 9.Tsiboli P, Konstantinidis G, Skendros Y, Katsani A, Choli-Papadopoulou T. Identification of post-translational modified amino acids. Amino Acids. 1997;13:13–23. [Google Scholar]
- 10.Yan JX, Sanchez JC, Binz PA, Williams KL, Hochstrasser DF. Method for identification and quantitative analysis of protein lysine methylation using matrix-assisted laser desorption/ionization--time-of-flight mass spectrometry and amino acid analysis. Electrophoresis. 1999;20:749–754. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<749::AID-ELPS749>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 11.Hirschey MD, Zhao Y. Metabolic Regulation by Lysine Malonylation, Succinylation, and Glutarylation. Mol Cell Proteomics. 2015;14:2308–2315. doi: 10.1074/mcp.R114.046664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Streich FC, Jr, Lima CD. Structural and functional insights to ubiquitin-like protein conjugation. Annu Rev Biophys. 2014;43:357–379. doi: 10.1146/annurev-biophys-051013-022958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watterson DM, Mendel PA, Vanaman TC. Comparison of calcium-modulated proteins from vertebrate brains. Biochemistry. 1980;19:2672–2676. doi: 10.1021/bi00553a020. [DOI] [PubMed] [Google Scholar]
- 14.Young PR, Grynspan F. Analysis of methylated amino acids by high-performance liquid chromatography: methylation of myelin basic protein. J Chromatogr. 1987;421:130–135. doi: 10.1016/0378-4347(87)80387-0. [DOI] [PubMed] [Google Scholar]
- 15.Brostoff S, Eylar EH. Localization of methylated arginine in the A1 protein from myelin. Proc Natl Acad Sci U S A. 1971;68:765–769. doi: 10.1073/pnas.68.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deibler GE, Martenson RE. Determination of methylated basic amino acids with the amino acid analyzer. Application to total acid hydrolyzates of myelin basic proteins. J Biol Chem. 1973;248:2387–2391. [PubMed] [Google Scholar]
- 17.Kakimoto Y, Matsuoka Y, Miyake M, Konishi H. Methylated amino acid residues of proteins of brain and other organs. J Neurochem. 1975;24:893–902. doi: 10.1111/j.1471-4159.1975.tb03653.x. [DOI] [PubMed] [Google Scholar]
- 18.Friedman M. Applications of the ninhydrin reaction for analysis of amino acids, peptides, and proteins to agricultural and biomedical sciences. J Agric Food Chem. 2004;52:385–406. doi: 10.1021/jf030490p. [DOI] [PubMed] [Google Scholar]
- 19.García Alvarez-Coque MC, Medina Hernández MJ, Villanueva Camañas RM, Mongay Fernández C. Formation and instability of o-phthalaldehyde derivatives of amino acids. Anal Biochem. 1989;178:1–7. doi: 10.1016/0003-2697(89)90346-1. [DOI] [PubMed] [Google Scholar]
- 20.Richardson SL, Hanjra P, Zhang G, Mackie BD, Peterson DL, Huang R. A direct, ratiometric, and quantitative MALDI-MS assay for protein methyltransferases and acetyltransferases. Anal Biochem. 2015;478:59–64. doi: 10.1016/j.ab.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jentoft N, Dearborn DG. Labeling of proteins by reductive methylation using sodium cyanoborohydride. J Biol Chem. 1979;254:4359–4365. [PubMed] [Google Scholar]
- 22.McKeon TA, Lyman ML. Calcium ion improves electrophoretic transfer of calmodulin and other small proteins. Anal Biochem. 1991;193:125–130. doi: 10.1016/0003-2697(91)90051-t. [DOI] [PubMed] [Google Scholar]
- 23.McClure T, Cocuron JC, Osmark V, McHale LK, Alonso AP. Impact of Environment on the Biomass Composition of Soybean (Glycine max) seeds. J Agric Food Chem. 2017;65:6753–6761. doi: 10.1021/acs.jafc.7b01457. [DOI] [PubMed] [Google Scholar]
- 24.Matsuo ES, Shin RW, Billingsley ML, Van deVoorde A, O’Connor M, Trojanowski JQ, Lee VM. Biopsy-derived adult human brain tau is phosphorylated at many of the same sites as Alzheimer’s disease paired helical filament tau. Neuron. 1994;13:989–1002. doi: 10.1016/0896-6273(94)90264-x. [DOI] [PubMed] [Google Scholar]
- 25.Taylor IA, Webb M. Chemical modification of lysine by reductive methylation. A probe for residues involved in DNA binding. Methods Mol Biol. 2001;148:301–314. doi: 10.1385/1-59259-208-2:301. [DOI] [PubMed] [Google Scholar]
- 26.Sternlicht H, Ringel I, Szasz J. Theory for modeling the copolymerization of tubulin and tubulin-colchicine complex. Biophys J. 1983;42:255–267. doi: 10.1016/S0006-3495(83)84393-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cocuron JC, Tsogtbaatar E, Alonso AP. High-throughput quantification of the levels and labeling abundance of free amino acids by liquid chromatography tandem mass spectrometry. J Chromatogr A. 2017;1490:148–155. doi: 10.1016/j.chroma.2017.02.028. [DOI] [PubMed] [Google Scholar]
- 28.Cocuron JC, Anderson B, Boyd A, Alonso AP. Targeted metabolomics of Physaria fendleri, an industrial crop producing hydroxy fatty acids. Plant Cell Physiol. 2014;55:620–633. doi: 10.1093/pcp/pcu011. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Tang WH, Cho L, Brennan DM, Hazen SL. Targeted metabolomic evaluation of arginine methylation and cardiovascular risks: potential mechanisms beyond nitric oxide synthase inhibition. Arterioscler Thromb Vac Biol. 2009;29:1383–1391. doi: 10.1161/ATVBAHA.109.185645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vial J, Jardy A. Experimental comparison of the different approaches to estimate LOD and LOQ of an HPLC method. Anal Chem. 1999;71:2672–2677. [Google Scholar]
- 31.Goedert M, Spillantini MG, Jakes R, Rutherford D, Crowther RA. Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron. 1989;3:519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 32.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Di Lorenzo A, Bedford MT. Histone arginine methylation. FEBS Lett. 2011;585:2024–2031. doi: 10.1016/j.febslet.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paik WK, Kim S. Epsilon-N-Dimethyllysine in Histones. Biochem Biophys Res Commun. 1967;27:479–483. [Google Scholar]
- 35.Delange RJ, Smith EL, Bonner J. Calf Thymus Histone 3 - Sequences of Amino-Terminal and Carboxyl-Terminal Regions and of Regions Containing Lysyl Residues Modified by Acetylation and Methylation. Biochem Biophys Res Commun. 1970;40:989–993. doi: 10.1016/0006-291x(70)91001-6. [DOI] [PubMed] [Google Scholar]
- 36.Horiuchi KY. Challenges in profiling and lead optimization of drug discovery for methyltransferases. Drug discovery today Technologies. 2015;18:62–68. doi: 10.1016/j.ddtec.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Goedert M, Jakes R. Expression of separate isoforms of human tau protein: correlation with the tau pattern in brain and effects on tubulin polymerization. EMBO J. 1990;9:4225–4230. doi: 10.1002/j.1460-2075.1990.tb07870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMillan P, Korvatska E, Poorkaj P, Evstafjeva Z, Robinson L, Greenup L, Leverenz J, Schellenberg GD, D’Souza I. Tau isoform regulation is region- and cell-specific in mouse brain. J Comp Neurol. 2008;511:788–803. doi: 10.1002/cne.21867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson GV, Seubert P, Cox TM, Motter R, Brown JP, Galasko D. The tau protein in human cerebrospinal fluid in Alzheimer’s disease consists of proteolytically derived fragments. J Neurochem. 1997;68:430–433. doi: 10.1046/j.1471-4159.1997.68010430.x. [DOI] [PubMed] [Google Scholar]
- 40.Meredith JE, Jr, Sankaranarayanan S, Guss V, Lanzetti AJ, Berisha F, Neely RJ, Slemmon JR, Portelius E, Zetterberg H, Blennow K, Soares H, Ahlijanian M, Albright CF. Characterization of novel CSF Tau and ptau biomarkers for Alzheimer’s disease. PLoS One. 2013;8:e76523. doi: 10.1371/journal.pone.0076523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamauchi M, Shiiba M. Lysine hydroxylation and cross-linking of collagen. Methods Mol Biol. 2008;446:95–108. doi: 10.1007/978-1-60327-084-7_7. [DOI] [PubMed] [Google Scholar]
- 42.Rabbani N, Ashour A, Thornalley PJ. Mass spectrometric determination of early and advanced glycation in biology. Glycoconj J. 2016;33:553–568. doi: 10.1007/s10719-016-9709-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amin B, Voelter W. Human Deiminases: Isoforms, Substrate Specificities, Kinetics, and Detection. Prog Chem Org Nat Prod. 2017;106:203–240. doi: 10.1007/978-3-319-59542-9_2. [DOI] [PubMed] [Google Scholar]
- 44.Tutturen AE, Fleckenstein B, de Souza GA. Assessing the citrullinome in rheumatoid arthritis synovial fluid with and without enrichment of citrullinated peptides. J Proteome Res. 2014;13:2867–2873. doi: 10.1021/pr500030x. [DOI] [PubMed] [Google Scholar]
- 45.Jin Z, Fu Z, Yang J, Troncosco J, Everett AD, Van Eyk JE. Identification and characterization of citrulline-modified brain proteins by combining HCD and CID fragmentation. Proteomics. 2013;13:2682–2691. doi: 10.1002/pmic.201300064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clarke SG. Protein methylation at the surface and buried deep: thinking outside the histone box. Trends Biochem Sci. 2013;38:243–252. doi: 10.1016/j.tibs.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kleene SJ, Toews ML, Adler J. Isolation of glutamic acid methyl ester from an Escherichia coli membrane protein involved in chemotaxis. J Biol Chem. 1977;252:3214–3218. [PubMed] [Google Scholar]
- 48.Kuhl DA, Methvin JT, Dickerson RN. Standardization of acid hydrolysis procedure for urinary 3-methylhistidine determination by high-performance liquid chromatography. J Chromatogr B Biomed Appl. 1996;681:390–394. doi: 10.1016/0378-4347(96)00052-7. [DOI] [PubMed] [Google Scholar]
- 49.Alix JH, Hayes D, Lontie JF, Colson C, Glatigny A, Lederer F. Methylated amino acids in ribosomal proteins from Escherichia coli treated with ethionine and from a mutant lacking methylation of protein L11. Biochimie. 1979;61:671–679. doi: 10.1016/s0300-9084(79)80165-0. [DOI] [PubMed] [Google Scholar]